Application of Priming Strategy for Enhanced Paclitaxel Biosynthesis in Taxus × Media Hairy Root Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Hairy Root Cultures and Treatment
2.3. Quantitative Real-Time PCR (qRT-PCR)
2.4. Paclitaxel Quantitative Determination Using HPLC-UV-DAD Method
3. Results
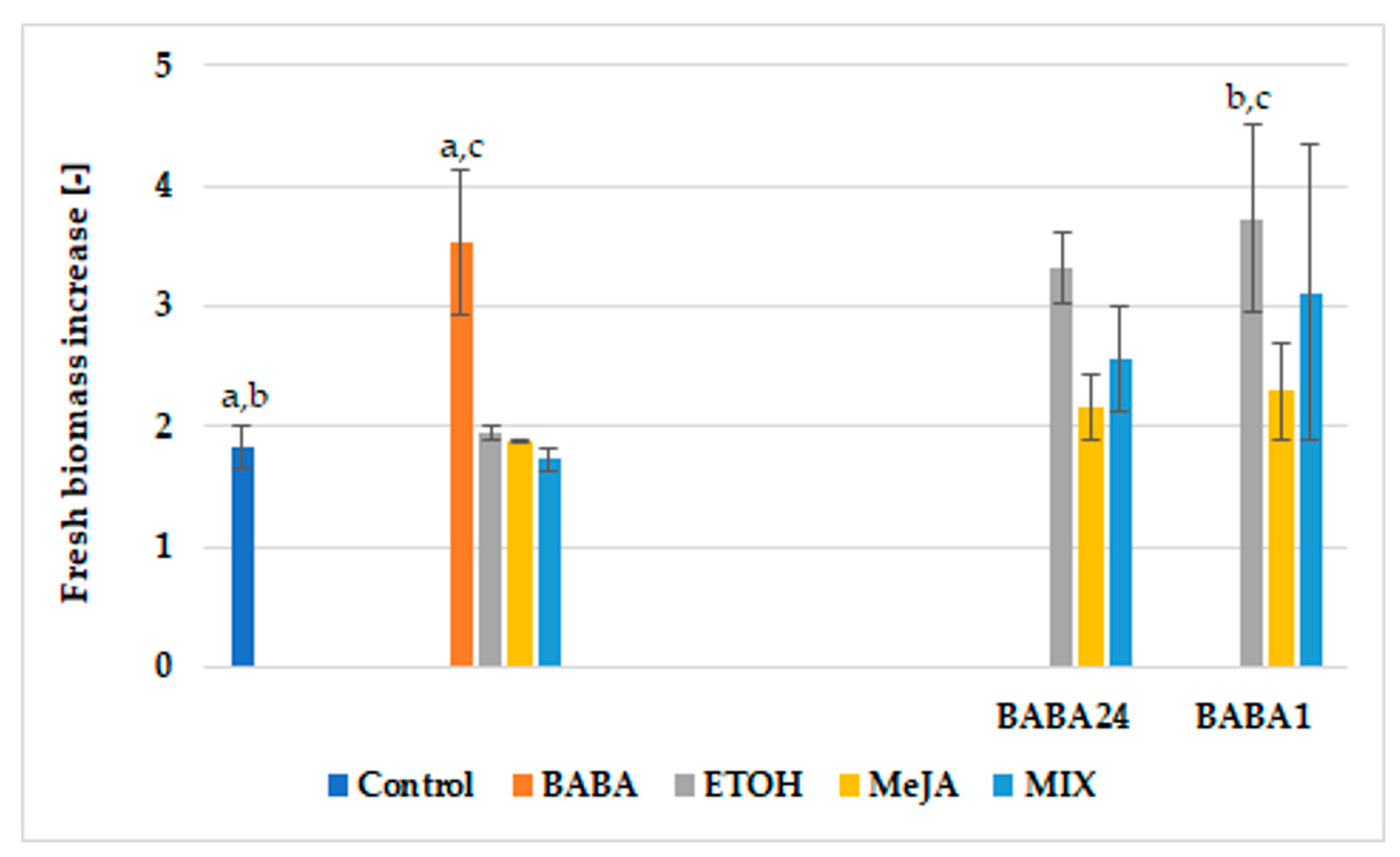
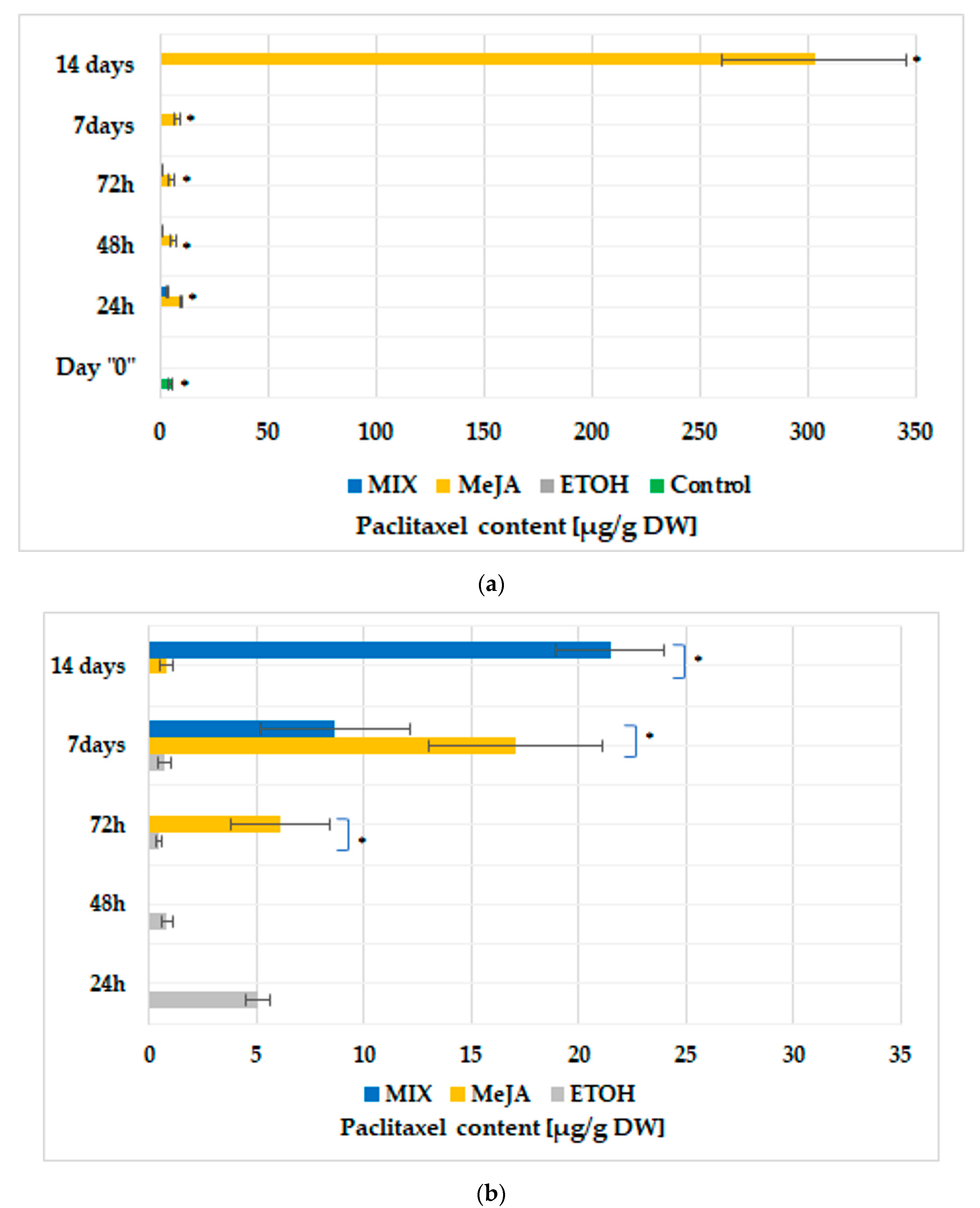
3.1. Hairy Root Cultures and Paclitaxel Biosynthesis
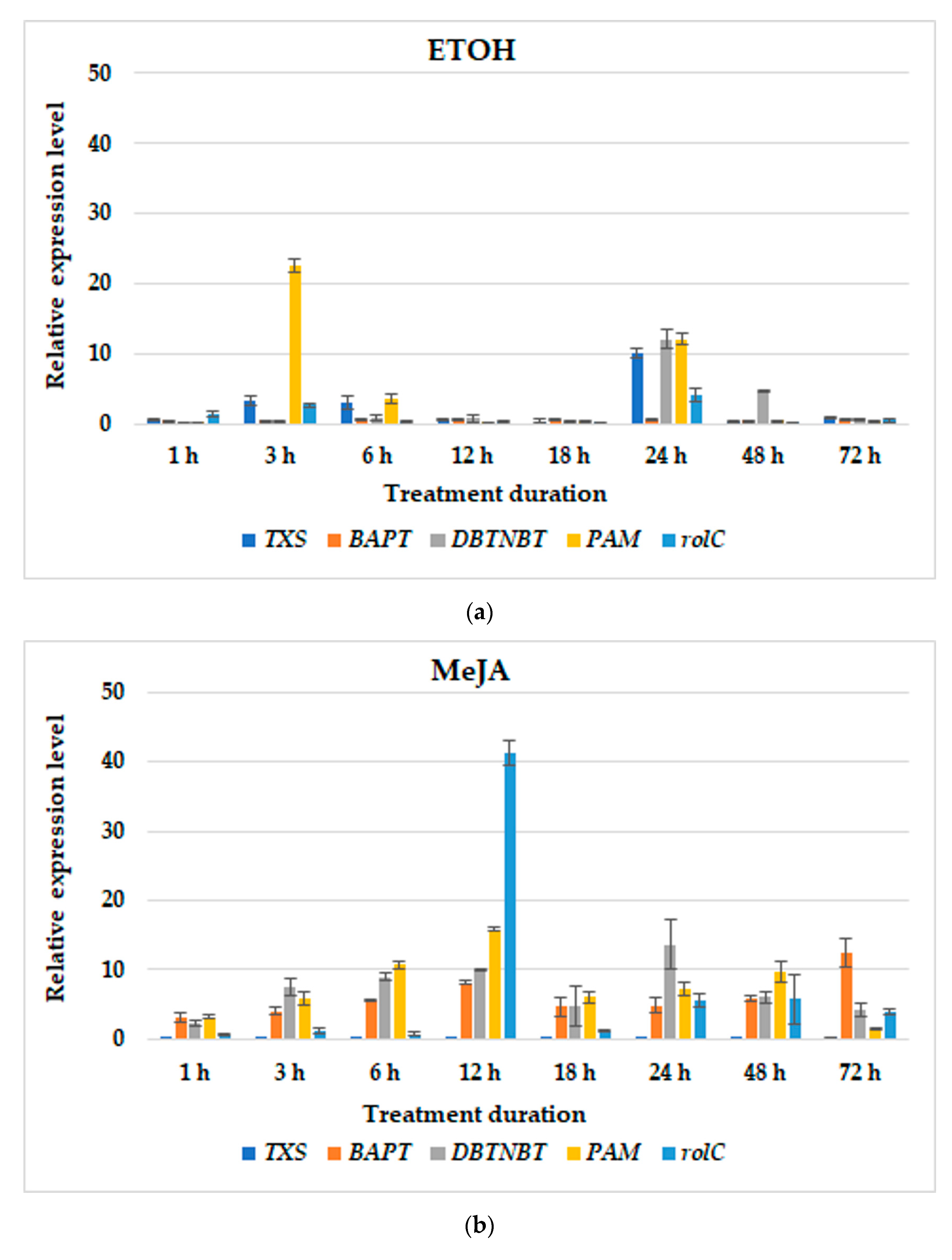
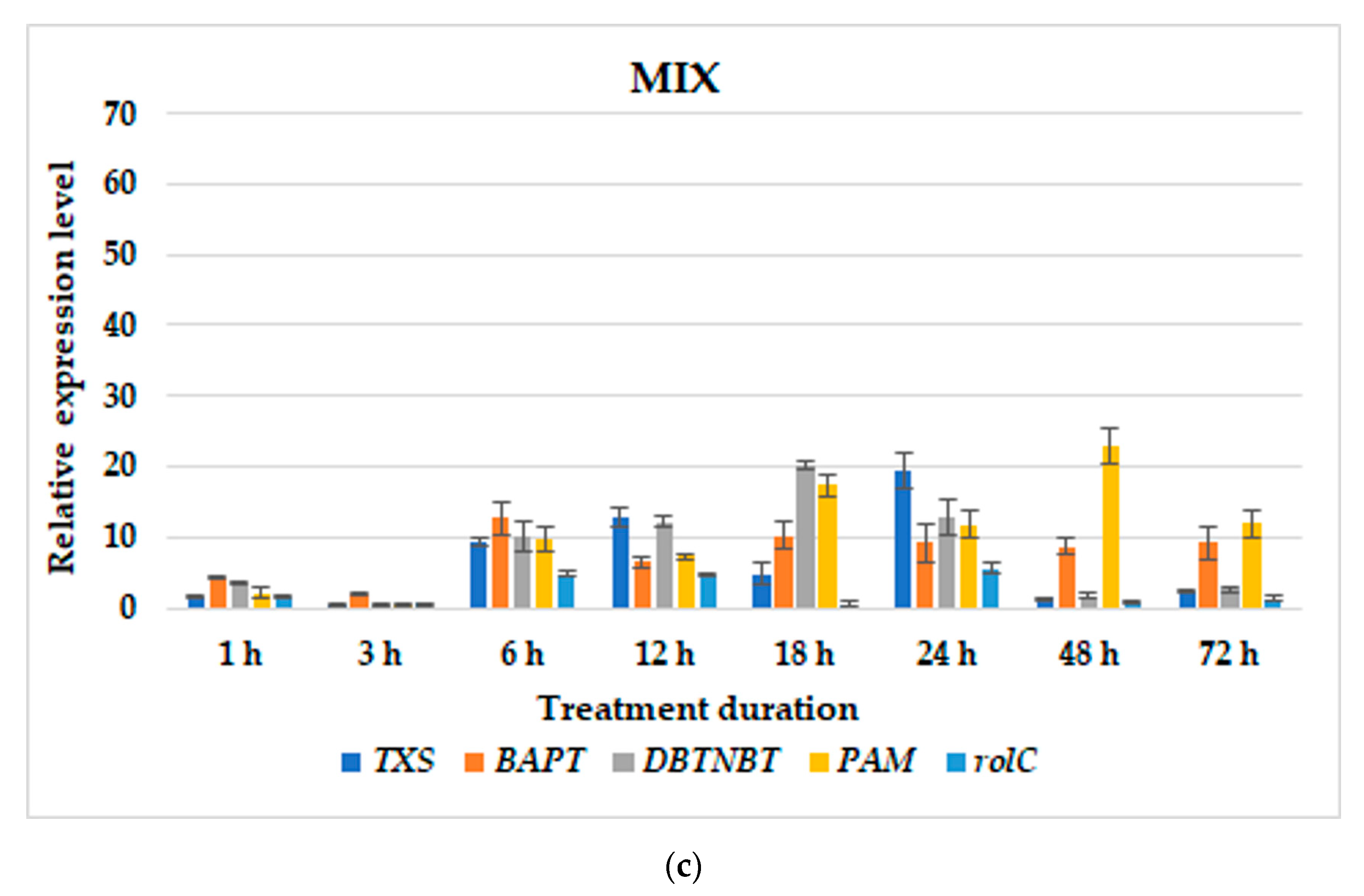
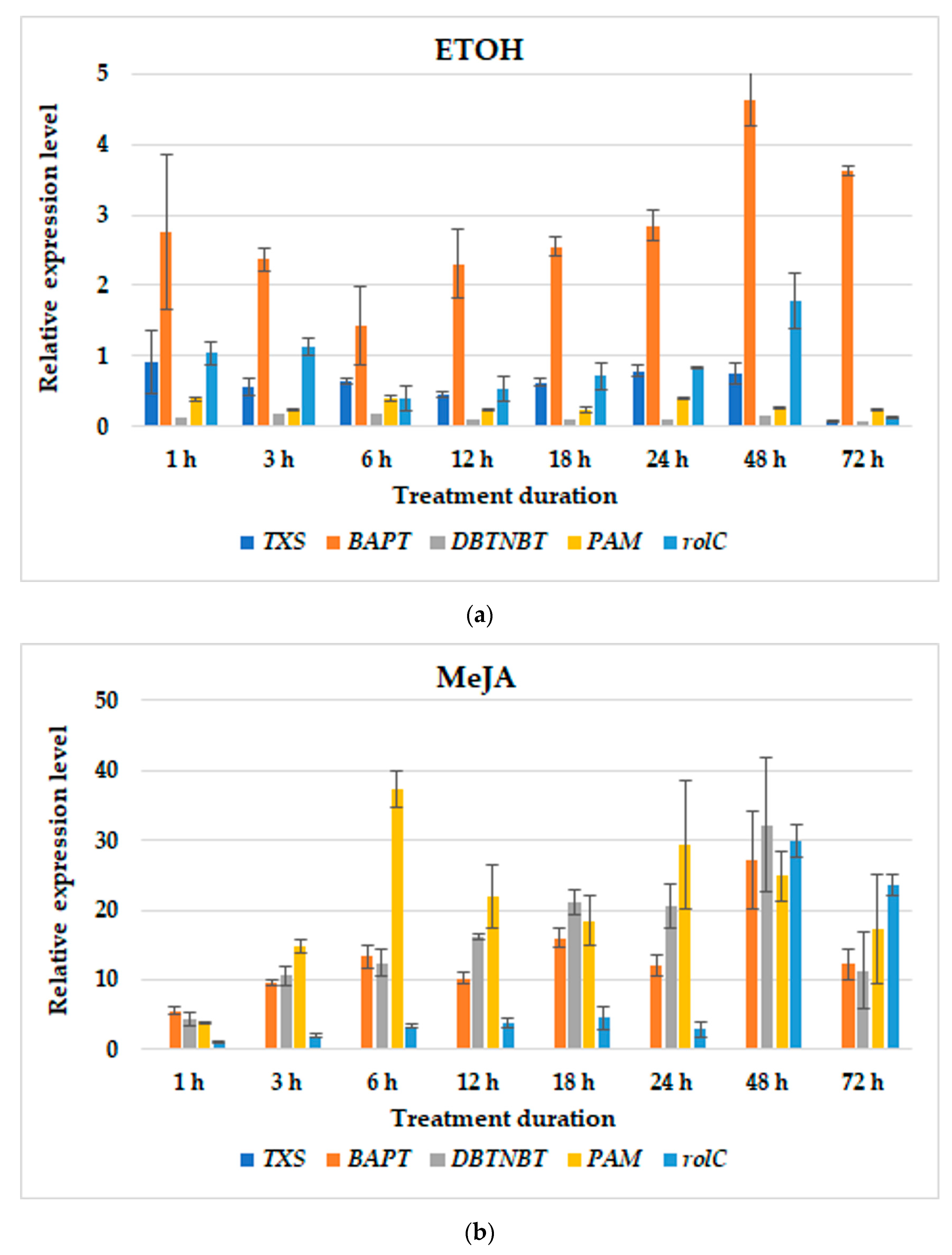
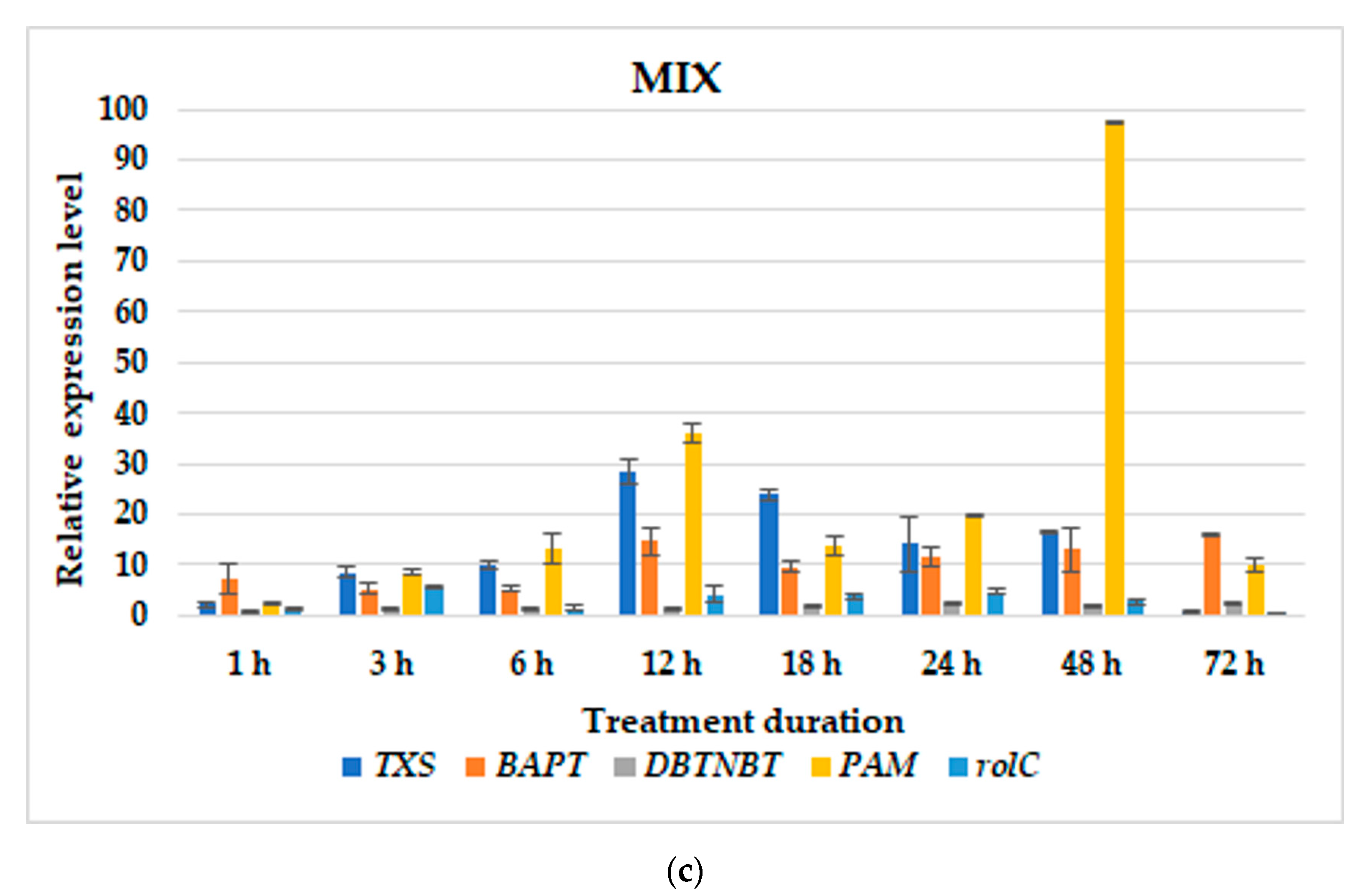
3.2. Gene Expression Profiles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fauzee, N.J.S.; Dong, Z.; Wang, Y. Taxanes: Promising anti-cancer drugs. Asian Pac. J. Cancer Prev. 2011, 12, 837–851. [Google Scholar] [PubMed]
- Yared, J.A.; Tkaczuk, K.H.R. Update on taxane development: New analogs and new formulations. Drug Des. Devel. Ther. 2012, 6, 371–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, D.; Bendiske, J.; Michaelis, M.L.; Karanian, D.A.; Bahr, B.A. Microtubule-stabilizing agent prevents protein accumulation-induced loss of synaptic markers. Eur. J. Pharmacol. 2007, 562, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Yang, J.; Cui, K.; Rao, Q.; Yin, T.; Tan, L.; Zhang, Y.; Li, Z.; Wang, G. Controlled slow-release drug-eluting stents for the prevention of coronary restenosis: Recent progress and future prospects. ACS Appl. Mater. Interfaces 2015, 7, 11695–11712. [Google Scholar] [CrossRef]
- Cusido, R.M.; Onrubia, M.; Sabater-Jara, A.B.; Moyano, E.; Bonfill, M.; Goossens, A.; Angeles Pedreño, M.; Palazon, J. A rational approach to improving the biotechnological production of taxanes in plant cell cultures of Taxus spp. Biotechnol. Adv. 2014, 32, 1157–1167. [Google Scholar] [CrossRef]
- Pieterse, M.J.; Van Der Does, D.; Zamioudis, C.; Leon-reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U.; Pieterse, C.M.J.; Mauch-Mani, B. Priming in plant–pathogen interactions. Trends Plant Sci. 2002, 7, 210–216. [Google Scholar] [CrossRef] [Green Version]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef]
- Halder, M.; Sarkar, S.; Jha, S. Elicitation: A biotechnological tool for enhanced production of secondary metabolites in hairy root cultures. Eng. Life Sci. 2019, 19, 880–895. [Google Scholar] [CrossRef] [Green Version]
- Cohen, Y. β-Aminobutyric acid-induced resistance against plant pathogens. Plant Dis. 2002, 86, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Wu, J. Induction and potentiation of diterpenoid tanshinone accumulation in Salvia miltiorrhiza hairy roots by β-aminobutyric acid. PubMed. 2005, 68, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Furmanowa, M.; Sykłowska-Baranek, K. Hairy root cultures of Taxus × media var. Hicksii Rehd. as a new source of paclitaxel and 10-deacetylbaccatin III. Biotechnol. Lett. 2000, 22, 683–686. [Google Scholar] [CrossRef]
- Syklowska-Baranek, K.; Pietrosiuk, A.; Kokoszka, A.; Furmanowa, M. Enhancement of taxane production in hairy root culture of Taxus × media var. Hicksii. J. Plant Physiol. 2009, 166, 1950–1954. [Google Scholar] [CrossRef] [PubMed]
- Jossen, V.; Muoio, F.; Panella, S.; Harder, Y.; Tallone, T.; Eibl, R. An approach towards a gmp compliant in-vitro expansion of human adipose stem cells for autologous therapies. Bioengineering 2020, 7, 77. [Google Scholar] [CrossRef] [PubMed]
- Nims, E.; Dubois, C.P.; Roberts, S.C.; Walker, E.L. Expression profiling of genes involved in paclitaxel biosynthesis for targeted metabolic engineering. Metab. Eng. 2006, 8, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Vongpaseuth, K.; Roberts, S.C. Advancements in the understanding of paclitaxel metabolism in tissue culture. Curr. Pharm. Biotechnol. 2007, 8, 219–236. [Google Scholar] [CrossRef]
- Sykłowska-Baranek, K.; Rymaszewski, W.; Gaweł, M.; Rokicki, P.; Pilarek, M.; Grech-Baran, M.; Hennig, J.; Pietrosiuk, A. Comparison of elicitor-based effects on metabolic responses of Taxus × media hairy roots in perfluorodecalin-supported two-phase culture system. Plant Cell Rep. 2019, 38, 85–99. [Google Scholar] [CrossRef] [Green Version]
- Sabater-Jara, A.B.; Onrubia, M.; Moyano, E.; Bonfill, M.; Palazón, J.; Pedreño, M.A.; Cusidó, R.M. Synergistic effect of cyclodextrins and methyl jasmonate on taxane production in Taxus × media cell cultures. Plant Biotechnol. J. 2014, 12, 1075–1084. [Google Scholar] [CrossRef]
- Theodoridis, G.; de Jong, C.; Laskaris, G.; Verpoorte, R. Application of SPE for the HPLC analysis of taxanes from Taxus cell cultures. Chromatographia 1998, 47, 25–34. [Google Scholar] [CrossRef]
- Sykłowska-Baranek, K.; Szala, K.; Pilarek, M.; Orzechowski, R.; Pietrosiuk, A. A cellulase-supported two-phase in situ system for enhanced biosynthesis of paclitaxel in Taxus × media hairy roots. Acta Physiol. Plant. 2018, 40, 201. [Google Scholar] [CrossRef] [Green Version]
- Theodoridis, G.; Laskaris, G.; De Jong, C.F.; Verpoorte, R. Determination of paclitaxel and related diterpenoids in plant extracts by high-performance liquid chromatography with UV detection in high-performance liquid chromatography–mass spectrometry. J. Chromatogr. A 1998, 802, 297–305. [Google Scholar] [CrossRef]
- Balmer, A.; Pastor, V.; Gamir, J.; Flors, V.; Mauch-Mani, B. The “prime-ome”: Towards a holistic approach to priming. Trends Plant Sci. 2015, 20, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Lenka, S.K.; Nims, N.E.; Vongpaseuth, K.; Boshar, R.A.; Roberts, S.C.; Walker, E.L. Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 2015, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenka, S.K.; Boutaoui, N.; Paulose, B.; Vongpaseuth, K.; Normanly, J.; Roberts, S.C.; Walker, E.L. Identification and expression analysis of methyl jasmonate responsive ESTs in paclitaxel producing Taxus cuspidata suspension culture cells. BMC Genomics 2012, 13, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Zhang, P.; Zhang, M.; Fu, C.; Yu, L. Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. 2013, 15, 19–26. [Google Scholar] [CrossRef]
- Li, S.; Zhang, P.; Zhang, M.; Fu, C.; Zhao, C.; Dong, Y.; Guo, A.; Yu, L. Transcriptional profile of Taxus chinensis cells in response to methyl jasmonate. BMC Genomics 2012, 13, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, G.; Yang, Y.; Xie, F.; Wen, J.F.; Wu, J.; Wilson, I.W.; Tang, Q.; Liu, H.; Qiu, D. Deep sequencing reveals transcriptome re-programming of Taxus × media cells to the elicitation with methyl jasmonate. PLoS ONE 2013, 8, e1005510. [Google Scholar] [CrossRef] [Green Version]
- Sykłowska-Baranek, K.; Grech-Baran, M.; Naliwajski, M.R.; Bonfill, M.; Pietrosiuk, A. Paclitaxel production and PAL activity in hairy root cultures of Taxus × media var. Hicksii carrying a taxadiene synthase transgene elicited with nitric oxide and methyl jasmonate. Acta Physiol. Plant. 2015, 37, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Sykłowska-Baranek, K.; Łysik, K.; Jeziorek, M.; Wencel, A.; Gajcy, M.; Pietrosiuk, A. Lignan accumulation in two-phase cultures of Taxus × media hairy roots. Plant Cell Tissue Organ Cult. 2018, 133, 371–384. [Google Scholar] [CrossRef] [Green Version]
- Selim, S.; Akhtar, N.; El Azab, E.; Warrad, M.; Alhassan, H.H.; Abdel-Mawgoud, M.; Al Jaouni, S.K.; Abdelgawad, H. Innovating the synergistic assets of β-amino butyric acid (BABA) and selenium nanoparticles (SeNOS) in improving the growth, nitrogen metabolism, biological activities, nutritive value of Medicago interexta sprouts. Plants 2022, 11, 306. [Google Scholar] [CrossRef]
- Exposito, O.; Syklowska-Baranek, K.; Moyano, E.; Onrubia, M.; Bonfill, M.; Palazon, J.; Cusido, R.M. Metabolic responses of Taxus media transformed cell cultures to the addition of methyl jasmonate. Biotechnol. Prog. 2010, 26, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Onrubia, M.; Moyano, E.; Bonfill, M.; Expósito, O.; Palazón, J.; Cusidó, R.M. An approach to the molecular mechanism of methyl jasmonate and vanadyl sulphate elicitation in Taxus baccata cell cultures: The role of txs and bapt gene expression. Biochem. Eng. J. 2010, 53, 104–111. [Google Scholar] [CrossRef]
- Onrubia, M.; Moyano, E.; Bonfill, M.; Cusidó, R.M.; Goossens, A.; Palazón, J. Coronatine, a more powerful elicitor for inducing taxane biosynthesis in Taxus media cell cultures than methyl jasmonate. J. Plant Physiol. 2013, 170, 211–219. [Google Scholar] [CrossRef]
- Sanchez-Muñoz, R.; Bonfill, M.; Cusidó, R.M.; Palazón, J.; Moyano, E. Advances in the regulation of in vitro paclitaxel production: Methylation of a Y-patch promoter region alters BAPT gene expression in Taxus cell cultures. Plant Cell Physiol. 2018, 59, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Li, L.; Wu, W.; Li, M.; Yu, X.; Yu, L. Assessment of genetic and epigenetic variation during long-term Taxus cell culture. Plant Cell Rep. 2012, 31, 1321–1331. [Google Scholar] [CrossRef]
- Li, L.Q.; Li, X.L.; Fu, C.H.; Zhao, C.F.; Yu, L.J. Sustainable use of Taxus media cell cultures through minimal growth conservation and manipulation of genome methylation. Process Biochem. 2013, 48, 525–531. [Google Scholar] [CrossRef]
- Sykłowska-Baranek, K.; Sygitowicz, G.; Pietrosiuk, A. Development of Taxus spp. hairy root cultures for enhanced taxane production. In Plant Cell and Tissue Differentiation and Secondary Metabolites; Ramawat, K., Ekiert, H., Goyal, S., Eds.; Springer: Cham, Switzerland, 2019; pp. 1–19. ISBN 9783030112530. [Google Scholar]
- Ge, X.; Wu, J. Tanshinone production and isoprenoid pathways in Salvia miltiorrhiza hairy roots induced by Ag+ and yeast elicitor. Plant Sci. 2005, 168, 487–491. [Google Scholar] [CrossRef]
- Li, T.; Fan, P.; Yun, Z.; Jiang, G.; Zhang, Z.; Jiang, Y. β-aminobutyric acid priming acquisition and defense response of mango fruit to Colletotrichum gloeosporioides infection based on quantitative proteomics. Cells 2019, 8, 1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, S. Natural plant genetic engineer Agrobacterium rhizogenes: Role of T-DNA in plant secondary metabolism. Biotechnol. Lett. 2012, 34, 407–415. [Google Scholar] [CrossRef]
- Bulgakov, V.P. Functions of rol genes in plant secondary metabolism. Biotechnol. Adv. 2008, 26, 318–324. [Google Scholar] [CrossRef]




| Taxus Spp. | Type of Culture | Elicitor/s | Peak in Gene Expression | Reference |
|---|---|---|---|---|
| T. cuspidata | cell suspension | MeJA | TXS–18 h; BAPT–6 h; DBTNBT and PAM–6 h till | [15] |
| T. × media | cell suspension; two lines: TXS—carrying TXS transgene and ROL genes of A. rhizogenes; ROLC—with ROL genes of A. rhizogenes | MeJA | TXS: TXS line–12 h; ROLC line–48 h | [31] |
| T. baccata | cell suspension | MeJA; vanadyl sulphate (VS) | MeJA: TXS–24 h; BAPT–12h and at day 20;VS-TXS–8 day; BAPT–at 4 day;MeJA and VS: TXS–4 h; BAPT–12h | [32] |
| T. × media | cell suspension; line TXS line-carrying TXS transgene | MeJA; coronatine (COR) | MeJA: TXS–24 h; BAPT and DBTNBT–4 day; PAM–1h and 4 day;COR: TXS–24 h; BAPT–24 h; DBTNBT–48 h; PAM–48 h | [33] |
| T. × media | cell suspension; line TXS line-carrying TXS transgene | MeJA, randomly methylated-β-cyclodextrin (M-β-CD) separately or combined | MeJA: TXS–72 h; BAPT–96 h; DBTNBT–72 h; M-β-CD: TXS–72 h; BAPT–1-4 h; DBTNBT–72 h;MeJA+M-β-CD: TXS–72 h; BAPT–4 h; DBTNBT–72 h; | [18] |
| T. × media | two lines of hairy roots: KT and ATMA | MeJA, sodium nitroprusside, L-phenylalanine, degassed perfluorodecalin additional sucrose | KT line: TXS–12 h; BAPT–6 h and at parallel level till 24 h; DBTNBT–12 h and 48 h;ATMA line: TXS–48 h; BAPT–48 h; DBTNBT–6 h and 48 h; | [17] |
| T. × media | hairy roots of line KT | BABA; MeJA; MIX; BABA24+MeJA or MIX; BABA1+MeJA or MIX | BABA: TXS–1 h and 48 h; BAPT–6 h, at 5 and 10 day; DBTNBT–at 5 day; PAM–24 h; rolC–24 h;MeJA: TXS–72 h; BAPT–72 h; DBTNBT–24 h; PAM–12 h; rolC–12 h;MIX: TXS–18 h; BAPT–72 h; DBTNBT–24 h; PAM–12 h; rolC–18 h;BABA24MeJA-TXS–12 h; BAPT–6 h and 48 h; DBTNBT–24 h; PAM–48 h; rolC–24 h;BABA24MIX-TXS–12 h and 24 h; BAPT–6 and at parallel level till 72 h; DBTNBT–6 h and at parallel level till 24 h; PAM–48 h; rolC–24 h;BABA1MeJA: TXS–48 h; BAPT–48 h; DBTNBT–48 h; PAM–6 h; rolC–48 h;BABA1MIX: TXS–12 h; BAPT–12 h; DBTNBT–48 h; PAM–48 h; rolC–3 h; | present study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sykłowska-Baranek, K.; Sygitowicz, G.; Maciejak-Jastrzębska, A.; Pietrosiuk, A.; Szakiel, A. Application of Priming Strategy for Enhanced Paclitaxel Biosynthesis in Taxus × Media Hairy Root Cultures. Cells 2022, 11, 2062. https://doi.org/10.3390/cells11132062
Sykłowska-Baranek K, Sygitowicz G, Maciejak-Jastrzębska A, Pietrosiuk A, Szakiel A. Application of Priming Strategy for Enhanced Paclitaxel Biosynthesis in Taxus × Media Hairy Root Cultures. Cells. 2022; 11(13):2062. https://doi.org/10.3390/cells11132062
Chicago/Turabian StyleSykłowska-Baranek, Katarzyna, Grażyna Sygitowicz, Agata Maciejak-Jastrzębska, Agnieszka Pietrosiuk, and Anna Szakiel. 2022. "Application of Priming Strategy for Enhanced Paclitaxel Biosynthesis in Taxus × Media Hairy Root Cultures" Cells 11, no. 13: 2062. https://doi.org/10.3390/cells11132062







