Narrow-Gap Rheometry: A Novel Method for Measuring Cell Mechanics
Abstract
:1. Introduction
2. Single Cell Studies vs. Average Rheological Properties
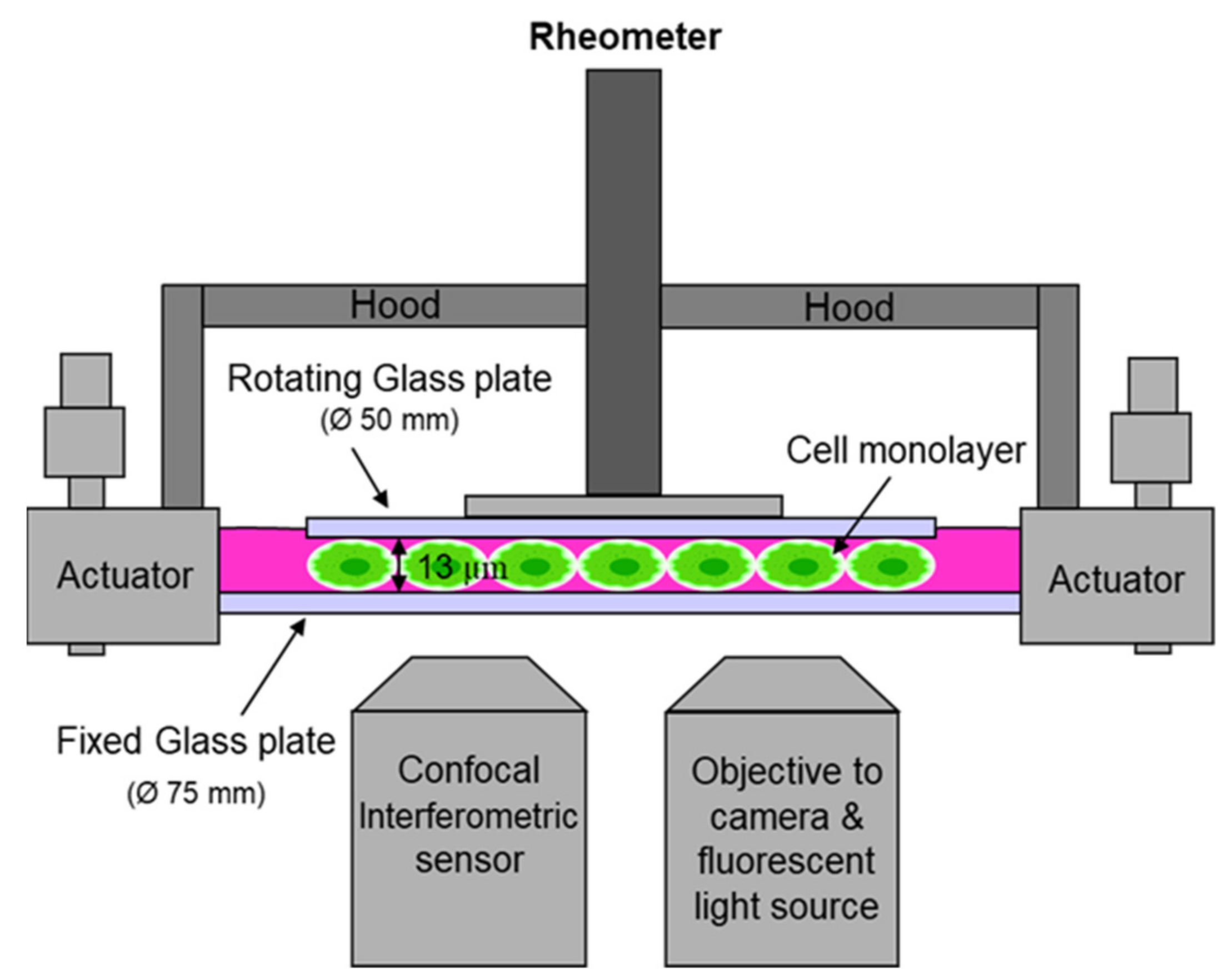
3. Narrow-Gap Rheology
4. Summary and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Kollmannsberger, P.; Fabry, B. Active Soft Glassy Rheology of Adherent Cells. Soft Matter 2009, 5, 1771–1774. [Google Scholar] [CrossRef]
- Wu, P.H.; Aroush, D.R.B.; Asnacios, A.; Chen, W.C.; Dokukin, M.E.; Doss, B.L.; Durand-Smet, P.; Ekpenyong, A.; Guck, J.; Guz, N.V.; et al. A Comparison of Methods to Assess Cell Mechanical Properties. Nat. Methods 2018, 15, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Janmey, P.A.; Fletcher, D.A.; Reinhart-King, C.A. Stiffness Sensing by Cells. Physiol. Rev. 2020, 100, 695–724. [Google Scholar] [CrossRef] [PubMed]
- Discher, D.E.; Janmey, P.; Wang, Y.L. Tissue Cells Feel and Respond to the Stiffness of Their Substrate. Science 2005, 310, 1139–1143. [Google Scholar] [CrossRef] [Green Version]
- Bertaud, J.; Qin, Z.; Buehler, M.J. Intermediate Filament-Deficient Cells Are Mechanically Softer at Large Deformation: A Multi-Scale Simulation Study. Acta Biomater. 2010, 6, 2457–2466. [Google Scholar] [CrossRef]
- Kollmannsberger, P.; Fabry, B. Linear and Nonlinear Rheology of Living Cells. Annu. Rev. Mater. Res. 2011, 41, 75–97. [Google Scholar] [CrossRef] [Green Version]
- Jordan, M.A.; Wilson, L. Microtubules as a Target for Anticancer Drugs. Nat. Rev. Cancer 2004, 4, 253–265. [Google Scholar] [CrossRef]
- Di Carlo, D. A Mechanical Biomarker of Cell State in Medicine. J. Lab. Autom. 2012, 17, 32–42. [Google Scholar] [CrossRef]
- Weinberg, R.A. The Biology of Cancer, 2nd ed.; W. W. Norton & Company: New York, NY, USA, 2013; pp. 1–876. [Google Scholar] [CrossRef]
- Yallapu, M.M.; Katti, K.S.; Katti, D.R.; Mishra, S.R.; Khan, S.; Jaggi, M.; Chauhan, S.C. The Roles of Cellular Nanomechanics in Cancer. Med. Res. Rev. 2015, 35, 198–223. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.Y.H.; Lim, C.T. Biomechanics Approaches to Studying Human Diseases. Trends Biotechnol. 2007, 25, 111–118. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Z.; Li, L.; Chen, B.C.; Revyakin, A.; Hajj, B.; Legant, W.; Dahan, M.; Lionnet, T.; Betzig, E.; et al. Single-Molecule Dynamics of Enhanceosome Assembly in Embryonic Stem Cells. Cell 2014, 156, 1274–1285. [Google Scholar] [CrossRef] [Green Version]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D.; et al. Optical Deformability as an Inherent Cell Marker for Testing Malignant Transformation and Metastatic Competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef] [Green Version]
- Fritsch, A.; Höckel, M.; Kiessling, T.; Nnetu, K.D.; Wetzel, F.; Zink, M.; Käs, J.A. Are Biomechanical Changes Necessary for Tumour Progression? Nat. Phys. 2010, 6, 730–732. [Google Scholar] [CrossRef]
- Swaminathan, V.; Mythreye, K.; Tim O’Brien, E.; Berchuck, A.; Blobe, G.C.; Superfine, R. Mechanical Stiffness Grades Metastatic Potential in Patient Tumor Cells and in Cancer Cell Lines. Cancer Res. 2011, 71, 5075–5080. [Google Scholar] [CrossRef] [Green Version]
- Mierke, C.T.; Kollmannsberger, P.; Zitterbart, D.P.; Diez, G.; Koch, T.M.; Marg, S.; Ziegler, W.H.; Goldmann, W.H.; Fabry, B. Vinculin Facilitates Cell Invasion into Three-Dimensional Collagen Matrices. J. Biol. Chem. 2010, 285, 13121–13130. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Cai, Y.; Qi, C.; Hansen, R.; Ding, C.; Thomas, C.; Yan, J. Orally Administered Particular β-Glucan Modulates Tumor-Capturing Dendritic Cells and Improves Anti-Tumor T Cell Responses in Cancer. Clin. Cancer Res. 2010, 16, 5153–5164. [Google Scholar] [CrossRef] [Green Version]
- Gal, N.; Weihs, D. Intracellular Mechanics and Activity of Breast Cancer Cells Correlate with Metastatic Potential. Cell Biochem. Biophys. 2012, 63, 199–209. [Google Scholar] [CrossRef]
- Suresh, S. Biomechanics and Biophysics of Cancer Cells. Acta Biomater. 2007, 3, 413–438. [Google Scholar] [CrossRef]
- Dakhil, H.; Gilbert, D.F.; Malhotra, D.; Limmer, A.; Engelhardt, H.; Amtmann, A.; Hansmann, J.; Hübner, H.; Buchholz, R.; Friedrich, O.; et al. Measuring Average Rheological Quantities of Cell Monolayers in the Linear Viscoelastic Regime. Rheol. Acta 2016, 55, 527–536. [Google Scholar] [CrossRef]
- Voulgari, A.; Pintzas, A. Epithelial–Mesenchymal Transition in Cancer Metastasis: Mechanisms, Markers and Strategies to Overcome Drug Resistance in the Clinic. Biochim. Biophys. Acta—Rev. Cancer 2009, 1796, 75–90. [Google Scholar] [CrossRef]
- Ceppi, P.; Peter, M.E. MicroRNAs Regulate Both Epithelial-to-Mesenchymal Transition and Cancer Stem Cells. Oncogene 2014, 33, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Koch, T.M.; Münster, S.; Bonakdar, N.; Butler, J.P.; Fabry, B. 3D Traction Forces in Cancer Cell Invasion. PLoS ONE 2012, 7, e33476. [Google Scholar] [CrossRef] [Green Version]
- Bhat, S.; Jun, D.; Paul, B.C.; Dahms, T.E.S. Viscoelasticity in Biological Systems: A Special Focus on Microbes; Chapter 6; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, R.; Rybenkov, V.V. A Guide to Magnetic Tweezers and Their Applications. Front. Phys. 2016, 4, 48. [Google Scholar] [CrossRef]
- Brill-Karniely, Y. Mechanical Measurements of Cells Using AFM: 3D or 2D Physics? Front. Bioeng. Biotechnol. 2020, 8, 605153. [Google Scholar] [CrossRef]
- Urbanska, M.; Muñoz, H.E.; Shaw Bagnall, J.; Otto, O.; Manalis, S.R.; Di Carlo, D.; Guck, J. A Comparison of Microfluidic Methods for High-Throughput Cell Deformability Measurements. Nat. Methods 2020, 17, 587–593. [Google Scholar] [CrossRef]
- Gil-Redondo, J.C.; Weber, A.; Zbiral, B.; Vivanco, M.d.M.; Toca-Herrera, J.L. Substrate Stiffness Modulates the Viscoelastic Properties of MCF-7 Cells. J. Mech. Behav. Biomed. Mater. 2022, 125, 104979. [Google Scholar] [CrossRef]
- Hiratsuka, S.; Mizutani, Y.; Tsuchiya, M.; Kawahara, K.; Tokumoto, H.; Okajima, T. The Number Distribution of Complex Shear Modulus of Single Cells Measured by Atomic Force Microscopy. Ultramicroscopy 2009, 109, 937–941. [Google Scholar] [CrossRef]
- Davidovits, P. Physics in Biology and Medicine, 4th ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 1–331. [Google Scholar] [CrossRef]
- Guz, N.; Dokukin, M.; Kalaparthi, V.; Sokolov, I. If Cell Mechanics Can Be Described by Elastic Modulus: Study of Different Models and Probes Used in Indentation Experiments. Biophys. J. 2014, 107, 564–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staunton, J.R.; Doss, B.L.; Lindsay, S.; Ros, R. Correlating Confocal Microscopy and Atomic Force Indentation Reveals Metastatic Cancer Cells Stiffen during Invasion into Collagen i Matrices. Sci. Rep. 2016, 6, 19686. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Trepat, X.; Butler, J.P.; Millet, E.; Morgan, K.G.; Weitz, D.A.; Fredberg, J.J. Fast and Slow Dynamics of the Cytoskeleton. Nat. Mater. 2006, 5, 636–640. [Google Scholar] [CrossRef]
- Zhang, Y.; Wei, F.; Poh, Y.C.; Jia, Q.; Chen, J.; Chen, J.; Luo, J.; Yao, W.; Zhou, W.; Huang, W.; et al. Interfacing 3D Magnetic Twisting Cytometry with Confocal Fluorescence Microscopy to Image Force Responses in Living Cells. Nat. Protoc. 2017, 12, 1437–1450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kollmannsberger, P.; Fabry, B. High-Force Magnetic Tweezers with Force Feedback for Biological Applications. Rev. Sci. Instrum. 2007, 78, 114301. [Google Scholar] [CrossRef] [PubMed]
- Vilfan, I.D.; Lipfert, J.; Koster, D.A.; Lemay, S.G.; Dekker, N.H. Magnetic Tweezers for single-molecule experiments. In Handbook of Single-Molecule Biophysics; Springer: Berlin/Heidelberg, Germany, 2009; pp. 371–396. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Fu, H.; Zhu, X.; Cong, P.; Nakamura, F.; Yan, J. Improved High-Force Magnetic Tweezers for Stretching and Refolding of Proteins and Short DNA. Biophys. J. 2011, 100, 517–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balland, M.; Desprat, N.; Icard, D.; Féréol, S.; Asnacios, A.; Browaeys, J.; Hénon, S.; Gallet, F. Power Laws in Microrheology Experiments on Living Cells: Comparative Analysis and Modeling. Phys. Rev. E.—Stat. Nonlinear Soft Matter Phys. 2006, 74, 021911. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Liu, K.K. Optical Tweezers for Single Cells. J. R. Soc. Interface 2008, 5, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Ayala, Y.A.; Pontes, B.; Ether, D.S.; Pires, L.B.; Araujo, G.R.; Frases, S.; Romão, L.F.; Farina, M.; Moura-Neto, V.; Viana, N.B.; et al. Rheological Properties of Cells Measured by Optical Tweezers. BMC Biophys. 2016, 9, 5. [Google Scholar] [CrossRef] [Green Version]
- Desprat, N.; Guiroy, A.; Asnacios, A. Microplates-Based Rheometer for a Single Living Cell. Rev. Sci. Instrum. 2006, 77, 055111. [Google Scholar] [CrossRef]
- Desprat, N.; Richert, A.; Simeon, J.; Asnacios, A. Creep Function of a Single Living Cell. Biophys. J. 2005, 88, 2224–2233. [Google Scholar] [CrossRef] [Green Version]
- Fernández, P.; Pullarkat, P.A.; Ott, A. A Master Relation Defines the Nonlinear Viscoelasticity of Single Fibroblasts. Biophys. J. 2006, 90, 3796. [Google Scholar] [CrossRef] [Green Version]
- Bufi, N.; Durand-Smet, P.; Asnacios, A. Single-Cell Mechanics: The Parallel Plates Technique. Methods Cell Biol. 2015, 125, 187–209. [Google Scholar] [CrossRef]
- Yamada, S.; Wirtz, D.; Kuo, S.C. Mechanics of Living Cells Measured by Laser Tracking Microrheology. Biophys. J. 2000, 78, 1736–1747. [Google Scholar] [CrossRef] [Green Version]
- Wirtz, D. Particle-Tracking Microrheology of Living Cells: Principles and Applications. Annu. Rev. Biophys. 2009, 38, 301–326. [Google Scholar] [CrossRef] [Green Version]
- McGlynn, J.A.; Wu, N.; Schultz, K.M. Multiple Particle Tracking Microrheological Characterization: Fundamentals, Emerging Techniques and Applications. J. Appl. Phys. 2020, 127, 201101. [Google Scholar] [CrossRef]
- Hochmuth, R.M. Micropipette Aspiration of Living Cells. J. Biomech. 2000, 33, 15–22. [Google Scholar] [CrossRef]
- Guilak, F.; Alexopoulos, L.G.; Haider, M.A.; Ting-Beall, H.P.; Setton, L.A. Zonal Uniformity in Mechanical Properties of the Chondrocyte Pericellular Matrix: Micropipette Aspiration of Canine Chondrons Isolated by Cartilage Homogenization. Ann. Biomed. Eng. 2005, 33, 1312–1318. [Google Scholar] [CrossRef]
- Rodriguez, M.L.; McGarry, P.J.; Sniadecki, N.J. Review on Cell Mechanics: Experimental and Modeling Approaches. Appl. Mech. Rev. 2013, 65, 060801. [Google Scholar] [CrossRef] [Green Version]
- Gangotra, A.; Willmott, G.R. Cellular and Sub-Cellular Mechanics: Measurement of Material Properties. In Reference Module in Materials Science and Materials Engineering, 2nd ed.; Elsavier: Amsterdam, The Netherlands, 2019; Volume 3, pp. 227–244. [Google Scholar] [CrossRef]
- Guck, J.; Ananthakrishnan, R.; Mahmood, H.; Moon, T.J.; Cunningham, C.C.; Käs, J. The Optical Stretcher: A Novel Laser Tool to Micromanipulate Cells. Biophys. J. 2001, 81, 767. [Google Scholar] [CrossRef] [Green Version]
- Lincoln, B.; Wottawah, F.; Schinkinger, S.; Ebert, S.; Guck, J. High-Throughput Rheological Measurements with an Optical Stretcher. Methods Cell Biol. 2007, 83, 397–423. [Google Scholar] [CrossRef]
- Yang, T.; Bragheri, F.; Minzioni, P. A Comprehensive Review of Optical Stretcher for Cell Mechanical Characterization at Single-Cell Level. Micromachines 2016, 7, 90. [Google Scholar] [CrossRef] [Green Version]
- Mierke, C.T. The Role of the Optical Stretcher Is Crucial in the Investigation of Cell Mechanics Regulating Cell Adhesion and Motility. Front. Cell Dev. Biol. 2019, 7, 184. [Google Scholar] [CrossRef] [Green Version]
- Fernández, P.; Heymann, L.; Ott, A.; Aksel, N.; Pullarkat, P.A. Shear Rheology of a Cell Monolayer. New J. Phys. 2007, 9, 149. [Google Scholar] [CrossRef]
- Sander, M.; Flesch, J.; Ott, A. Using Cell Monolayer Rheology to Probe Average Single Cell Mechanical Properties. Biorheology 2015, 52, 269–278. [Google Scholar] [CrossRef]
- Kiran, A.; Shekhar, C.; Sabapathy, M.; Mishra, M.; Kumar, L.; Kumar, N.; Mehandia, V. Effect of Serum Starvation on Rheology of Cell Monolayers. Phys. Fluids 2021, 33, 071908. [Google Scholar] [CrossRef]
- Dahl, J.B.; Lin, J.M.G.; Muller, S.J.; Kumar, S. Microfluidic Strategies for Understanding the Mechanics of Cells and Cell-Mimetic Systems. Annu. Rev. Chem. Biomol. Eng. 2015, 6, 293–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halldorsson, S.; Lucumi, E.; Gómez-Sjöberg, R.; Fleming, R.M.T. Advantages and Challenges of Microfluidic Cell Culture in Polydimethylsiloxane Devices. Biosens. Bioelectron. 2015, 63, 218–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, T.; Fan, L.; Zhu, R.; Sun, D. Microfluidic Single-Cell Manipulation and Analysis: Methods and Applications. Micromachines 2019, 10, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, P.; Mizutani, Y.; Tsuchiya, M.; Maloney, J.M.; Fabry, B.; Van Vliet, K.J.; Okajima, T. Quantifying Cell-to-Cell Variation in Power-Law Rheology. Biophys. J. 2013, 105, 1093–1102. [Google Scholar] [CrossRef] [Green Version]
- Mierke, C.T.; Kollmannsberger, P.; Zitterbart, D.P.; Smith, J.; Fabry, B.; Goldmann, W.H. Mechano-Coupling and Regulation of Contractility by the Vinculin Tail Domain. Biophys. J. 2008, 94, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Lange, J.R.; Steinwachs, J.; Kolb, T.; Lautscham, L.A.; Harder, I.; Whyte, G.; Fabry, B. Microconstriction Arrays for High-Throughput Quantitative Measurements of Cell Mechanical Properties. Biophys. J. 2015, 109, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Herbig, M.; Tessmer, K.; Nötzel, M.; Nawaz, A.A.; Santos-Ferreira, T.; Borsch, O.; Gasparini, S.J.; Guck, J.; Ader, M. Label-Free Imaging Flow Cytometry for Analysis and Sorting of Enzymatically Dissociated Tissues. Sci. Rep. 2022, 12, 963. [Google Scholar] [CrossRef]
- Paguirigan, A.L.; Beebe, D.J. From the Cellular Perspective: Exploring Differences in the Cellular Baseline in Macroscale and Microfluidic Cultures. Integr. Biol. 2009, 1, 182. [Google Scholar] [CrossRef] [Green Version]
- Elkins, C.M.; Shen, W.-J.; Khor, V.K.; Kraemer, F.B.; Fuller, G.G. Quantification of Stromal Vascular Cell Mechanics with a Linear Cell Monolayer Rheometer. J. Rheol. 2015, 59, 33–50. [Google Scholar] [CrossRef]
- Pérez-Domínguez, S.; Kulkarni, S.G.; Rianna, C.; Radmacher, M. Atomic Force Microscopy for Cell Mechanics and Diseases. Neuroforum 2020, 26, 101–109. [Google Scholar] [CrossRef]
- Lekka, M. Atomic Force Microscopy: A Tip for Diagnosing Cancer. Nat. Nanotechnol. 2012, 7, 691–692. [Google Scholar] [CrossRef]
- Lekka, M. Discrimination Between Normal and Cancerous Cells Using AFM. Bionanoscience 2016, 6, 65–80. [Google Scholar] [CrossRef] [Green Version]
- Rianna, C.; Radmacher, M. Cell Mechanics as a Marker for Diseases: Biomedical Applications of AFM. AIP Conf. Proc. 2016, 1760, 020057. [Google Scholar] [CrossRef]
- Rother, J.; Nöding, H.; Mey, I.; Janshoff, A. Atomic Force Microscopy-Based Microrheology Reveals Significant Differences in the Viscoelastic Response between Malign and Benign Cell Lines. Open Biol. 2014, 4, 140046. [Google Scholar] [CrossRef]
- Nematbakhsh, Y.; Pang, K.T.; Lim, C.T. Correlating the Viscoelasticity of Breast Cancer Cells with Their Malignancy. Converg. Sci. Phys. Oncol. 2017, 3, 034003. [Google Scholar] [CrossRef]
- Smolyakov, G.; Thiebot, B.; Campillo, C.; Labdi, S.; Severac, C.; Pelta, J.; Dague, É. Elasticity, Adhesion, and Tether Extrusion on Breast Cancer Cells Provide a Signature of Their Invasive Potential. ACS Appl. Mater. Interfaces 2016, 8, 27426–27431. [Google Scholar] [CrossRef] [Green Version]
- Gavara, N. A Beginner’s Guide to Atomic Force Microscopy Probing for Cell Mechanics. Microsc. Res. Tech. 2017, 80, 75–84. [Google Scholar] [CrossRef]
- Zemła, J.; Danilkiewicz, J.; Orzechowska, B.; Pabijan, J.; Seweryn, S.; Lekka, M. Atomic Force Microscopy as a Tool for Assessing the Cellular Elasticity and Adhesiveness to Identify Cancer Cells and Tissues. Semin. Cell Dev. Biol. 2018, 73, 115–124. [Google Scholar] [CrossRef]
- Efremov, Y.M.; Okajima, T.; Raman, A. Measuring Viscoelasticity of Soft Biological Samples Using Atomic Force Microscopy. Soft Matter 2019, 16, 64–81. [Google Scholar] [CrossRef]
- Darling, E.M.; Carlo, D. Di High-Throughput Assessment of Cellular Mechanical Properties. Annu. Rev. Biomed. Eng. 2015, 17, 35–62. [Google Scholar] [CrossRef]
- Liu, N.; Du, P.; Xiao, X.; Liu, Y.; Peng, Y.; Yang, C.; Yue, T. Microfluidic-Based Mechanical Phenotyping of Androgen-Sensitive and Non-Sensitive Prostate Cancer Cells Lines. Micromachines 2019, 10, 602. [Google Scholar] [CrossRef] [Green Version]
- Choi, G.; Tang, Z.; Guan, W. Microfluidic High-Throughput Single-Cell Mechanotyping: Devices and Applications. Nami Jishu Yu Jingmi Gongcheng/Nanotechnol. Precis. Eng. 2021, 4, 045002. [Google Scholar] [CrossRef]
- Van Citters, K.M.; Hoffman, B.D.; Massiera, G.; Crocker, J.C. The Role of F-Actin and Myosin in Epithelial Cell Rheology. Biophys. J. 2006, 91, 3946–3956. [Google Scholar] [CrossRef] [Green Version]
- Stamenović, D. Rheological Behavior of Mammalian Cells. Cell. Mol. Life Sci. 2008, 65, 3592–3605. [Google Scholar] [CrossRef]
- Chowdhury, F.; Huang, B.; Wang, N. Cytoskeletal Prestress: The Cellular Hallmark in Mechanobiology and Mechanomedicine. Cytoskeleton 2021, 78, 249–276. [Google Scholar] [CrossRef]
- Macosko, C.W. Rheology: Principles, Measurements, and Applications (Advances in Interfacial Engineering); NY Weinheim Cambridge VCH: New York, NY, USA, 1994; pp. 1–568. [Google Scholar]
- Dakhil, H.; Wierschem, A. Measuring Low Viscosities and High Shear Rates with a Rotational Rheometer in a Thin-Gap Parallel-Disk Configuration. Appl. Rheol. 2014, 24, 1–6. [Google Scholar] [CrossRef]
- Pipe, C.J.; Majmudar, T.S.; McKinley, G.H. High Shear Rate Viscometry. Rheol. Acta 2008, 47, 621–642. [Google Scholar] [CrossRef]
- Dakhil, H.; Auhl, D.; Wierschem, A. Infinite-Shear Viscosity Plateau of Salt-Free Aqueous Xanthan Solutions. J. Rheol. 2019, 63, 63–69. [Google Scholar] [CrossRef]
- Dakhil, H.; Basu, S.K.; Steiner, S.; Gerlach, Y.; Soller, A.; Pan, S.; Germann, N.; Leidenberger, M.; Kappes, B.; Wierschem, A. Buffered λ-DNA Solutions at High Shear Rates. J. Rheol. 2021, 65, 159–169. [Google Scholar] [CrossRef]
- Dakhil, H.; Do, H.; Hübner, H.; Wierschem, A. Measuring the Adhesion Limit of Fibronectin for Fibroblasts with a Narrow-Gap Rotational Rheometer. Bioprocess Biosyst. Eng. 2018, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kokkinos, D.; Dakhil, H.; Wierschem, A.; Briesen, H.; Braun, A. Deformation and Rupture of Dunaliella Salina at High Shear Rates without the Use of Thickeners. Biorheology 2016, 53, 1–11. [Google Scholar] [CrossRef]
- Dakhil, H.; Basu, S.K.; Lee, S.; Bashir, K.M.I.; Wierschem, A. Quantitative Measurement of Rheological Cell Properties and the Impact of Drugs. In Proceedings of the ANBRE19 Conference, Jeju, Korea, 17–21 September 2019; pp. 17–21. [Google Scholar]
- Dakhil, H.; Do, H.; Wierschem, A. Adhesion Limit of Fibroblasts Measured in a Narrow-Gap Rotational Rheometer. In Proceedings of the Experimentelle Strömungsmechanik Conference, Cottbus, Germany, 6–8 September 2016; pp. 2–7. [Google Scholar]
- Lee, S.; Bashir, K.M.I.; Jung, D.H.; Basu, S.K.; Seo, G.; Cho, M.-G.; Wierschem, A. Measuring the Linear Viscoelastic Regime of MCF-7 Cells with a Monolayer Rheometer in the Presence of Microtubule-Active Anticancer Drugs at High Concentrations. Interface Focus 2022. Manuscript in review. [Google Scholar]
- Mammoto, T.; Ingber, D.E. Mechanical Control of Tissue and Organ Development. Development 2010, 137, 1407–1420. [Google Scholar] [CrossRef] [Green Version]
- Chan, C.J.; Heisenberg, C.P.; Hiiragi, T. Coordination of Morphogenesis and Cell-Fate Specification in Development. Curr. Biol. 2017, 27, R1024–R1035. [Google Scholar] [CrossRef] [Green Version]
- Guck, J.; Chilvers, E.R. Mechanics Meets Medicine. Sci. Transl. Med. 2013, 5, 212fs41. [Google Scholar] [CrossRef]
- Nematbakhsh, Y.; Lim, C.T. Cell Biomechanics and Its Applications in Human Disease Diagnosis. Acta Mech. Sinica 2015, 31, 268–273. [Google Scholar] [CrossRef]
- Lautenschläger, F.; Paschke, S.; Schinkinger, S.; Bruel, A.; Beil, M.; Guck, J. The Regulatory Role of Cell Mechanics for Migration of Differentiating Myeloid Cells. Proc. Natl. Acad. Sci. USA 2009, 106, 15696–15701. [Google Scholar] [CrossRef] [Green Version]
- Lammerding, J.; Schulze, P.C.; Takahashi, T.; Kozlov, S.; Sullivan, T.; Kamm, R.D.; Stewart, C.L.; Lee, R.T. Lamin A/C Deficiency Causes Defective Nuclear Mechanics and Mechanotransduction. J. Clin. Investig. 2004, 113, 370–378. [Google Scholar] [CrossRef] [Green Version]
- Wirtz, D.; Konstantopoulos, K.; Searson, P.C. The Physics of Cancer: The Role of Physical Interactions and Mechanical Forces in Metastasis. Nat. Rev. Cancer 2011, 11, 512. [Google Scholar] [CrossRef] [Green Version]
- Phillip, J.M.; Aifuwa, I.; Walston, J.; Wirtz, D. The Mechanobiology of Aging. Annu. Rev. Biomed. Eng. 2015, 17, 113–141. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Kaneko, M. On-Chip Cell Manipulation and Applications to Deformability Measurements. ROBOMECH J. 2020, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ojima, I.; Wang, X.; Jing, Y.; Wang, C. Quest for Efficacious Next-Generation Taxoid Anticancer Agents and Their Tumor-Targeted Delivery. J. Nat. Prod. 2018, 81, 703–721. [Google Scholar] [CrossRef] [Green Version]


| Method | Experimental Condition | Measured Moduli | Advantages | Limitations | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| Tissue | Single Cell | Adherent Cells | Suspended Cells | Perturbations in Real Time | |||||
 Atomic force microscopy | x | x | x | - | x | E |
|
| [2,29,30,31,32] |
 Magnetic twisting cytometry | x | x | x | - | x | G |
|
| [33,34] |
 Magnetic tweezers | x | x | x | - | x | G |
|
| [6,25,35,36,37] |
 Optical tweezers | x | x | x | - | x | G |
|
| [38,39,40] |
 Microplate rheometer | x | x | x | - | x | E |
|
| [41,42,43,44] |
 Particle-tracking microrheology/ Nanorheology | x | x | x | - | x | G |
|
| [45,46,47] |
 Micropipette aspiration | x | x | x | - | x | E |
|
| [48,49,50,51] |
 Optical stretching | - | x | - | x | x | E |
|
| [38,52,53,54,55] |
 Cell monolayer rheology | x | - | x | - | x | G |
|
| [11,20,56,57,58] |
 Microfluidic techniques | - | x | x | x | x | G |
|
| [27,59,60,61] |
| Type of Cells | Measured Moduli | Gap Width (µm) | Gap-Width Precision (µm) | Characteristic Frequency | Characteristic Features | Reference |
|---|---|---|---|---|---|---|
| 3T3 fibroblasts | G′ and G″ | 10 | ±1 | Amplitude sweep: 5 Hz Frequency sweep: 0.1–10 Hz |
| [56] |
| Stromal vascular cells | Gr | 5 | ±0.25 | - |
| [67] |
| 3T3 fibroblasts | Adhesion strength and G* | 6.89 | - | - |
| [57] |
| 3T6 fibroblasts 1, Human fibroblasts, HeLa cells 2 | G′ and G″ | 1 5 2 14 | ±0.7 | Amplitude sweep: 1 Hz Frequency sweep: 0.1–10 Hz |
| [92] |
| 3T6 fibroblasts | Adhesion limit | 40 | ±1 | - |
| [89] |
| MCF-7 | G′ and G″ | 15 | - | Amplitude sweep: 0.5 Hz Frequency sweep: 0.1–10 Hz |
| [2] |
| Murine 3T6 fibroblasts | G′ and G″ | 5 | ±1 | Amplitude sweep: 1 Hz Frequency sweep: 0.1–10 Hz |
| [91] |
| MDCK-II epithelial cells | Adhesion strength, G′ and G″ | 160–200 | - | Amplitude sweep: 1, 5 and 10 rad/s Frequency sweep: 1–100 rad/s |
| [58] |
| MCF-7 | G′ and G″ | 13 | ±0.4 | Amplitude sweep: 1 rad/s Frequency sweep: 0.1–30 rad/s |
| [93] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bashir, K.M.I.; Lee, S.; Jung, D.H.; Basu, S.K.; Cho, M.-G.; Wierschem, A. Narrow-Gap Rheometry: A Novel Method for Measuring Cell Mechanics. Cells 2022, 11, 2010. https://doi.org/10.3390/cells11132010
Bashir KMI, Lee S, Jung DH, Basu SK, Cho M-G, Wierschem A. Narrow-Gap Rheometry: A Novel Method for Measuring Cell Mechanics. Cells. 2022; 11(13):2010. https://doi.org/10.3390/cells11132010
Chicago/Turabian StyleBashir, Khawaja Muhammad Imran, Suhyang Lee, Dong Hee Jung, Santanu Kumar Basu, Man-Gi Cho, and Andreas Wierschem. 2022. "Narrow-Gap Rheometry: A Novel Method for Measuring Cell Mechanics" Cells 11, no. 13: 2010. https://doi.org/10.3390/cells11132010






