Differential Transcriptional Responses in Two Old World Bemisia tabaci Cryptic Species Post Acquisition of Old and New World Begomoviruses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of Plants and Insect Rearing
2.2. Feeding Assay, DNA & RNA Isolation, and RNA Sequencing
2.3. RNA Sequencing, Transcriptome Assembly, and Analysis
2.4. Validation of RNA Sequencing Data
3. Results
3.1. Summary of RNA Sequencing
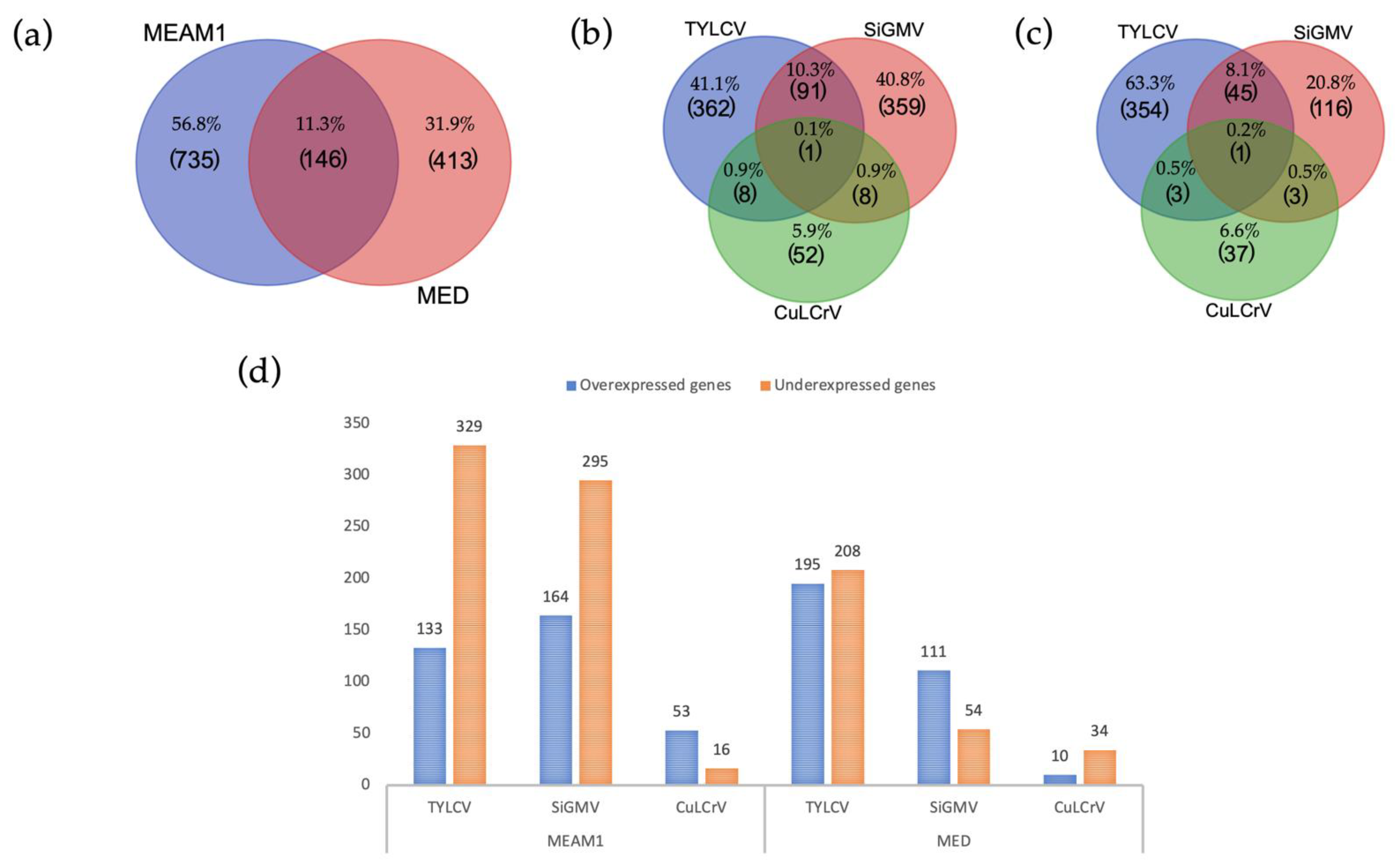
3.2. Overview of Differentially Expressed Genes in B. tabaci MEAM1 and MED Adults
3.3. Common DEGs among B. tabaci MEAM1 and MED Adults
3.4. Unique DEGs in B. tabaci MEAM1 and MED Adults
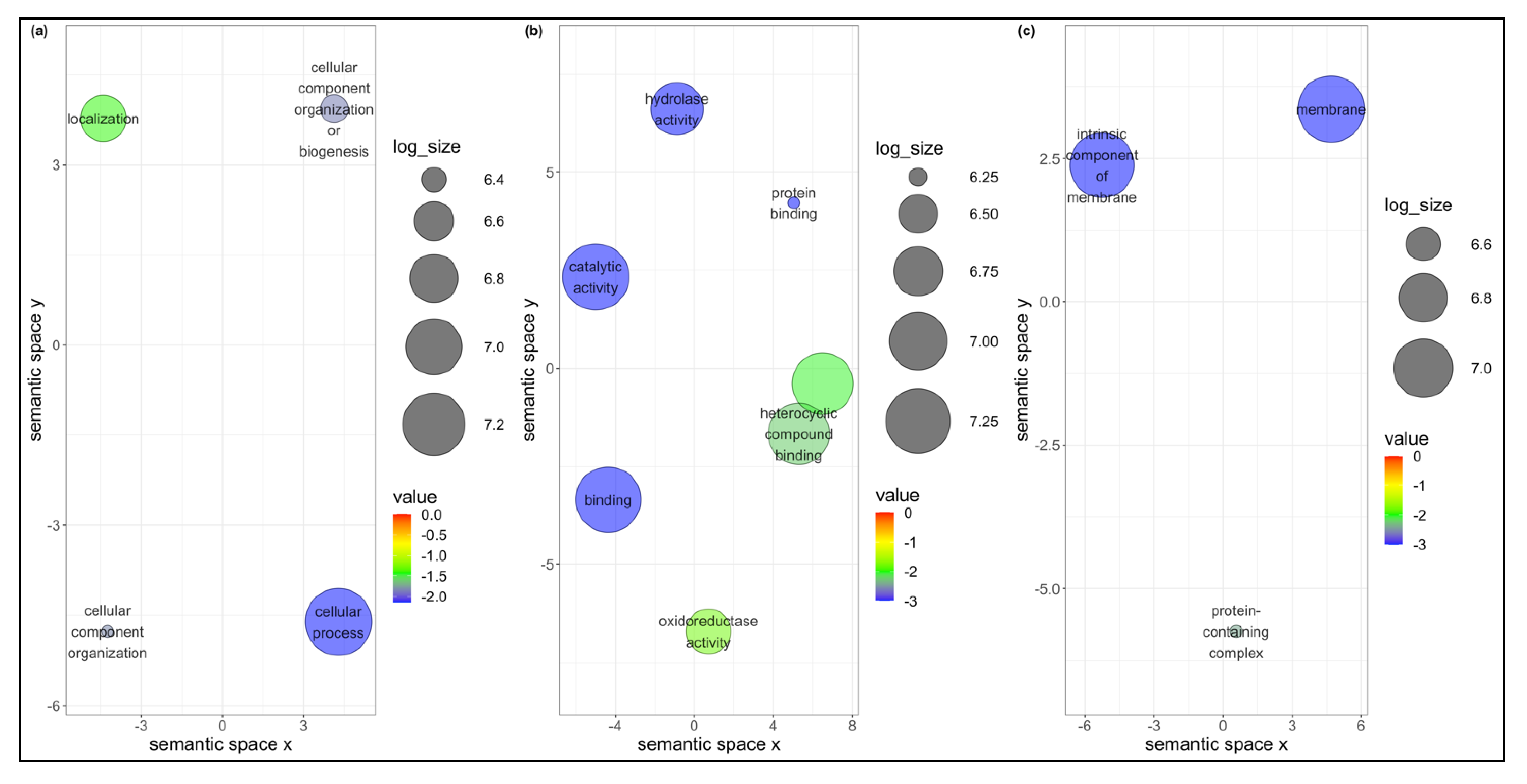
3.5. Co-Expression Networks from B. tabaci MEAM1 and MED Adults
3.6. DEGs among B. tabaci MEAM1 and MED Adults Associated with Virus–Vector Interactions
3.6.1. Virus Infection
3.6.2. Signal Transduction
3.6.3. Signaling Molecules and Virus Interaction
3.6.4. Immune Systems
3.6.5. Cellular Processes (Apoptosis, Lysosome, and Phagosome)
3.7. DEGs among B. tabaci MEAM1 and MED Adults Associated with Fitness
3.7.1. Reproduction
3.7.2. Longevity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brown, J.K.; Frohlich, D.R.; Rosell, R.C. The sweetpotato or silverleaf whiteflies: Biotypes of Bemisia tabaci or species complex? Annu. Rev. Entomol. 1995, 40, 511–534. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.-S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Ghanim, M. Factors Determining Transmission of Persistent Viruses by Bemisia tabaci and Emergence of New Virus–Vector Relationships. Viruses 2021, 13, 1808. [Google Scholar] [CrossRef]
- Liu, G.; Yu, F.X.; Kim, Y.C.; Meng, Z.; Naipauer, J.; Looney, D.J.; Liu, X.; Gutkind, J.S.; Mesri, E.A.; Guan, K.L. Kaposi sarcoma-associated herpesvirus promotes tumorigenesis by modulating the Hippo pathway. Oncogene 2015, 34, 3536–3546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Rivera-Bustamante, R.; Roumagnac, P.; Varsani, A. ICTV virus taxonomy profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Byrne, D.N.; Bellows, T.S.J. Whitefly biology. Annu. Rev. Entomol. 1991, 36, 431–457. [Google Scholar] [CrossRef]
- Sseruwagi, P.; Legg, J.P.; Maruthi, M.N.; Colvin, J.; Rey, M.E.C.; Brown, J.K. Genetic diversity of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) populations and presence of the B biotype and a non-B biotype that can induce silverleaf symptoms in squash, in Uganda. Ann. Appl. Biol. 2005, 147, 253–265. [Google Scholar] [CrossRef]
- Brown, J.K. Phylogenetic biology of the Bemisia tabaci sibling species group. In Bemisia: Bionomics and Management of a Global Pest; Springer: Dordrecht, The Netherland, 2010; pp. 31–67. [Google Scholar] [CrossRef]
- Gill, R.J.; Brown, J.K. Systematics of Bemisia and Bemisia Relatives: Can Molecular Techniques Solve the Bemisia tabaci Complex Conundrum—A Taxonomist’s Viewpoint. In Bemisia: Bionomics and Management of a Global Pest; Springer: Dordrecht, The Netherland, 2010; pp. 5–29. [Google Scholar] [CrossRef]
- de Moya, R.S.; Brown, J.K.; Sweet, A.D.; Kimberly, K.K.; Paredes-Montero, J.R.; Waterhouse, R.M.; Johnson, K.P. Nuclear orthologs derived from whole genome sequencing indicate cryptic diversity in the Bemisia tabaci (Insecta: Aleyrodidae) complex of whiteflies. Diversity 2019, 11, 151. [Google Scholar] [CrossRef] [Green Version]
- Dinsdale, A.; Cook, L.; Riginos, C.; Buckley, Y.M.; De Barro, P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann. Entomol. Soc. Am. 2010, 103, 196–208. [Google Scholar] [CrossRef]
- Liu, S.-S.; Colvin, J.; De Barro, P.J. Species concepts as applied to the whitefly Bemisia tabaci systematics: How many species are there? J. Integr. Agric. 2012, 11, 176–186. [Google Scholar] [CrossRef]
- Mugerwa, H.; Colvin, J.; Alicai, T.; Omongo, C.A.; Kabaalu, R.; Visendi, P.; Sseruwagi, P.; Seal, S.E. Genetic diversity of whitefly (Bemisia spp.) on crop and uncultivated plants in Uganda: Implications for the control of this devastating pest species complex in Africa. J. Pest Sci. 2021, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.R. Plant viruses transmitted by whiteflies. Eur. J. Plant Pathol. 2003, 109, 195–219. [Google Scholar] [CrossRef]
- Dennehy, T.J.; Degain, B.A.; Harpold, V.S.; Brown, J.K.; Morin, S.; Fabrick, J.A.; Byrne, F.J.; Nichols, R.L. New Challenges to Management of Whitefly Resistance to Insecticides in Arizona; University of Arizona Cooperative Extension: Tucson, AZ, USA, 2005. [Google Scholar]
- Liu, S.-S.; De Barro, P.J.; Xu, J.; Luan, J.-B.; Zang, L.-S.; Ruan, Y.-M.; Wan, F.-H. Asymmetric mating interactions drive widespread invasion and displacement in a whitefly. Science 2007, 318, 1769–1772. [Google Scholar] [CrossRef]
- Chi, Y.; Pan, L.-L.; Bouvaine, S.; Fan, Y.-Y.; Liu, Y.-Q.; Liu, S.-S.; Seal, S.; Wang, X.-W. Differential transmission of Sri Lankan cassava mosaic virus by three cryptic species of the whitefly Bemisia tabaci complex. Virology 2020, 540, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Mugerwa, H.; Wang, H.; Sseruwagi, P.; Seal, S.; Colvin, J. Whole-genome single nucleotide polymorphism and mating compatibility studies reveal the presence of distinct species in sub-Saharan Africa Bemisia tabaci whiteflies. Insect Sci. 2020, 28, 1553–1566. [Google Scholar] [CrossRef] [PubMed]
- Adkins, S.; Polston, J.E.; Turechek, W.W. Cucurbit leaf crumple virus identified in common bean in Florida. Plant Dis. 2009, 93, 320. [Google Scholar] [CrossRef]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 71, 123–134. [Google Scholar] [CrossRef]
- Durham, T.C.; Baker, C.; Jones, L.; Snyder, L.U. First report of Sida golden mosaic virus infecting snap bean (Phaseolus vulgaris) in Florida. Plant Dis. 2010, 94, 487. [Google Scholar] [CrossRef]
- Czosnek, H.; Hariton-Shalev, A.; Sobol, I.; Gorovits, R.; Ghanim, M. The incredible journey of Begomoviruses in their whitefly vector. Viruses 2017, 9, 273. [Google Scholar] [CrossRef]
- Gotz, M.; Popovski, S.; Kollenberg, M.; Gorovits, R.; Brown, J.K.; Cicero, J.M.; Czosnek, H.; Winter, S.; Ghanim, M. Implication of Bemisia tabaci Heat Shock Protein 70 in Begomovirus-Whitefly Interactions. J. Virol. 2012, 86, 13241–13252. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.Z.; Shi, M.; Huang, Y.C.; Wang, X.W.; Stanley, D.; Chen, X.X. A peptidoglycan recognition protein acts in whitefly (Bemisia tabaci) immunity and involves in Begomovirus acquisition. Sci. Rep. 2016, 6, 37806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, V.S.; Popli, S.; Saurav, G.K.; Raina, H.S.; Chaubey, R.; Ramamurthy, V.V.; Rajagopal, R. A Bemisia tabaci midgut protein interacts with begomoviruses and plays a role in virus transmission. Cell. Microbiol. 2016, 18, 663–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanakala, S.; Ghanim, M. Implication of the whitefly Bemisia tabaci cyclophilin B protein in the transmission of tomato yellow leaf curl virus. Front. Plant Sci. 2016, 7, 1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, V.S.; Singh, S.T.; Priya, N.G.; Kumar, J.; Rajagopal, R. Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci. PLoS ONE 2012, 7, e42168. [Google Scholar] [CrossRef]
- Costa, H.S.; Brown, J.; Sivasupramaniam, S.; Bird, J. Regional distribution, insecticide resistance and reciprocal crosses between the A-biotype and B-biotype of Bemisia tabaci. Int. J. Trop. Insect Sci. 1993, 14, 255–266. [Google Scholar] [CrossRef]
- Mckenzie, C.L.; Osborne, L.S. Bemisia tabaci MED (Q biotype) (Hemiptera: Aleyrodidae) in Florida is on the Move to Residential Landscapes and May Impact Open-Field Agriculture. Fla. Entomol. 2017, 100, 481–484. [Google Scholar] [CrossRef] [Green Version]
- Gautam, S.; Crossley, M.S.; Dutta, B.; Coolong, T.; Simmons, A.M.; da Silva, A.; Snyder, W.E.; Srinivasan, R. Low genetic variability in Bemisia tabaci MEAM1 populations within farmscapes of Georgia, USA. Insects 2020, 11, 834. [Google Scholar] [CrossRef]
- McKenzie, C.L.; Sparks, A.N.; Roberts, P.; Oetting, R.D.; Osborne, L.S. Survey of Bemisia tabaci (Hemiptera: Aleyrodidae) in agricultural ecosystems in Georgia1. J. Entomol. Sci. 2020, 55, 163–170. [Google Scholar] [CrossRef]
- Dutta, B.; Myer, B.; Coolong, T.; Srinivasan, R.; Sparks, A. Whitefly-Transmitted Plant Viruses in South Georgia. UGA Coop. Ext. Bull. 2018, 1507, 1–7. [Google Scholar]
- Agarwal, G.; Kavalappara, S.R.; Gautam, S.; da Silva, A.; Simmons, A.; Srinivasan, R.; Dutta, B. Field screen and genotyping of Phaseolus vulgaris against two Begomoviruses in Georgia, USA. Insects 2021, 12, 49. [Google Scholar] [CrossRef]
- Li, M.; Hu, J.; Xu, F.-C.; Liu, S.-S. Transmission of Tomato Yellow Leaf Curl Virus by two invasive biotypes and a Chinese indigenous biotype of the whitefly Bemisia tabaci. Int. J. Pest Manag. 2010, 56, 275–280. [Google Scholar] [CrossRef]
- Ning, W.; Shi, X.; Liu, B.; Pan, H.; Wei, W.; Zeng, Y.; Sun, X.; Xie, W.; Wang, S.; Wu, Q.; et al. Transmission of tomato yellow leaf curl virus by Bemisia tabaci as affected by whitefly sex and biotype. Sci. Rep. 2015, 5, 10744. [Google Scholar] [CrossRef] [PubMed]
- Gautam, S.; Mugerwa, H.; Buck, J.W.; Dutta, B.; Coolong, T.; Adkins, S.; Srinivasan, R. Differential Transmission of Old and New World Begomoviruses by Middle East-Asia Minor 1 (MEAM1) and Mediterranean (MED) Cryptic Species of Bemisia tabaci. Viruses 2022, 14, 1104. [Google Scholar] [CrossRef]
- Sisterson, M.S. Effects of Insect-Vector Preference for Healthy or Infected Plants on Pathogen Spread: Insights from a Model. J. Econ. Entomol. 2008, 101, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mauck, K.; Bosque-Pérez, N.A.; Eigenbrode, S.D.; De Moraes, C.M.; Mescher, M.C. Transmission mechanisms shape pathogen effects on host-vector interactions: Evidence from plant viruses. Funct. Ecol. 2012, 26, 1162–1175. [Google Scholar] [CrossRef]
- Legarrea, S.; Barman, A.; Marchant, W.; Diffie, S.; Srinivasan, R. Temporal effects of a begomovirus infection and host plant resistance on the preference and development of an insect vector, Bemisia tabaci, and implications for epidemics. PLoS ONE 2015, 10, e0142114. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Liu, J.; Liu, S.-S. Tomato yellow leaf curl virus infection of tomato does not affect the performance of the Q and ZHJ2 biotypes of the viral vector Bemisia tabaci. Insect Sci. 2011, 18, 40–49. [Google Scholar] [CrossRef]
- Liu, J.; Li, M.; Li, J.; Huang, C.; Zhou, X.; Xu, F.; Liu, S. Viral infection of tobacco plants improves performance of Bemisia tabaci but more so for an invasive than for an indigenous biotype of the whitefly. J. Zhejiang Univ. Sci. B 2010, 11, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Gautam, S.; Gadhave, K.R.; Buck, J.W.; Dutta, B.; Coolong, T.; Adkins, S.; Srinivasan, R. Virus-virus interactions in a plant host and in a hemipteran vector: Implications for vector fitness and virus epidemics. Virus Res. 2020, 286, 198069. [Google Scholar] [CrossRef]
- Catto, M.A.; Mugerwa, H.; Myers, B.K.; Pandey, S.; Dutta, B.; Srinivasan, R. A review on transcriptional responses of interactions between insect vectors and plant viruses. Cells 2022, 11, 693. [Google Scholar] [CrossRef]
- Luan, J.-B.; Li, J.-M.; Varela, N.; Wang, Y.-L.; Li, F.-F.; Bao, Y.-Y.; Zhang, C.-X.; Liu, S.-S.; Wang, X.-W. Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 2011, 85, 3330–3340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaur, N.; Chen, W.; Zheng, Y.; Hasegawa, D.K.; Ling, K.S.; Fei, Z.; Wintermantel, W.M. Transcriptome analysis of the whitefly, Bemisia tabaci MEAM1 during feeding on tomato infected with the crinivirus, Tomato chlorosis virus, identifies a temporal shift in gene expression and differential regulation of novel orphan genes. BMC Genom. 2017, 18, 380. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Chen, W.; Fei, Z.; Wintermantel, W.M. Differences in gene expression in whitefly associated with CYSDV-infected and virus-free melon, and comparison with expression in whiteflies fed on ToCV- and TYLCV-infected tomato. BMC Genom. 2019, 20, 654. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, D.K.; Chen, W.; Zheng, Y.; Kaur, N.; Wintermantel, W.M.; Simmons, A.M.; Fei, Z.; Ling, K.S. Comparative transcriptome analysis reveals networks of genes activated in the whitefly, Bemisia tabaci when fed on tomato plants infected with Tomato yellow leaf curl virus. Virology 2018, 513, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, J.; Su, Y.-L. Transcriptome Analysis of Gene Expression Profiles of Tomato Yellow Leaf Curl Virus-Infected Whiteflies over Different Viral Acquisition Access Periods. Insects 2020, 11, 297. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.-B.; Li, J.; Chen, E.-H.; Niu, J.-Z.; Chu, D. Transcriptome Profiling of the Whitefly Bemisia tabaci MED in Response to Single Infection of Tomato yellow leaf curl virus, Tomato chlorosis virus, and Their Co-infection. Front. Physiol. 2019, 10, 302. [Google Scholar] [CrossRef] [Green Version]
- Luan, J.B.; Wang, Y.L.; Wang, J.; Wang, X.W.; Liu, S.S. Detoxification activity and energy cost is attenuated in whiteflies feeding on Tomato yellow leaf curl China virus-infected tobacco plants. Insect Mol. Biol. 2013, 22, 597–607. [Google Scholar] [CrossRef]
- Chen, W.; Hasegawa, D.K.; Kaur, N.; Kliot, A.; Pinheiro, P.V.; Luan, J.; Stensmyr, M.C.; Zheng, Y.; Liu, W.; Sun, H.; et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016, 14, 110. [Google Scholar] [CrossRef]
- Geng, L.; Qian, L.X.; Shao, R.X.; Liu, Y.Q.; Liu, S.S.; Wang, X.W. Transcriptome profiling of whitefly guts in response to Tomato yellow leaf curl virus infection. Virol. J. 2018, 15, 14. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Wang, L.; Chen, F.; Yang, X.; Ding, M.; Zhang, Z.; Liu, S.S.; Wang, X.W.; Zhou, X. MicroRNA profiling of the whitefly Bemisia tabaci Middle East-Aisa Minor i following the acquisition of Tomato yellow leaf curl China virus Plant viruses. Virol. J. 2016, 13, 20. [Google Scholar] [CrossRef] [Green Version]
- Nekkanti, A.; Chakraborty, P.; Ghosh, A.; Iquebal, M.A.; Jaiswal, S.; Baranwal, V.K. Transcriptomic Changes of Bemisia tabaci Asia II 1 Induced by Chilli Leaf Curl Virus Trigger Infection and Circulation in Its Vector. Front. Microbiol. 2022, 13, 1310. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, D.R.; Torres-Jerez, I.D.; Bedford, D.; Markham, G.P.; Brown, J.K. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol. Ecol. 1999, 8, 1683–1691. [Google Scholar] [CrossRef] [PubMed]
- Mugerwa, H.; Seal, S.; Wang, H.-L.; Patel, M.V.; Kabaalu, R.; Omongo, C.A.; Alicai, T.; Tairo, F.; Ndunguru, J.; Sseruwagi, P.; et al. African ancestry of New World, Bemisia tabaci-whitefly species. Sci. Rep. 2018, 8, 2734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kent, W.J. BLAT-The BLAST-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Chen, C.; Yang, Z.; Guo, L.; Yang, X.; Wang, D.; Chen, M.; Huang, J.; Wen, Y.; Zeng, Y.; et al. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. Gigascience 2017, 6, gix018. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Gingeras, T.R. Mapping RNA-seq Reads with STAR. Curr. Protoc. Bioinform. 2015, 51, 11.14.1–11.14.19. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [Green Version]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- R Core Team, R. R: A Language and Environment for Statistical Computing; R Foubdation for Statistical Computing: Vienna, Austria, 2013; Volume 1, ISBN 3900051070. [Google Scholar]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Sinisterra, X.H.; McKenzie, C.L.; Hunter, W.B.; Powell, C.A.; Shatters, R.G. Differential transcriptional activity of plant-pathogenic begomoviruses in their whitefly vector (Bemisia tabaci, Gennadius: Hemiptera Aleyroadidae). J. Gen. Virol. 2005, 86, 1525–1532. [Google Scholar] [CrossRef]
- Li, J.-M.; Ruan, Y.-M.; Li, F.-F.; Liu, S.-S.; Wang, X.-W. Gene expression profiling of the whitefly (Bemisia tabaci) Middle East—Asia Minor 1 feeding on healthy and Tomato yellow leaf curl China virus-infected tobacco. Insect Sci. 2011, 18, 11–22. [Google Scholar] [CrossRef]
- Bedford, I.D.; Briddon, R.W.; Brown, J.K.; Rosell, R.C.; Markham, P.G. Geminivirus transmission and biological characterisation of Bemisia tabaci (Gennadius) biotypes from different geographic regions. Ann. Appl. Biol. 1994, 125, 311–325. [Google Scholar] [CrossRef]
- Hogenhout, S.A.; Ammar, E.; Whitfield, A.E.; Redinbaugh, M.G. Insect Vector Interactions with Persistently Transmitted Viruses. Annu. Rev. Phytopathol. 2008, 46, 327–359. [Google Scholar] [CrossRef] [Green Version]
- Idris, A.M.; Smith, S.E.; Brown, J.K. Ingestion, Transmission, and Persistence of Chino del Tomate Virus (CdTV), a New World Begomovirus, by Old and New World Biotypes of the Whitefly Vector Bemisia tabaci. Ann. Appl. Biol. 2001, 139, 145–154. [Google Scholar] [CrossRef]
- Bello, V.H.; Watanabe, L.F.M.; Santos, B.R.; Marubayashi, J.M.; Yuki, V.A.; De Marchi, B.R.; Pavan, M.A.; Krause-Sakate, R. Evidence for increased efficiency of virus transmission by populations of Mediterranean species of Bemisia tabaci with high Hamiltonella prevalence. Phytoparasitica 2019, 47, 293–300. [Google Scholar] [CrossRef]
- Maynard, N.D.; Gutschow, M.V.; Birch, E.W.; Covert, M.W. The virus as metabolic engineer. Biotechnol. J. 2010, 5, 686–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesquita, I.; Estaquier, J. Viral manipulation of the host metabolic network. Exp. Suppl. 2018, 109, 377–401. [Google Scholar] [PubMed]
- Shrestha, A.; Champagne, D.E.; Culbreath, A.K.; Rotenberg, D.; Whitfield, A.E.; Srinivasan, R. Transcriptome changes associated with Tomato spotted wilt virus infection in various life stages of its thrips vector, Frankliniella fusca (Hinds). J. Gen. Virol. 2017, 98, 2156–2170. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, P.; Li, W.; Zhang, J.; Huang, F.; Yang, J.; Bei, Y.; Lu, Y. De novo transcriptome sequencing in Frankliniella occidentalis to identify genes involved in plant virus transmission and insecticide resistance. Genomics 2013, 101, 296–305. [Google Scholar] [CrossRef] [Green Version]
- Becker, N.; Rimbaud, L.; Chiroleu, F.; Reynaud, B.; Thebaud, G.; Lett, J.M. Rapid accumulation and low degradation: Key parameters of Tomato yellow leaf curl virus persistence in its insect vector Bemisia tabaci. Sci. Rep. 2015, 5, 17696. [Google Scholar] [CrossRef]
- Sánchez-Campos, S.; Rodríguez-Negrete, E.A.; Cruzado, L.; Grande-Pérez, A.; Bejarano, E.R.; Navas-Castillo, J.; Moriones, E. Tomato yellow leaf curl virus: No evidence for replication in the insect vector Bemisia tabaci. Sci. Rep. 2016, 6, 30942. [Google Scholar] [CrossRef] [Green Version]
- He, Y.Z.; Wang, Y.M.; Yin, T.Y.; Fiallo-Olivé, E.; Liu, Y.Q.; Hanley-Bowdoin, L.; Wang, X.W. A plant DNA virus replicates in the salivary glands of its insect vector via recruitment of host DNA synthesis machinery. Proc. Natl. Acad. Sci. USA 2020, 117, 16928–16937. [Google Scholar] [CrossRef]
- Han, J.; Rotenberg, D. Integration of transcriptomics and network analysis reveals co-expressed genes in Frankliniella occidentalis larval guts that respond to tomato spotted wilt virus infection. BMC Genom. 2021, 22, 810. [Google Scholar] [CrossRef]
- Tian, J.; Zhan, H.; Dewer, Y.; Zhang, B.; Qu, C.; Luo, C.; Li, F.; Yang, S. Whitefly Network Analysis Reveals Gene Modules Involved in Host Plant Selection, Development and Evolution. Front. Physiol. 2021, 12, 656649. [Google Scholar] [CrossRef]
- Zhang, F.; Chen, Y.; Shen, J.; Zhang, J. The Ubiquitin Conjugating Enzyme UbcD1 is Required for Notch Signaling Activation during Drosophila Wing Development. Front. Genet. 2021, 12, 2030. [Google Scholar] [CrossRef]
- Lin, H.; Xia, X.; Yu, L.; Vasseur, L.; Gurr, G.M.; Yao, F.; Yang, G.; You, M. Genome-wide identification and expression profiling of serine proteases and homologs in the diamondback moth, Plutella xylostella (L.). BMC Genom. 2015, 16, 1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.J.; Chen, C.X.; Yan, Y.; Xu, K.K.; Li, C. Clip-Domain Serine Protease Gene (LsCLIP3) Is Essential for Larval–Pupal Molting and Immunity in Lasioderma serricorne. Front. Physiol. 2020, 10, 1631. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.C.; Wang, Q.H.; Bhowmick, B.; Li, Y.X.; Han, Q. Functional characterization of two clip domain serine proteases in innate immune responses of Aedes aegypti. Parasites Vectors 2021, 14, 584. [Google Scholar] [CrossRef] [PubMed]
- Shakeel, M.; Xu, X.; De Mandal, S.; Jin, F. Role of serine protease inhibitors in insect-host-pathogen interactions. Arch. Insect Biochem. Physiol. 2019, 102, e21556. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shen, Y.; Yang, R.; Wu, X.; Hu, W.; Shen, G. Bombyx mori nucleopolyhedrovirus (BmNPV) Bm64 is required for BV production and per os infection. Virol. J. 2015, 12, 173. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Guo, H.; Zhu-Salzman, K.; Ge, F.; Sun, Y. PEBP balances apoptosis and autophagy in whitefly upon arbovirus infection. Nat. Commun. 2022, 13, 846. [Google Scholar] [CrossRef]
- Shin, J.; Nile, A.; Oh, J.W. Role of adaptin protein complexes in intracellular trafficking and their impact on diseases. Bioengineered 2021, 12, 8259. [Google Scholar] [CrossRef]
- Ward, C.; Maselko, M.; Lupfer, C.; Prescott, M.; Pastey, M.K. Interaction of the Human Respiratory Syncytial Virus matrix protein with cellular adaptor protein complex 3 plays a critical role in trafficking. PLoS ONE 2017, 12, e0184629. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Dandekar, S. Targeting NF-κB signaling with protein Kinase C agonists as an emerging strategy for combating HIV latency. AIDS Res. Hum. Retrovir. 2015, 31, 4–12. [Google Scholar] [CrossRef]
- Kamakura, M.; Goshima, F.; Luo, C.; Kimura, H.; Nishiyama, Y. Herpes simplex virus induces the marked up-regulation of the zinc finger transcriptional factor INSM1, which modulates the expression and localization of the immediate early protein ICP0. Virol. J. 2011, 8, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Erlichman, C.; Mcdonald, C.J.; Ingle, J.N.; Zollman, P.; Iankov, I.; Russell, S.J.; Galanis, E.; Author, G.T. Heat shock protein inhibitors increase the efficacy of measles virotherapy NIH Public Access Author Manuscript. Gene Ther. 2008, 15, 1024–1034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Wang, J.; Shuai, L.; Ma, X.; Zhang, H.; Liu, R.; Chen, W.; Wang, X.; Ge, J.; Wen, Z.; et al. The serine/threonine kinase AP2-associated kinase 1 plays an important role in rabies virus entry. Viruses 2019, 12, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teng, Y.H.F.; Aquino, R.S.; Park, P.W. Molecular functions of syndecan-1 in disease. Matrix Biol. 2012, 31, 3–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, M.; Chandra, V.; Rahman, S.A.; Sehgal, D.; Jameel, S. Heparan sulfate proteoglycans are required for cellular binding of the Hepatitis E Virus ORF2 capsid protein and for viral Infection. J. Virol. 2009, 83, 12714–12724. [Google Scholar] [CrossRef] [Green Version]
- Saphire, A.C.S.; Bobardt, M.D.; Zhang, Z.; David, G.; Gallay, P.A. Syndecans serve as attachment receptors for Human Immunodeficiency Virus Type 1 on macrophages. J. Virol. 2001, 75, 9187–9200. [Google Scholar] [CrossRef] [Green Version]
- Shafti-keramat, S.; Handisurya, A.; Meneguzzi, G.; Slupetzky, K.; Kirnbauer, R.; Kriehuber, E. Different heparan sulfate proteoglycans serve as cellular receptors for human Papillomaviruses. J. Virol. 2003, 77, 13125–13135. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Sivakumar, S.; Sparks, W.O.; Miller, W.A.; Bonning, B.C. A peptide that binds the pea aphid gut impedes entry of Pea enation mosaic virus into the aphid hemocoel. Virology 2010, 401, 107–116. [Google Scholar] [CrossRef] [Green Version]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [Green Version]
- Kanost, M.R.; Jiang, H.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef]
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14. [Google Scholar] [CrossRef]
- Jiang, H.; Vilcinskas, A.; Kanost, M.R. Immunity in lepidopteran insects. Adv. Exp. Med. Biol. 2010, 708, 181–204. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Cellular and Molecular Mechanisms of Insect Immunity. In Insect Physiology and Ecology; Oxford University Press: Oxford, UK, 2017. [Google Scholar]
- Kubo, Y.; Hayashi, H.; Matsuyama, T.; Sato, H.; Yamamoto, N. Retrovirus entry by endocytosis and cathepsin proteases. Adv. Virol. 2012, 2012, 640894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sim, S.; Ramirez, J.L.; Dimopoulos, G. Dengue virus infection of the aedes aegypti salivary gland and chemosensory apparatus induces genes that modulate infection and blood-feeding behavior. PLoS Pathog. 2012, 8, e1002631. [Google Scholar] [CrossRef] [Green Version]
- Huntriss, J.; Lu, J.; Hemmings, K.; Bayne, R.; Anderson, R.; Rutherford, A.; Balen, A.; Elder, K.; Picton, H.M. Isolation and expression of the human gametocyte-specific factor 1 gene (GTSF1) in fetal ovary, oocytes, and preimplantation embryos. J. Assist. Reprod. Genet. 2017, 34, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, T.; Toyoda, S.; Kuramochi-Miyagawa, S.; Miyazaki, T.; Miyazaki, S.; Tashiro, F.; Yamato, E.; Nakano, T.; Miyazaki, J. ichi Gtsf1/Cue110, a gene encoding a protein with two copies of a CHHC Zn-finger motif, is involved in spermatogenesis and retrotransposon suppression in murine testes. Dev. Biol. 2009, 335, 216–227. [Google Scholar] [CrossRef] [Green Version]
- Jiu, M.; Zhou, X.-P.; Tong, L.; Xu, J.; Yang, X.; Wan, F.-H.; Liu, S.-S. Vector-virus mutualism accelerates population increase of an invasive whitefly. PLoS ONE 2007, 2, e182. [Google Scholar] [CrossRef] [Green Version]
- Sidhu, J.S.; Mann, R.S.; Butter, N.S. Deleterious effects of cotton leaf curl virus on longevity and fecundity of whitefly, Bemisia tabaci (Gennadius). J. Entomol. 2009, 6, 62–66. [Google Scholar] [CrossRef]
- Pan, H.; Chu, D.; Liu, B.; Shi, X.; Guo, L.; Xie, W.; Carrière, Y.; Li, X.; Zhang, Y. Differential effects of an exotic plant virus on its two closely related vectors. Sci. Rep. 2013, 3, 2230. [Google Scholar] [CrossRef] [Green Version]







| Whitefly (wf) | Sample Description | No. Raw Read Pairs | No. Final Cleaned Read Pairs | Mapped to B. tabaci MEAM1 Genome | |
|---|---|---|---|---|---|
| No. Mapped | % Mapped | ||||
| MEAM1 | TYLCV viruliferous wf rep 1 | 27,270,100 | 25,637,467 | 24,019,254 | 93.69 |
| TYLCV viruliferous wf rep 2 | 33,377,647 | 31,071,277 | 29,119,197 | 93.72 | |
| TYLCV viruliferous wf rep 3 | 24,223,840 | 21,083,268 | 19,512,995 | 92.55 | |
| TYLCV viruliferous wf rep 4 | 22,603,100 | 20,914,166 | 19,565,243 | 93.55 | |
| TYLCV viruliferous wf rep 5 | 27,080,141 | 25,645,471 | 23,601,484 | 92.03 | |
| TYLCV non-viruliferous wf rep 1 | 23,986,058 | 20,725,007 | 19,011,846 | 91.73 | |
| TYLCV non-viruliferous wf rep 2 | 22,543,749 | 21,484,751 | 20,078,449 | 93.45 | |
| TYLCV non-viruliferous wf rep 3 | 26,130,542 | 25,088,690 | 23,641,100 | 94.23 | |
| TYLCV non-viruliferous wf rep 4 | 19,634,831 | 18,563,885 | 16,658,491 | 89.74 | |
| SiGMV viruliferous wf rep 1 | 24,481,178 | 22,306,202 | 20,999,224 | 94.14 | |
| SiGMV viruliferous wf rep 2 | 25,457,659 | 25,032,825 | 23,802,144 | 95.08 | |
| SiGMV viruliferous wf rep 3 | 29,098,951 | 26,525,917 | 24,902,267 | 93.88 | |
| SiGMV non-viruliferous wf rep 1 | 24,332,531 | 24,082,074 | 22,848,884 | 94.88 | |
| SiGMV non-viruliferous wf rep 2 | 25,933,031 | 25,672,553 | 24,281,335 | 94.58 | |
| SiGMV non-viruliferous wf rep 3 | 22,759,943 | 22,505,467 | 21,186,253 | 94.14 | |
| CuLCrV viruliferous wf rep 1 | 21,397,610 | 19,723,901 | 18,448,967 | 93.54 | |
| CuLCrV viruliferous wf rep 2 | 22,301,938 | 20,532,734 | 19,168,732 | 93.36 | |
| CuLCrV viruliferous wf rep 3 | 21,582,185 | 19,996,867 | 18,700,091 | 93.52 | |
| CuLCrV viruliferous wf rep 4 | 21,650,343 | 20,031,545 | 18,739,720 | 93.55 | |
| CuLCrV viruliferous wf rep 5 | 19,784,311 | 18,353,651 | 17,045,909 | 92.87 | |
| CuLCrV non-viruliferous wf rep 1 | 23,300,250 | 21,146,623 | 19,768,756 | 93.48 | |
| CuLCrV non-viruliferous wf rep 2 | 22,140,101 | 20,294,419 | 19,013,129 | 93.69 | |
| CuLCrV non-viruliferous wf rep 3 | 22,523,401 | 20,735,034 | 19,428,624 | 93.70 | |
| CuLCrV non-viruliferous wf rep 4 | 21,194,688 | 19,576,275 | 18,327,411 | 93.62 | |
| CuLCrV non-viruliferous wf rep 5 | 22,698,997 | 20,790,783 | 19,433,502 | 93.47 | |
| MED | TYLCV viruliferous wf rep 1 | 23,977,947 | 21,535,882 | 19,304,336 | 89.64 |
| TYLCV viruliferous wf rep 2 | 24,395,357 | 21,999,676 | 19,854,074 | 90.25 | |
| TYLCV viruliferous wf rep 3 | 20,9762,11 | 18,879,921 | 17,061,784 | 90.37 | |
| TYLCV viruliferous wf rep 4 | 26,209,320 | 23,610,513 | 21,313,232 | 90.27 | |
| TYLCV non-viruliferous wf rep 1 | 20,685,800 | 18,272,491 | 16,550,563 | 90.58 | |
| TYLCV non-viruliferous wf rep 2 | 22,763,950 | 20,476,851 | 18,633,954 | 91.00 | |
| TYLCV non-viruliferous wf rep 3 | 23,892,860 | 21,062,128 | 19,086,844 | 90.62 | |
| TYLCV non-viruliferous wf rep 4 | 22,570,010 | 19,970,201 | 18,086,769 | 90.57 | |
| TYLCV non-viruliferous wf rep 5 | 20,134,315 | 17,960,386 | 16,234,631 | 90.39 | |
| SiGMV viruliferous wf rep 1 | 22,732,648 | 20,756,619 | 18,734,819 | 90.26 | |
| SiGMV viruliferous wf rep 2 | 19,747,880 | 18,122,103 | 16,407,875 | 90.54 | |
| SiGMV viruliferous wf rep 3 | 22,988,951 | 20,864,408 | 18,730,928 | 89.77 | |
| SiGMV viruliferous wf rep 4 | 22,521,272 | 20,582,499 | 18,583,828 | 90.29 | |
| SiGMV viruliferous wf rep 5 | 29,236,819 | 26,690,404 | 24,037,689 | 90.06 | |
| SiGMV non-viruliferous wf rep 1 | 26,310,239 | 24,295,385 | 21,962,427 | 90.40 | |
| SiGMV non-viruliferous wf rep 2 | 23,960,072 | 22,029,656 | 19,493,192 | 88.49 | |
| SiGMV non-viruliferous wf rep 3 | 24,484,549 | 22,676,536 | 20,522,495 | 90.50 | |
| SiGMV non-viruliferous wf rep 4 | 22,282,789 | 20,521,023 | 18,454,698 | 89.93 | |
| SiGMV non-viruliferous wf rep 5 | 29,952,155 | 27,649,370 | 25,014,401 | 90.47 | |
| CuLCrV viruliferous wf rep 1 | 22,647,116 | 19,812,026 | 17,824,979 | 89.97 | |
| CuLCrV viruliferous wf rep 2 | 22,271,955 | 19,857,105 | 17,933,941 | 90.31 | |
| CuLCrV viruliferous wf rep 3 | 21,412,367 | 17,573,543 | 15,635,887 | 88.97 | |
| CuLCrV viruliferous wf rep 4 | 21,739,525 | 19,470,833 | 17,583,080 | 90.30 | |
| CuLCrV viruliferous wf rep 5 | 22,185,444 | 18,994,180 | 16,993,665 | 89.47 | |
| CuLCrV non-viruliferous wf rep 1 | 21,346,943 | 18,119,382 | 16,207,549 | 89.45 | |
| CuLCrV non-viruliferous wf rep 2 | 21,931,996 | 18,517,714 | 16,503,842 | 89.12 | |
| CuLCrV non-viruliferous wf rep 3 | 28,625,279 | 24,307,191 | 21,703,554 | 89.29 | |
| Gene ID | Annotation | FC in Whitefly & Virus Treatment |
|---|---|---|
| DEGs shared by MEAM1 and MED | ||
| Bta03437 | Zinc finger protein | −2.69 for B-TYLCV, −1.38 for Q-SiGMV |
| Bta02903 | Heat shock protein 70 | −5.55 for B-CuLCrV, 1.83 for Q-CuLCrV |
| Bta09867 | 70 kDa heat shock protein | −4.54 for B-CuLCrV, −1.50 for Q- TYLCV, −2.63 for Q-SiGMV |
| DEGs specific to MEAM1 | ||
| Bta00981 | Cyclic AMP-responsive element-binding protein 3-like protein 2 | 1.60 for SiGMV |
| Bta03000 | Heat shock protein 70 | −5.91 for CuLCrV |
| Bta03004 | Suppressor of hairless protein | 1.07 for TYLCV, 1.29 for SiGMV |
| Bta03628 | Zinc finger protein 569 | −1.30 for SiGMV |
| Bta04025 | Serine/threonine-protein phosphatase 2A regulatory subunit B subunit beta | 1.02 for SiGMV |
| Bta04026 | Serine/threonine-protein phosphatase 2A regulatory subunit B subunit beta | −1.67 for TYLCV |
| Bta04167 | Ras-like GTP-binding protein RHO | 1.22 for SiGMV |
| Bta04484 | Guanine nucleotide-binding protein subunit alpha-like protein | 1.53 for TYLCV |
| Bta04715 | Ras-related C3 botulinum toxin substrate 1 | 1.69 for TYLCV, 1.49 for SiGMV |
| Bta04818 | Type I serine/threonine kinase receptor | 1.47 for SiGMV |
| Bta06076 | Heat shock protein | −1.39 for TYLCV |
| Bta06234 | Guanine nucleotide-binding protein G(O) subunit alpha | 1.34 for SiGMV |
| Bta08159 | Transcription factor 7-like 1-A | 1.50 for SiGMV |
| Bta10215 | Ran-specific GTPase-activating protein | 1.29 for TYLCV |
| Bta10683 | AP-1 complex subunit sigma-2 | 1.56 for TYLCV, 1.59 for SiGMV |
| Bta11558 | Ras-related C3 botulinum toxin substrate 1 | 1.22 for TYLCV, 1.29 for SiGMV |
| Bta11785 | Zinc finger protein | 0.86 for CuLCrV |
| Bta12195 | Phosphoinositide phospholipase C | 1.15 for SiGMV |
| Bta13153 | Sensory neuron membrane protein 1 | 1.04 for SiGMV |
| Bta13437 | Actin | −1.84 for TYLCV |
| Bta14253 | Protein kinase C | 1.54 for SiGMV |
| Bta14532 | 70 kDa heat shock protein | −2.20 for CuLCrV |
| Bta14568 | Guanine nucleotide-binding protein G(S) subunit alpha | 1.12 for TYLCV |
| Bta14627 | Mothers against decapentaplegic homolog | 1.15 for SiGMV |
| Bta15240 | F-box/WD repeat-containing protein 1A | 1.19 for SiGMV |
| Bta15478 | Ras-like GTP-binding protein RHO | 1.45 for SiGMV |
| Bta15565 | Partitioning defective protein 6 | 1.37 for SiGMV |
| DEGs specific to MED | ||
| Bta00008 | 70 kDa heat shock protein | −2.02 for SiGMV |
| Bta00607 | SAM domain and HD domain-containing protein 1 | 0.71 for TYLCV |
| Bta01535 | Zinc finger protein 208 | 1.28 for SIGMV |
| Bta01860 | Calreticulin | 0.72 for TYLCV |
| Bta02113 | Protein disulfide-isomerase | 0.76 for TYLCV |
| Bta06159 | Ribosomal protein L19 | −3.19 for TYLCV |
| Bta12772 | Serine/threonine-protein phosphatase | −0.73 for TYLCV |
| Gene ID | Annotation | FC in Whitefly & Virus Treatment |
|---|---|---|
| DEGs shared by MEAM1 and MED | ||
| Bta03437 | Zinc finger protein | −2.69 for B-TYLCV, −1.38 for Q-SiGMV |
| Bta02903 | Heat shock protein 70 | −5.55 for B-CuLCrV, 1.83 for Q-CuLCrV |
| Bta07645 | Prickle, putative | −1.71 for B- TYLCV, −1.26 for Q-TYLCV |
| Bta09867 | 70 kDa heat shock protein | −4.54 for B-CuLCrV, −1.50 for Q-TYLCV, −2.63 for Q-SiGMV |
| DEGs specific to MEAM1 | ||
| Bta00915 | Homeobox protein 9 | 1.29 for SiGMV |
| Bta00981 | Cyclic AMP-responsive element-binding protein 3-like protein 2 | 1.60 for SiGMV |
| Bta03000 | Heat shock protein 70 | −5.91 for CuLCrV |
| Bta03004 | Suppressor of hairless protein | 1.07 for TYLCV, 1.29 for SiGMV |
| Bta03110 | 5-hydroxytryptamine receptor 2A | −1.62 for TYLCV |
| Bta03159 | Ras-like protein 2 | 1.17 for SiGMV |
| Bta03261 | Homeobox protein prospero | 1.13 for SiGMV |
| Bta03841 | Casein kinase | 1.11 for SiGMV |
| Bta04025 | Serine/threonine-protein phosphatase 2A regulatory subunit B subunit beta | 1.02 for SiGMV |
| Bta04026 | Serine/threonine-protein phosphatase 2A regulatory subunit B subunit beta | −1.67 for TYLCV |
| Bta04098 | Acyl-CoA Z9 desaturase | −1.26 for SiGMV |
| Bta04167 | Ras-like GTP-binding protein RHO | 1.22 for SiGMV |
| Bta04210 | Protein kinase | 1.60 for SiGMV |
| Bta04351 | Transcriptional enhancer factor TEF-1 | −1.62 for TYLCV |
| Bta04481 | Calcium release-activated calcium channel protein 1 | 1.63 for SiGMV |
| Bta04484 | Guanine nucleotide-binding protein subunit alpha-like protein | 1.53 for TYLCV |
| Bta04715 | Ras-related C3 botulinum toxin substrate 1 | 1.69 for TYLCV, 1.49 for SiGMV |
| Bta04818 | Type I serine/threonine kinase receptor | 1.47 for SiGMV |
| Bta05836 | Protein prickle | −1.79 for TYLCV |
| Bta05989 | Inositol hexakisphosphate kinase 2 | 1.11 for SiGMV |
| Bta06076 | Heat shock protein | −1.39 for TYLCV |
| Bta06089 | Calcium/calmodulin-dependent protein kinase type II subunit delta | 1.51 for TYLCV |
| Bta06234 | Guanine nucleotide-binding protein G(O) subunit alpha | 1.34 for SiGMV |
| Bta07544 | Alkaline phosphatase | −1.86 for TYLCV |
| Bta07953 | Raw, isoform A | 1.24 for SiGMV |
| Bta07992 | Protein kinase C | 1.58 for SiGMV |
| Bta08159 | Transcription factor 7-like 1-A | 1.50 for SiGMV |
| Bta08259 | Inositol-trisphosphate 3-kinase B | 1.36 for SiGMV |
| Bta08332 | Arrestin 1c | −1.89 for TYLCV |
| Bta08932 | Acyl-CoA desaturase 1 | −1.63 for TYLCV |
| Bta08933 | Acyl-CoA desaturase | 3.55 for SiGMV |
| Bta09047 | Phosphoenolpyruvate carboxykinase [GTP] | −1.25 for TYLCV, −1.21 for SiGMV |
| Bta09151 | Protein kinase | −1.51 for TYLCV |
| Bta09834 | Sodium/potassium-transporting ATPase subunit beta-2, putative | 1.01 for SiGMV |
| Bta10421 | E3 ubiquitin-protein ligase CBL, putative | 1.22 for SiGMV |
| Bta11558 | Ras-related C3 botulinum toxin substrate 1 | 1.22 for TYLCV, 1.29 for SiGMV |
| Bta12096 | Phosphatidate cytidylyltransferase | 1.08 for SiGMV |
| Bta12195 | Phosphoinositide phospholipase C | 1.15 for SiGMV |
| Bta13074 | Sodium/calcium exchanger 1 | 1.84 for SiGMV |
| Bta13437 | Actin | −1.84 for TYLCV |
| Bta13534 | Ceramide synthase 6 | 1.28 for SiGMV |
| Bta14253 | Protein kinase C | 1.54 for SiGMV |
| Bta14395 | Alkaline phosphatase | −1.53 for TYLCV, -0.98 for SiGMV |
| Bta14493 | E3 ubiquitin-protein ligase | −2.14 for TYLCV |
| Bta14532 | 70 kDa heat shock protein | −2.20 for CuLCrV |
| Bta14568 | Guanine nucleotide-binding protein G(S) subunit alpha | 1.12 for TYLCV |
| Bta14627 | Mothers against decapentaplegic homolog | 1.15 for SiGMV |
| Bta15240 | F-box/WD repeat-containing protein 1A | 1.19 for SiGMV |
| Bta15478 | Ras-like GTP-binding protein RHO | 1.45 for SiGMV |
| Bta15489 | Transporter | 1.08 for SiGMV |
| Bta15565 | Partitioning defective protein 6 | 1.37 for SiGMV |
| DEGs specific to MED | ||
| Bta00008 | 70 kDa heat shock protein | −2.02 for SiGMV |
| Bta01763 | Acyl-CoA Delta(11) desaturase | 0.95 for TYLCV |
| Bta02612 | Protein kinase C | −0.82 for TYLCV |
| Bta04657 | Speckle-type POZ protein B | −0.79 for TYLCV |
| Bta05672 | Acyl-CoA desaturase | −1.62 for TYLCV |
| Bta09402 | Phospholipase A2 | 1.81 for TYLCV |
| Bta09287 | Chaperone protein HtpG | 0.73 for TYLCV |
| Bta12301 | Nucleoside diphosphate kinase | 0.83 for SiGMV |
| Bta12772 | Serine/threonine-protein phosphatase | −0.73 for TYLCV |
| Bta14581 | Calcium-transporting ATPase | −1.79 for CuLCrV |
| Bta14821 | Wnt inhibitory factor 1 | −1.24 for TYLCV |
| Gene ID | Annotation | FC in Whitefly & Virus Treatment |
|---|---|---|
| DEGs shared by MEAM1 and MED | ||
| Bta02886 | Protein spaetzle | 1.13 for B-CuLCrV, −1.35 for Q-TYLCV |
| Bta02903 | Heat shock protein 70 | −5.55 for B-CuLCrV, 1.83 for Q-CuLCrV |
| Bta08035 | Cathepsin B | −1.55 for B-TYLCV, −1.19 for Q-TYLCV |
| Bta09314 | Cathepsin B | −2.17 for B-SiGMV, 0.89 for Q-TYLCV, 1.18 for Q-SiGMV |
| Bta09772 | Cathepsin B | −1.11 for B-SiGMV, 1.02 for Q-TYLCV, −1.67 for Q-SiGMV |
| Bta09867 | 70 kDa heat shock protein | −4.54 for B-CuLCrV, −1.50 for Q-TYLCV, −2.63 for Q-SiGMV |
| Bta10364 | Cathepsin B | 1.46 for B-SiGMV, 3.67 for B-CuLCrV, −2.79 for Q-TYLCV |
| Bta10767 | Cathepsin B | 1.37 for B-CuLCrV, −1.35 for Q-TYLCV |
| Bta12604 | Cathepsin B | −1.19 for B-SiGMV, −1.09 for Q-TYLCV |
| Bta14750 | Cathepsin B | 2.84 for B-CuLCrV −2.25 for Q-TYLCV |
| DEGs specific to MEAM1 | ||
| Bta01559 | Aminopeptidase N, putative | −1.70 for TYLCV |
| Bta03000 | Heat shock protein 70 | −5.91 for CuLCrV |
| Bta03004 | Suppressor of hairless protein | 1.07 for TYLCV, 1.29 for SiGMV |
| Bta03159 | Ras-like protein 2 | 1.17 for SiGMV |
| Bta04167 | Ras-like GTP-binding protein RHO | 1.22 for SiGMV |
| Bta04210 | Protein kinase | 1.60 for SiGMV |
| Bta04481 | Calcium release-activated calcium channel protein 1 | 1.63 for SiGMV |
| Bta04640 | Actin-related protein 2/3 complex subunit 2 | 1.38 for SiGMV |
| Bta04715 | Ras-related C3 botulinum toxin substrate 1 | 1.69 for TYLCV, 1.49 for SiGMV |
| Bta04792 | Aminopeptidase N-like protein | 1.40 for TYLCV |
| Bta04818 | Type I serine/threonine kinase receptor | 1.47 for SiGMV |
| Bta06076 | Heat shock protein | −1.39 for TYLCV |
| Bta07992 | Protein kinase C | 1.58 for SiGMV |
| Bta08332 | Arrestin 1c | −1.89 for TYLCV |
| Bta08697 | Cathepsin B | −1.17 for TYLCV |
| Bta09313 | Cathepsin B | −2.34 for SiGMV |
| Bta09601 | Unknown protein | −1.51 for TYLCV |
| Bta10766 | Cathepsin B | 1.02 for CuLCrV |
| Bta11052 | Gelsolin | −1.19 for TYLCV |
| Bta11333 | Peptidoglycan-recognition protein | −1.74 for TYLCV |
| Bta11558 | Ras-related C3 botulinum toxin substrate 1 | 1.22 for TYLCV, 1.29 for SiGMV |
| Bta12195 | Phosphoinositide phospholipase C | 1.15 for SiGMV |
| Bta12605 | Cathepsin B | 1.31 for CuLCrV |
| Bta12986 | Cathepsin B | −2.24 for TYLCV |
| Bta13437 | Actin | −1.84 for TYLCV |
| Bta14253 | Protein kinase C | 1.54 for SiGMV |
| Bta14532 | 70 kDa heat shock protein | −2.20 for CuLCrV |
| Bta14568 | Guanine nucleotide-binding protein G(S) subunit alpha | 1.12 for TYLCV |
| Bta14627 | Mothers against decapentaplegic homolog | 1.15 for SiGMV |
| Bta14721 | Cathepsin B | 0.85 for CuLCrV |
| Bta14751 | Cathepsin B, partial | 1.32 for CuLCrV |
| Bta15240 | F-box/WD repeat-containing protein 1A | 1.19 for SiGMV |
| Bta15478 | Ras-like GTP-binding protein RHO | 1.45 for SiGMV |
| DEGs specific to MED | ||
| Bta00008 | 70 kDa heat shock protein | −2.02 for SiGMV |
| Bta01860 | Calreticulin | 0.72 for TYLCV |
| Bta02113 | Protein disulfide-isomerase | 0.74 for TYLCV |
| Bta02546 | Aminopeptidase-like protein | −0.72 for TYLCV |
| Bta02612 | Protein kinase C | −0.82 for TYLCV |
| Bta03883 | Cathepsin B | −1.42 for TYLCV |
| Bta03885 | Cathepsin B | −1.36 for TYLCV |
| Bta04776 | Cathepsin B | −1.26 for TYLCV |
| Bta06903 | Cathepsin L | 0.74 for TYLCV |
| Bta08022 | CG13675, isoform D | −1.02 for SiGMV |
| Bta08951 | Cathepsin B | −0.89 for TYLCV |
| Bta09287 | Chaperone protein HtpG | 0.73 for TYLCV |
| Bta09613 | Cathepsin L, partial | 2.92 for SiGMV |
| Bta10363 | Cathepsin B | −0.93 for TYLCV |
| Bta11419 | Cathepsin B | −1.34 for TYLCV |
| Bta11420 | Cathepsin B | −1.45 for TYLCV |
| Bta11544 | Heat shock 70 kDa protein 5 | 0.71 for TYLCV |
| Bta12285 | Cathepsin B | −2.20 for TYLCV, −1.66 for SiGMV |
| Bta12772 | Serine/threonine-protein phosphatase | −0.73 for TYLCV |
| Bta14718 | Cathepsin B | −0.79 for TYLCV |
| Gene ID | Annotation | FC in Whitefly & Virus Treatment |
|---|---|---|
| DEGs specific to MEAM1 | ||
| Bta01964 | Acid phosphatase-like protein | 1.62 for TYLCV |
| Bta02098 | Transport protein Sec61 subunit gamma | 2.28 for SiGMV |
| Bta02143 | Cathepsin F-like protease | −1.77 for TYLCV, −1.09 for SiGMV |
| Bta03900 | Tubulin alpha-3 chain | −1.72 for TYLCV, −1.17 for SiGMV |
| Bta04715 | Ras-related C3 botulinum toxin substrate 1 | 1.69 for TYLCV, 1.49 for SiGMV |
| Bta04774 | Arylsulfatase | −1.44 for TYLCV |
| Bta04870 | Cathepsin F | −1.89 for TYLCV |
| Bta05911 | Cathepsin F-like protease | 3.37 for CuLCrV |
| Bta05912 | Cathepsin F-like protease | −2.06 for SiGMV |
| Bta06085 | Protein kinase | −2.28 for TYLCV |
| Bta06086 | Protein kinase C | −1.85 for TYLCV |
| Bta06403 | Vesicular glutamate transporter 1 | −1.44 for TYLCV |
| Bta06690 | Cathepsin F | 1.15 for CuLCrV |
| Bta06693 | Cathepsin F | −1.05 for SiGMV |
| Bta06964 | DNAation factor subunit beta | 1.36 for SiGMV |
| Bta07988 | Syntaxin-18 | 1.35 for SiGMV |
| Bta07992 | Protein kinase C | 1.58 for SiGMV |
| Bta08697 | Cathepsin B | −1.17 for TYLCV |
| Bta09313 | Cathepsin B | −2.34 for SiGMV |
| Bta09676 | Cathepsin F | 0.94 for CuLCrV |
| Bta10683 | AP-1 complex subunit sigma-2 | 1.56 for TYLCV, 1.59 for SiGMV |
| Bta10766 | Cathepsin B | 1.02 for CuLCrV |
| Bta11284 | Tetraspanin | −1.12 for TYLCV |
| Bta11558 | Ras-related C3 botulinum toxin substrate 1 | 1.22 for TYLCV, 1.29 for SiGMV |
| Bta11871 | Cathepsin F | −1.15 for TYLCV |
| Bta12131 | Beta-hexosaminidase | −1.61 for TYLCV |
| Bta12605 | Cathepsin B | 1.31 for CuLCrV |
| Bta12986 | Cathepsin B | −2.24 for TYLCV |
| Bta13153 | Sensory neuron membrane protein 1 | 1.04 for SiGMV |
| Bta13437 | Actin | −1.84 for TYLCV |
| Bta14721 | Cathepsin B | 0.85 for CuLCrV |
| Bta14751 | Cathepsin B, partial | 1.32 for CuLCrV |
| Bta14774 | Beta-hexosaminidase | −2.13 for TYLCV |
| Bta20004 | Cathepsin F | 0.77 for CuLCrV |
| DEGs specific to MED | ||
| Bta01413 | Lipase | 1.19 for TYLCV |
| Bta01860 | Calreticulin | 0.72 for TYLCV |
| Bta03221 | Alpha-mannosidase | −0.72 for TYLCV |
| Bta03883 | Cathepsin B | −1.42 for TYLCV |
| Bta03885 | Cathepsin B | −1.36 for TYLCV |
| Bta04776 | Cathepsin B | −1.26 for TYLCV |
| Bta06903 | Cathepsin L | 0.74 for TYLCV |
| Bta07114 | Cathepsin F | −0.87 for TYLCV |
| Bta07355 | N(4)-(Beta-N-acetylglucosaminyl)-L-asparaginase | 1.06 for CuLCrV |
| Bta08951 | Cathepsin B | −0.89 for TYLCV |
| Bta09613 | Cathepsin L, partial | 2.92 for SiGMV |
| Bta10363 | Cathepsin B | −0.93 for TYLCV |
| Bta10727 | Protein transport protein Sec61 subunit alpha isoform 2 | 0.92 for TYLCV |
| Bta10829 | Acid phosphatase-1 | −1.71 for TYLCV, 1.78 for SiGMV |
| Bta11205 | Cathepsin F-like protease | −1.21 for CuLCrV |
| Bta11419 | Cathepsin B | −1.34 for TYLCV |
| Bta11420 | Cathepsin B | −1.45 for TYLCV |
| Bta12285 | Cathepsin B | −2.20 for TYLCV, −1.66 for SiGMV |
| Bta14718 | Cathepsin B | −0.79 for TYLCV |
| Gene ID | Annotation | FC in Whitefly & Virus Treatment |
|---|---|---|
| DEGs specific to MEAM1 | ||
| Bta00981 | Cyclic AMP-responsive element-binding protein 3-like protein 2 | 1.60 for SiGMV |
| Bta01299 | Forkhead box A | −2.21 for TYLCV |
| Bta03000 | Heat shock protein 70 | −5.91 for CuLCrV |
| Bta03177 | Glutathione S-transferase | −1.31 for SiGMV |
| Bta04098 | Acyl-CoA Z9 desaturase | −1.26 for SiGMV |
| Bta06076 | Heat shock protein | −1.39 for TYLCV |
| Bta06196 | Sestrin-like protein | 1.19 for TYLCV, 1.30 for SiGMV |
| Bta08630 | Fatty acyl-CoA reductase 1 | 1.30 for CuLCrV |
| Bta08932 | Acyl-CoA desaturase 1 | −1.63 for TYLCV |
| Bta08933 | Acyl-CoA desaturase | 3.55 for SiGMV |
| Bta09151 | Protein kinase | −1.51 for TYLCV |
| Bta13853 | Heat shock factor-binding protein, putative | 2.42 for SiGMV |
| Bta14222 | Fatty acyl-CoA reductase 1 | −1.02 for TYLCV |
| Bta14532 | 70 kDa heat shock protein | −2.20 for CuLCrV |
| Bta15447 | Glutathione s-transferase d1 | −0.87 for SiGMV |
| DEGs specific to MED | ||
| Bta00008 | 70 kDa heat shock protein | −2.02 for SiGMV |
| Bta01763 | Acyl-CoA Delta (11) desaturase | 0.95 for TYLCV |
| Bta05672 | Acyl-CoA desaturase | −1.62 for TYLCV |
| Bta11628 | Fatty acyl-CoA reductase 1 | −1.58 for CuLCrV |
| Bta13675 | Fatty acyl-CoA reductase 1 | 1.00 for TYLCV |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mugerwa, H.; Gautam, S.; Catto, M.A.; Dutta, B.; Brown, J.K.; Adkins, S.; Srinivasan, R. Differential Transcriptional Responses in Two Old World Bemisia tabaci Cryptic Species Post Acquisition of Old and New World Begomoviruses. Cells 2022, 11, 2060. https://doi.org/10.3390/cells11132060
Mugerwa H, Gautam S, Catto MA, Dutta B, Brown JK, Adkins S, Srinivasan R. Differential Transcriptional Responses in Two Old World Bemisia tabaci Cryptic Species Post Acquisition of Old and New World Begomoviruses. Cells. 2022; 11(13):2060. https://doi.org/10.3390/cells11132060
Chicago/Turabian StyleMugerwa, Habibu, Saurabh Gautam, Michael A. Catto, Bhabesh Dutta, Judith K. Brown, Scott Adkins, and Rajagopalbabu Srinivasan. 2022. "Differential Transcriptional Responses in Two Old World Bemisia tabaci Cryptic Species Post Acquisition of Old and New World Begomoviruses" Cells 11, no. 13: 2060. https://doi.org/10.3390/cells11132060
APA StyleMugerwa, H., Gautam, S., Catto, M. A., Dutta, B., Brown, J. K., Adkins, S., & Srinivasan, R. (2022). Differential Transcriptional Responses in Two Old World Bemisia tabaci Cryptic Species Post Acquisition of Old and New World Begomoviruses. Cells, 11(13), 2060. https://doi.org/10.3390/cells11132060








