Hormetic Heat Shock Enhances Autophagy through HSF1 in Retinal Pigment Epithelium Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Hormetic Heat Shock Treatment
2.3. HSF1 Gain-of-Function
2.4. Immunoblotting
2.5. Immunoprecipitation
2.6. Quantitative PCR
2.7. Immunofluorescence
2.8. Autophagic Flux Detection
2.9. Statistical Analysis
3. Results
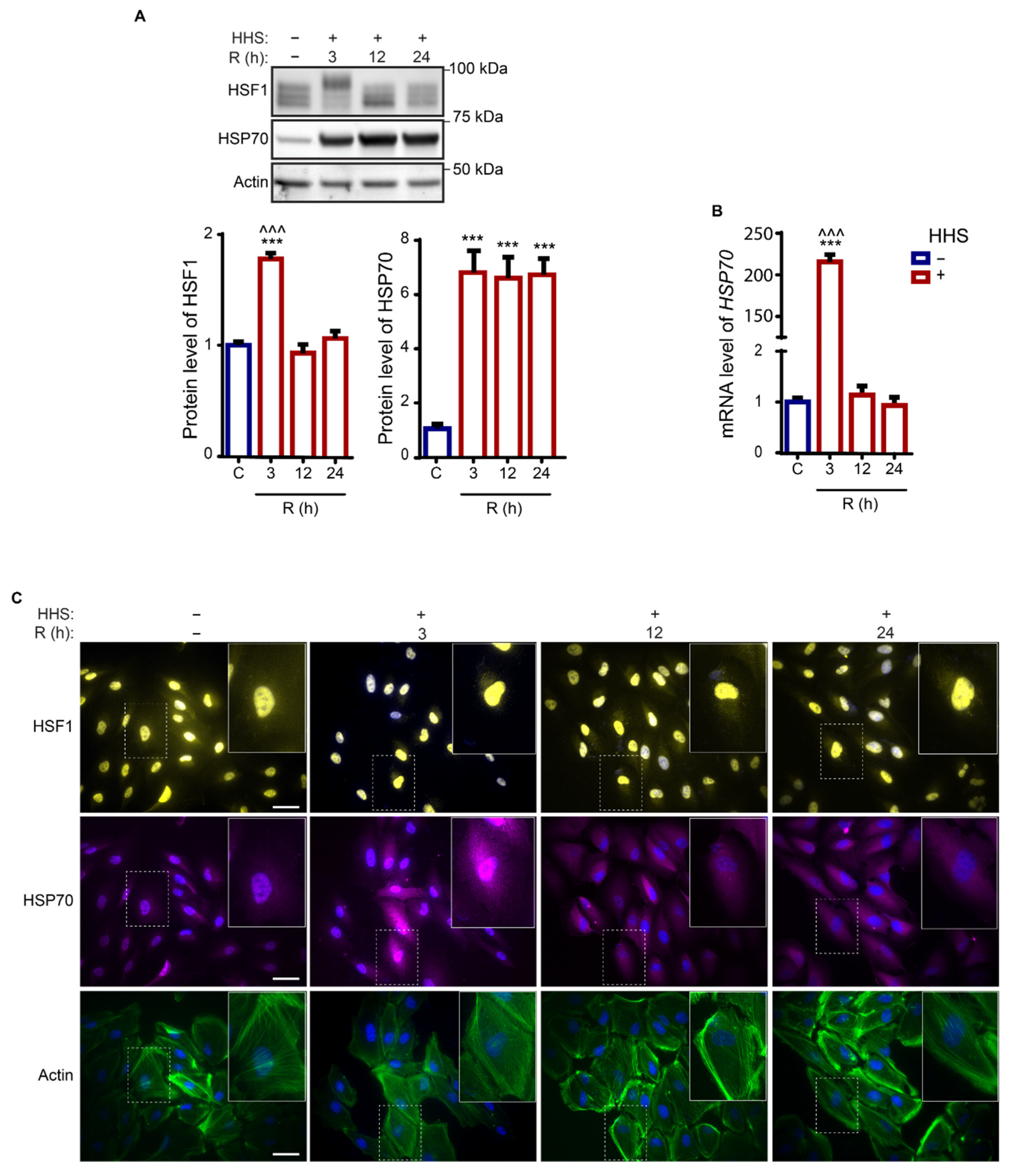
3.1. Hormetic Heat Shock Induces a Long-Lasting Heat Shock Response in ARPE-19 Cells
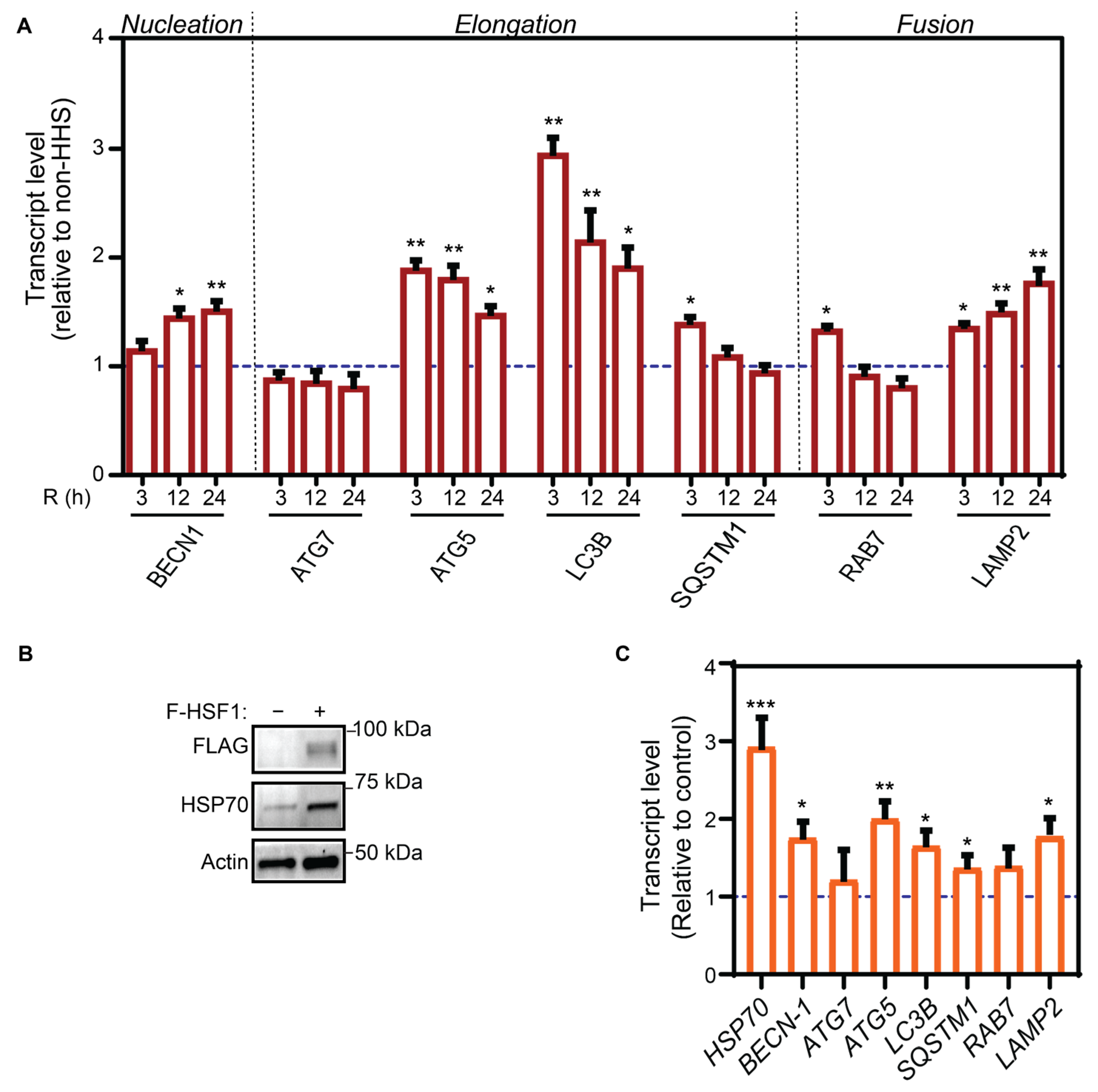
3.2. Hormetic Heat Shock Increase Autophagy Gene Expression by HSF1 Transcriptional Activation
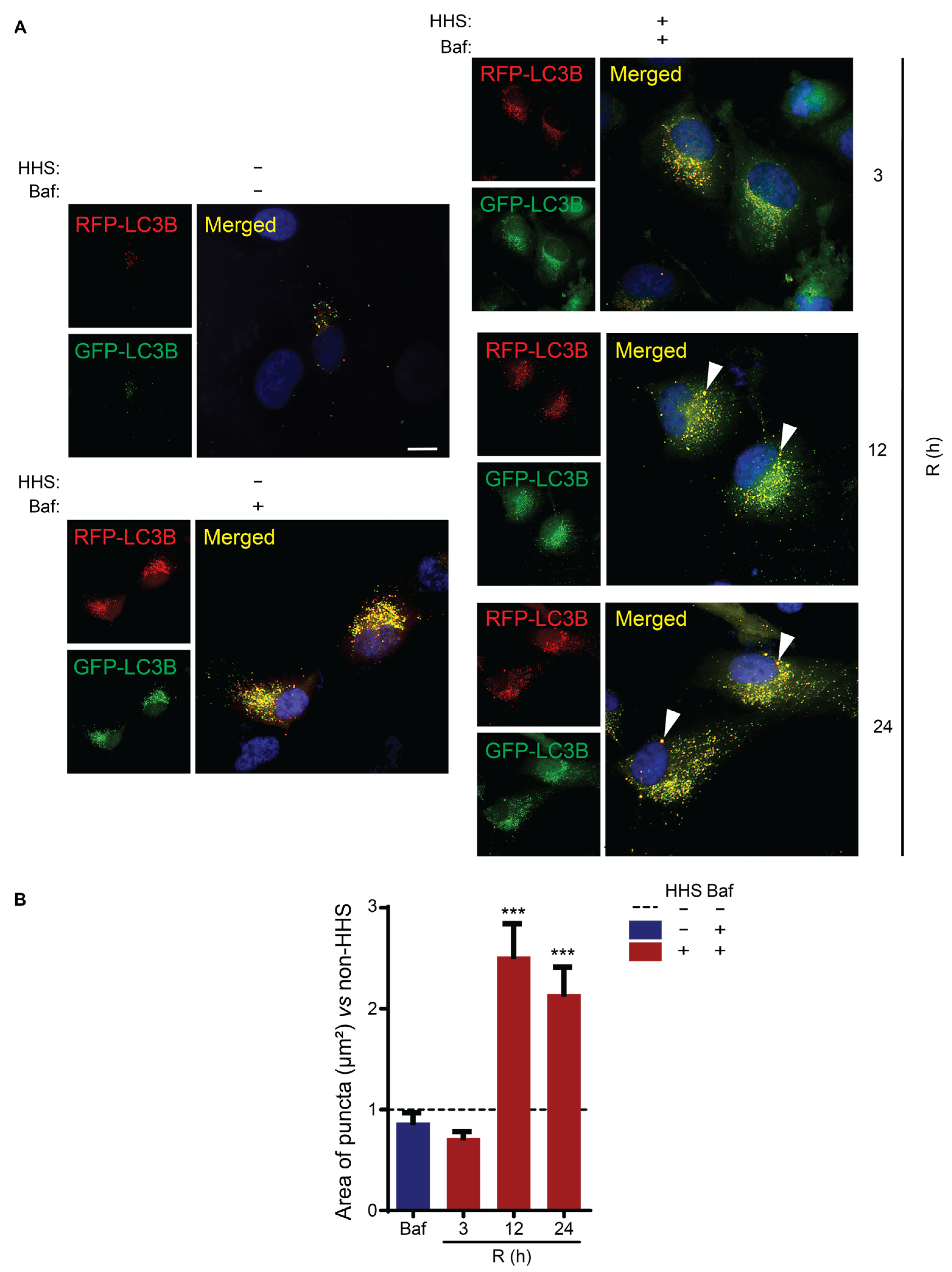
3.3. Hormetic Heat Shock Increases Autophagic Flux
3.4. HHS-Induced Increase in Autophagic Flux Does Not Depend on HSP70
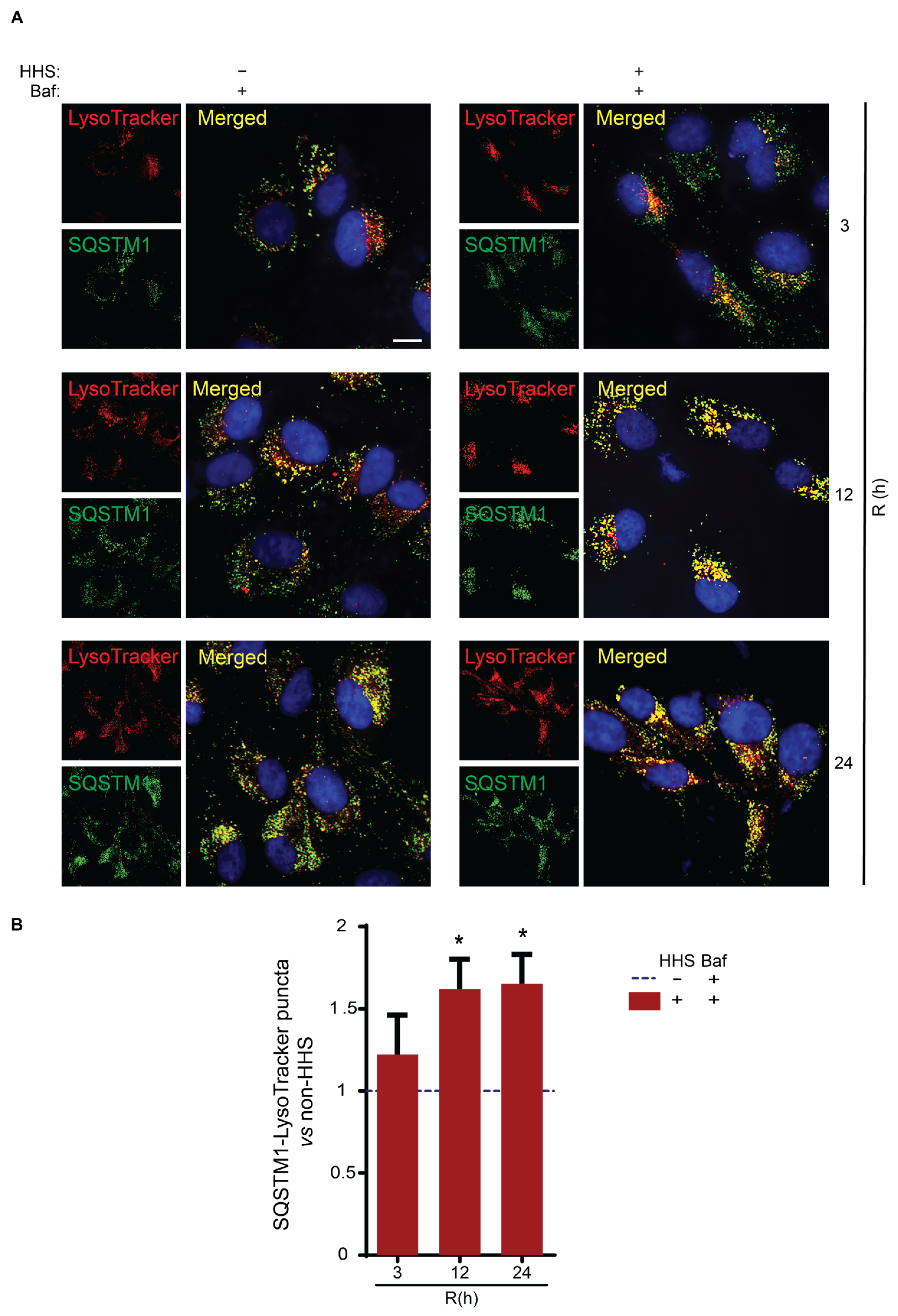
3.5. Hormetic Heat Shock Increases the Size of Autophagosomes and the Presentation of SQSTM1-Associated Cargo to Lysosomes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kuusisto, E.; Salminen, A.; Alafuzoff, I. Early Accumulation of P62 in Neurofibrillary Tangles in Alzheimer’s Disease: Possible Role in Tangle Formation. Neuropathol. Appl. Neurobiol. 2002, 28, 228–237. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Salminen, A.; Haapasalo, A.; Soininen, H.; Hiltunen, M. Age-Related Macular Degeneration (AMD): Alzheimer’s Disease in the Eye? J. Alzheimer’s Dis. 2011, 24, 615–631. [Google Scholar] [CrossRef] [Green Version]
- Braak, H.; Thal, D.R.; Del Tredici, K. Nerve Cells Immunoreactive for P62 in Select Hypothalamic and Brainstem Nuclei of Controls and Parkinson’s Disease Cases. J. Neural Transm. 2011, 118, 809–819. [Google Scholar] [CrossRef]
- Kaarniranta, K.; Uusitalo, H.; Blasiak, J.; Felszeghy, S.; Kannan, R.; Kauppinen, A.; Salminen, A.; Sinha, D.; Ferrington, D. Mechanisms of Mitochondrial Dysfunction and Their Impact on Age-Related Macular Degeneration. Prog. Retin. Eye Res. 2020, 79, 100858. [Google Scholar] [CrossRef]
- Naso, F.; Intartaglia, D.; Falanga, D.; Soldati, C.; Polishchuk, E.; Giamundo, G.; Tiberi, P.; Marrocco, E.; Scudieri, P.; Di Malta, C.; et al. Light-responsive MicroRNA MiR-211 Targets Ezrin to Modulate Lysosomal Biogenesis and Retinal Cell Clearance. EMBO J. 2020, 39, e102468. [Google Scholar] [CrossRef]
- Kuo, C.; Green, C.R.; Rupenthal, I.D.; Mugisho, O.O. Connexin43 Hemichannel Block Protects against Retinal Pigment Epithelial Cell Barrier Breakdown. Acta Diabetol. 2020, 57, 13–22. [Google Scholar] [CrossRef]
- Blasiak, J.; Pawlowska, E.; Szczepanska, J.; Kaarniranta, K. Interplay between Autophagy and the Ubiquitin-Proteasome System and Its Role in the Pathogenesis of Age-Related Macular Degeneration. Int. J. Mol. Sci. 2019, 20, 210. [Google Scholar] [CrossRef] [Green Version]
- Kroemer, G.; Mariño, G.; Levine, B. Autophagy and the Integrated Stress Response. Mol. Cell 2010, 40, 280–293. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.C.; Guan, K.L. MTOR: A Pharmacologic Target for Autophagy Regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Wong, P.M.; Puente, C.; Ganley, I.G.; Jiang, X. The ULK1 Complex Sensing Nutrient Signals for Autophagy Activation. Autophagy 2013, 9, 124–137. [Google Scholar] [CrossRef] [Green Version]
- Pesce, N.A.; Canovai, A.; Lardner, E.; Cammalleri, M.; Kvanta, A.; Andr, H.; Monte, M.D. Autophagy Involvement in the Postnatal Development of the Rat Retina Noemi. Cells 2021, 10, 177. [Google Scholar] [CrossRef]
- Wesselborg, S.; Stork, B. Autophagy Signal Transduction by ATG Proteins: From Hierarchies to Networks. Cell. Mol. Life Sci. 2015, 72, 4721–4757. [Google Scholar] [CrossRef] [Green Version]
- Metlagel, Z.; Otomo, C.; Takaesu, G.; Otomo, T. Structural Basis of ATG3 Recognition by the Autophagic Ubiquitin-like Protein ATG12. Proc. Natl. Acad. Sci. USA 2013, 110, 18844–18849. [Google Scholar] [CrossRef] [Green Version]
- Pyo, J.O.; Nah, J.; Jung, Y.K. Molecules and Their Functions in Autophagy. Exp. Mol. Med. 2012, 44, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Simonsen, A.; Tooze, S.A. Coordination of Membrane Events during Autophagy by Multiple Class III PI3-Kinase Complexes. J. Cell Biol. 2009, 186, 773–782. [Google Scholar] [CrossRef]
- Wurzer, B.; Zaffagnini, G.; Fracchiolla, D.; Turco, E.; Abert, C.; Romanov, J.; Martens, S. Oligomerization of P62 Allows for Selection of Ubiquitinated Cargo and Isolation Membrane during Selective Autophagy. eLife 2015, 4, e08941. [Google Scholar] [CrossRef]
- Itakura, E.; Mizushima, N. P62 Targeting to the Autophagosome Formation Site Requires Self-Oligomerization but Not LC3 Binding. J. Cell Biol. 2011, 192, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Fader, C.M.; Colombo, M.I. Autophagy and Multivesicular Bodies: Two Closely Related Partners. Cell Death Differ. 2009, 16, 70–78. [Google Scholar] [CrossRef] [Green Version]
- Saftig, P.; Klumperman, J. Lysosome Biogenesis and Lysosomal Membrane Proteins: Trafficking Meets Function. Nat. Rev. Mol. Cell Biol. 2009, 10, 623–635. [Google Scholar] [CrossRef]
- Anckar, J.; Sistonen, L. Regulation of HSF1 Function in the Heat Stress Response: Implications in Aging and Disease. Annu. Rev. Biochem. 2011, 80, 1089–1115. [Google Scholar] [CrossRef]
- Ciechanover, A.; Kwon, Y.T. Protein Quality Control by Molecular Chaperones in Neurodegeneration. Front. Neurosci. 2017, 11, 185. [Google Scholar] [CrossRef] [Green Version]
- Kaushik, S.; Cuervo, A.M. Chaperones in Autophagy. Pharmacol. Res. 2012, 66, 484–493. [Google Scholar] [CrossRef] [Green Version]
- Brader, H.S.; Young, L.H.Y. Subthreshold Diode Micropulse Laser: A Review. Semin. Ophthalmol. 2016, 31, 30–39. [Google Scholar] [CrossRef]
- Lavinsky, D.; Wang, J.; Huie, P.; Dalal, R.; Lee, S.J.; Lee, D.Y.; Palanker, D. Nondamaging Retinal Laser Therapy: Rationale and Applications to the Macula. Investig. Ophthalmol. Vis. Sci. 2016, 57, 2488–2500. [Google Scholar] [CrossRef]
- Sramek, C.; Mackanos, M.; Spitler, R.; Leung, L.S.; Nomoto, H.; Contag, C.H.; Palanker, D. Non-Damaging Retinal Phototherapy: Dynamic Range of Heat Shock Protein Expression. Investig. Ophthalmol. Vis. Sci. 2011, 52, 1780–1787. [Google Scholar] [CrossRef] [Green Version]
- Amirkavei, M.; Pitkänen, M.; Kaikkonen, O.; Kaarniranta, K.; André, H.; Koskelainen, A. Induction of Heat Shock Protein 70 in Mouse RPE as an In Vivo Model of Transpupillary Thermal Stimulation. Int. J. Mol. Sci. 2020, 21, 2063. [Google Scholar] [CrossRef] [Green Version]
- She, H.; Li, X.; Yu, W. Subthreshold Transpupillary Thermotherapy of the Retina and Experimental Choroidal Neovascularization in a Rat Model. Graefe’s Arch. Clin. Exp. Ophthalmol. 2006, 244, 1143–1151. [Google Scholar] [CrossRef]
- Desmettre, T.; Maurage, C.A.; Mordon, S. Heat Shock Protein Hyperexpression on Chorioretinal Layers after Transpupillary Thermotherapy. Investig. Ophthalmol. Vis. Sci. 2001, 42, 2976–2980. [Google Scholar]
- Wang, J.; Huie, P.; Dalal, R.; Lee, S.; Tan, G.; Lee, D.; Lavinksy, D.; Palanker, D. Heat Shock Protein Expression as Guidance for the Therapeutic Window of Retinal Laser Therapy. Ophthalmic Technol. XXVI 2016, 9693, 969319. [Google Scholar] [CrossRef]
- Inagaki, K.; Shuo, T.; Katakura, K.; Ebihara, N.; Murakami, A.; Ohkoshi, K. Sublethal Photothermal Stimulation with a Micropulse Laser Induces Heat Shock Protein Expression in ARPE-19 Cells. J. Ophthalmol. 2015, 2015, 729792. [Google Scholar] [CrossRef] [Green Version]
- Kern, K.; Mertineit, C.L.; Brinkmann, R.; Miura, Y. Expression of Heat Shock Protein 70 and Cell Death Kinetics after Different Thermal Impacts on Cultured Retinal Pigment Epithelial Cells. Exp. Eye Res. 2018, 170, 117–126. [Google Scholar] [CrossRef]
- Mackanos, M.A.; Contag, C.H. Pulse Duration Determines Levels of Hsp70 Induction in Tissues Following Laser Irradiation. J. Biomed. Opt. 2011, 16, 078002. [Google Scholar] [CrossRef] [Green Version]
- Viiri, J.; Amadio, M.; Marchesi, N.; Hyttinen, J.M.T.; Kivinen, N.; Sironen, R.; Rilla, K.; Akhtar, S.; Provenzani, A.; D’Agostino, V.G.; et al. Autophagy Activation Clears ELAVL1/HuR-Mediated Accumulation of SQSTM1/P62 during Proteasomal Inhibition in Human Retinal Pigment Epithelial Cells. PLoS ONE 2013, 8, e69563. [Google Scholar] [CrossRef] [Green Version]
- Golestaneh, N.; Chu, Y.; Xiao, Y.Y.; Stoleru, G.L.; Theos, A.C. Dysfunctional Autophagy in RPE, a Contributing Factor in Age-Related Macular Degeneration. Cell Death Dis. 2017, 8, e2537. [Google Scholar] [CrossRef]
- Mitter, S.K.; Song, C.; Qi, X.; Mao, H.; Rao, H.; Akin, D.; Lewin, A.; Grant, M.; Dunn, W.; Ding, J.; et al. Dysregulated Autophagy in the RPE Is Associated with Increased Susceptibility to Oxidative Stress and AMD. Autophagy 2014, 10, 1989–2005. [Google Scholar] [CrossRef] [Green Version]
- Dunn, K.C.; Aotaki-Keen, A.E.; Putkey, F.R.; Hjelmeland, L.M. ARPE-19, a Human Retinal Pigment Epithelial Cell Line with Differentiated Properties. Exp. Eye Res. 1996, 62, 155–170. [Google Scholar] [CrossRef]
- Hytti, M.; Korhonen, E.; Hongisto, H.; Kaarniranta, K.; Skottman, H.; Kauppinen, A. Differential Expression of Inflammasome-Related Genes in Induced Pluripotent Stem-Cell-Derived Retinal Pigment Epithelial Cells with or without History of Age-Related Macular Degeneration. Int. J. Mol. Sci. 2021, 22, 6800. [Google Scholar] [CrossRef]
- Plaza Reyes, A.; Petrus-Reurer, S.; Padrell Sánchez, S.; Kumar, P.; Douagi, I.; Bartuma, H.; Aronsson, M.; Westman, S.; Lardner, E.; André, H.; et al. Identification of Cell Surface Markers and Establishment of Monolayer Differentiation to Retinal Pigment Epithelial Cells. Nat. Commun. 2020, 11, 1609. [Google Scholar] [CrossRef] [Green Version]
- Sandqvist, A.; Björk, J.K.; Åkerfelt, M.; Chitikova, Z.; Grichine, A.; Vourc’h, C.; Jolly, C.; Salminen, T.A.; Nymalm, Y.; Sistonen, L. Heterotrimerization of Heat-Shock Factors 1 and 2 Provides a Transcriptional Switch in Response to Distinct Stimuli. Mol. Biol. Cell 2009, 20, 1340–1347. [Google Scholar] [CrossRef] [Green Version]
- Sarge, K.D.; Murphy, S.P.; Morimoto, R.I. Activation of Heat Shock Gene Transcription by Heat Shock Factor 1 Involves Oligomerization, Acquisition of DNA-Binding Activity, and Nuclear Localization and Can Occur in the Absence of Stress. Mol. Cell. Biol. 1993, 13, 1392–1407. [Google Scholar] [CrossRef]
- Sistonen, L.; Sarge, K.D.; Morimoto, R.I. Human Heat Shock Factors 1 and 2 Are Differentially Activated and Can Synergistically Induce Hsp70 Gene Transcription. Mol. Cell. Biol. 1994, 14, 2087–2099. [Google Scholar] [CrossRef]
- Nivon, M.; Richet, E.; Codogno, P.; Arrigo, A.P.; Kretz-Remy, C. Autophagy Activation by NFκB Is Essential for Cell Survival after Heat Shock. Autophagy 2009, 5, 766–783. [Google Scholar] [CrossRef] [Green Version]
- Lapierre, L.R.; Kumsta, C.; Sandri, M.; Ballabio, A.; Hansen, M. Transcriptional and Epigenetic Regulation of Autophagy in Aging. Autophagy 2015, 11, 867–880. [Google Scholar] [CrossRef] [Green Version]
- Wiegant, F.A.C.; de Poot, S.A.H.; Boers-Trilles, V.E.; Schreij, A.M.A. Hormesis and Cellular Quality Control: A Possible Explanation for the Molecular Mechanisms That Underlie the Benefits of Mild Stress. Dose-Response 2013, 11, 413–430. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy (4th Edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef]
- Rusmini, P.; Cristofani, R.; Galbiati, M.; Cicardi, M.E.; Meroni, M.; Ferrari, V.; Vezzoli, G.; Tedesco, B.; Messi, E.; Piccolella, M.; et al. The Role of the Heat Shock Protein B8 (HSPB8) in Motoneuron Diseases. Front. Mol. Neurosci. 2017, 10, 176. [Google Scholar] [CrossRef] [Green Version]
- Agholme, L.; Agnello, M.; Agostinis, P.; Aguirre-ghiso, J.A.; Ahn, H.J.; Ait-mohamed, O.; Brown, E.J.; Brumell, J.H.; Brunetti-pierri, N.; Brunk, U.T.; et al. Guidelines for the Use and Interpretation of Assays for Monitoring Autophagy. Autophagy 2012, 8, 445–544. [Google Scholar]
- Kivinen, N.; Hyttinen, J.; Viiri, J. Hsp70 Binds Reversibly to Proteasome Inhibitor-Induced Protein Aggregates and Evades Autophagic Clearance in ARPE-19 Cells. J. Biochem. 2014, 2, 1–7. [Google Scholar]
- Mahat, D.B.; Salamanca, H.H.; Duarte, F.M.; Danko, C.G.; Lis, J.T. Mammalian Heat Shock Response and Mechanisms Underlying Its Genome-Wide Transcriptional Regulation. Mol. Cell 2016, 62, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Kmiecik, S.W.; Mayer, M.P. Molecular Mechanisms of Heat Shock Factor 1 Regulation. Trends Biochem. Sci. 2022, 47, 218–234. [Google Scholar] [CrossRef]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat Shock Factors: Integrators of Cell Stress, Development and Lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef]
- Batista-Nascimento, L.; Neef, D.W.; Liu, P.C.C.; Rodrigues-Pousada, C.; Thiele, D.J. Deciphering Human Heat Shock Transcription Factor 1 Regulation via Post-Translational Modification in Yeast. PLoS ONE 2011, 6, e15976. [Google Scholar] [CrossRef] [Green Version]
- Kumsta, C.; Chang, J.T.; Schmalz, J.; Hansen, M. Hormetic Heat Stress and HSF-1 Induce Autophagy to Improve Survival and Proteostasis in C. Elegans. Nat. Commun. 2017, 8, 14337. [Google Scholar] [CrossRef] [Green Version]
- Komata, T.; Kanzawa, T.; Nashimoto, T.; Aoki, H.; Endo, S.; Nameta, M.; Takahashi, H.; Yamamoto, T.; Kondo, S.; Tanaka, R. Mild Heat Shock Induces Autophagic Growth Arrest, but Not Apoptosis in U251-MG and U87-MG Human Malignant Glioma Cells. J. Neuro-Oncol. 2004, 68, 101–111. [Google Scholar] [CrossRef]
- Oberley, T.D.; Swanlund, J.M.; Zhang, H.J.; Kregel, K.C. Aging Results in Increased Autophagy of Mitochondria and Protein Nitration in Rat Hepatocytes Following Heat Stress. J. Histochem. Cytochem. 2008, 56, 615–627. [Google Scholar] [CrossRef] [Green Version]
- Cuervo, A.M.; Wong, E. Autophagy Gone Awry in Neurodegenerative Diseases. Nat. Neurosci. 2012, 29, 997–1003. [Google Scholar] [CrossRef]
- Barna, J.; Csermely, P.; Vellai, T. Roles of Heat Shock Factor 1 beyond the Heat Shock Response. Cell. Mol. Life Sci. 2018, 75, 2897–2916. [Google Scholar] [CrossRef]
- Trinklein, N.D.; Murray, J.I.; Hartman, S.J.; Botstein, D.; Myers, R.M.M. The Role of Heat Shock Transcription Factor 1 in the Genome-Wide Regulation of the Mammalian Heat Shock Response. Mol. Biol. Cell 2004, 15, 1254–1261. [Google Scholar] [CrossRef]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. P62 Filaments Capture and Present Ubiquitinated Cargos for Autophagy. EMBO J. 2018, 37, e98308. [Google Scholar] [CrossRef]
- Sun, D.; Wu, R.; Zheng, J.; Li, P.; Yu, L. Polyubiquitin Chain-Induced P62 Phase Separation Drives Autophagic Cargo Segregation. Cell Res. 2018, 28, 405–415. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, S.; Gudmundsson, S.R.; Sou, Y.S.; Ichimura, Y.; Tamura, N.; Kazuno, S.; Ueno, T.; Miura, Y.; Noshiro, D.; Abe, M.; et al. P62/SQSTM1-Droplet Serves as a Platform for Autophagosome Formation and Anti-Oxidative Stress Response. Nat. Commun. 2021, 12, 16. [Google Scholar] [CrossRef]
- Dokladny, K.; Zuhl, M.N.; Mandell, M.; Bhattacharya, D.; Schneider, S.; Deretic, V.; Moseley, P.L. Regulatory Coordination between Two Major Intracellular Homeostatic Systems: Heat Shock Response and Autophagy. J. Biol. Chem. 2013, 288, 14959–14972. [Google Scholar] [CrossRef] [Green Version]
- Jin, M.; Klionsky, D.J. Regulation of Autophagy: Modulation of the Size and Number of Autophagosomes. FEBS Lett. 2009, 42, 115–125. [Google Scholar] [CrossRef]
- Xie, Z.; Nair, U.; Klionsky, D.J. Atg8 Controls Phagophore Expansion during Autophagosome Formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef] [Green Version]
- Kaikkonen, O.; Turunen, T.T.; Meller, A.; Ahlgren, J.; Koskelainen, A. Retinal Temperature Determination Based on Photopic Porcine Electroretinogram. IEEE Trans. Biomed. Eng. 2021, 69, 1–12. [Google Scholar] [CrossRef]
- Pitkänen, M.; Kaikkonen, O.; Koskelainen, A. In Vivo Monitoring of Mouse Retinal Temperature by ERG Photoresponses. Exp. Eye Res. 2019, 187, 107675. [Google Scholar] [CrossRef]
- Pitkänen, M.; Kaikkonen, O.; Koskelainen, A. A Novel Method for Mouse Retinal Temperature Determination Based on ERG Photoresponses. Ann. Biomed. Eng. 2017, 45, 2360–2372. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amirkavei, M.; Plastino, F.; Kvanta, A.; Kaarniranta, K.; André, H.; Koskelainen, A. Hormetic Heat Shock Enhances Autophagy through HSF1 in Retinal Pigment Epithelium Cells. Cells 2022, 11, 1778. https://doi.org/10.3390/cells11111778
Amirkavei M, Plastino F, Kvanta A, Kaarniranta K, André H, Koskelainen A. Hormetic Heat Shock Enhances Autophagy through HSF1 in Retinal Pigment Epithelium Cells. Cells. 2022; 11(11):1778. https://doi.org/10.3390/cells11111778
Chicago/Turabian StyleAmirkavei, Mooud, Flavia Plastino, Anders Kvanta, Kai Kaarniranta, Helder André, and Ari Koskelainen. 2022. "Hormetic Heat Shock Enhances Autophagy through HSF1 in Retinal Pigment Epithelium Cells" Cells 11, no. 11: 1778. https://doi.org/10.3390/cells11111778






