Proteomics, Phosphoproteomics and Mirna Analysis of Circulating Extracellular Vesicles through Automated and High-Throughput Isolation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Collection and Preparation of Urine Samples
2.3. Purification and Capture of EV Samples
2.4. Characterization of EVs Using Transmission Electron Microscopy
2.5. Resistive Pulse Sensing (RPS) Analysis
2.6. Western Blotting Analysis
2.7. Samples Preparation of EVs for qRT-PCR
2.8. Preparation of EV Samples for LC-MS
2.9. LC−MS/MS Analysis
2.10. LC−MS/MS Data Processing and Quantitation
3. Results and Discussion
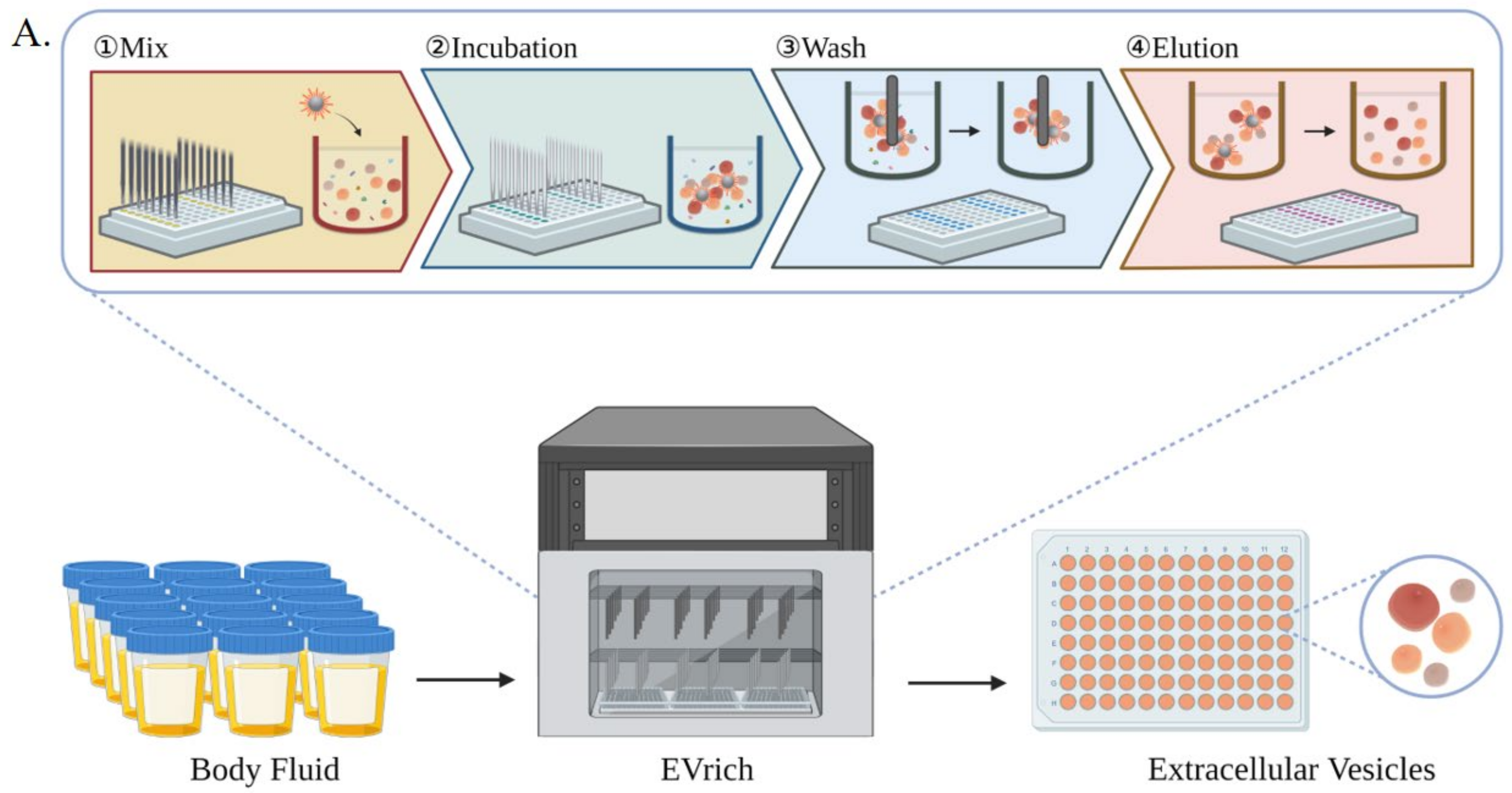
3.1. Implementing High-Throughput, Automated EV Isolation
3.2. Application of EVrich for Prostate Cancer Biomarker Discovery
3.3. Analysis of miRNAs in EVs from Prostate Cancer Urine Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Van Niel, G.; D’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Turiello, R.; Capone, M.; Morretta, E.; Monti, M.C.; Madonna, G.; Azzaro, R.; Del Gaudio, P.; Simeone, E.; Sorrentino, A.; Ascierto, P.A.; et al. Exosomal CD73 from serum of patients with melanoma suppresses lymphocyte functions and is associated with therapy resistance to anti-PD-1 agents. J. Immunother. Cancer 2022, 10, e004043. [Google Scholar] [CrossRef]
- Gupta, P.; Kadamberi, I.P.; Mittal, S.; Tsaih, S.W.; George, J.; Kumar, S.; Vijayan, D.K.; Geethadevi, A.; Parashar, D.; Topchyan, P.; et al. Tumor Derived Extracellular Vesicles Drive T Cell Exhaustion in Tumor Microenvironment through Sphingosine Mediated Signaling and Impacting Immunotherapy Outcomes in Ovarian Cancer. Adv. Sci. 2022, 9, e2104452. [Google Scholar] [CrossRef]
- Yin, Y.; Liu, B.; Cao, Y.; Yao, S.; Liu, Y.; Jin, G.; Qin, Y.; Chen, Y.; Cui, K.; Zhou, L.; et al. Colorectal Cancer-Derived Small Extracellular Vesicles Promote Tumor Immune Evasion by Upregulating PD-L1 Expression in Tumor-Associated Macrophages. Adv. Sci. 2022, 9, 2102620. [Google Scholar] [CrossRef]
- Xue, P.; Huang, S.; Han, X.; Zhang, C.; Yang, L.; Xiao, W.; Fu, J.; Li, H.; Zhou, Y. Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ. 2022, 29, 82–95. [Google Scholar] [CrossRef]
- Zhang, H.G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef] [Green Version]
- Morelli, A.E.; Larregina, A.T.; Shufesky, W.J.; Sullivan, M.L.; Stolz, D.B.; Papworth, G.D.; Zahorchak, A.F.; Logar, A.J.; Wang, Z.; Watkins, S.C.; et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood 2004, 104, 3257–3266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, Q.; Zhou, L.; Sui, H.; Yang, L.; Wu, X.; Song, Q.; Jia, R.; Li, R.; Sun, J.; Wang, Z.; et al. Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation. Nat. Commun. 2020, 11, 1211. [Google Scholar] [CrossRef] [Green Version]
- Fang, T.; Lv, H.; Lv, G.; Li, T.; Wang, C.; Han, Q.; Yu, L.; Su, B.; Guo, L.; Huang, S.; et al. Tumor-derived exosomal miR-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat. Commun. 2018, 9, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luga, V.; Zhang, L.; Viloria-Petit, A.M.; Ogunjimi, A.A.; Inanlou, M.R.; Chiu, E.; Buchanan, M.; Hosein, A.N.; Basik, M.; Wrana, J.L. Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 2012, 151, 1542–1556. [Google Scholar] [CrossRef] [Green Version]
- Atay, S.; Banskota, S.; Crow, J.; Sethi, G.; Rink, L.; Godwin, A.K. Oncogenic KIT-containing exosomes increase gastrointestinal stromal tumor cell invasion. Proc. Natl. Acad. Sci. USA 2014, 111, 711–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.W.; Lee, H.M.; Lee, T.H.; Kang, C.; Kleinman, H.K.; Gho, Y.S. Extracellular membrane vesicles from tumor cells promote angiogenesis via sphingomyelin. Cancer Res. 2002, 62, 6312–6317. [Google Scholar]
- Qu, J.L.; Qu, X.J.; Zhao, M.F.; Teng, Y.E.; Zhang, Y.; Hou, K.Z.; Jiang, Y.H.; Yang, X.H.; Liu, Y.P. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig. Liver Dis. 2009, 41, 875–880. [Google Scholar] [CrossRef]
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am. J. Physiol. Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef]
- Kosaka, N.; Yoshioka, Y.; Fujita, Y.; Ochiya, T. Versatile roles of extracellular vesicles in cancer. J. Clin. Investig. 2016, 126, 1163–1172. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Mantel, P.Y.; Halleck, A.E.; Trachtenberg, A.J.; Soria, C.E.; Oquin, S.; Bonebreak, C.M.; Saracoglu, E.; et al. Current methods for the isolation of extracellular vesicles. Biol. Chem. 2013, 394, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Mueller, S.K.; Nocera, A.L.; Bleier, B.S. Exosome function in aerodigestive mucosa. Nanomedicine 2018, 14, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Linares, R.; Tan, S.; Gounou, C.; Arraud, N.; Brisson, A.R. High-speed centrifugation induces aggregation of extracellular vesicles. J. Extracell. Vesicles 2015, 4, 29509. [Google Scholar] [CrossRef] [PubMed]
- Nocera, A.L.; Miyake, M.M.; Seifert, P.; Han, X.; Bleier, B.S. Exosomes mediate interepithelial transfer of functional P-glycoprotein in chronic rhinosinusitis with nasal polyps. Laryngoscope 2017, 127, E295–E300. [Google Scholar] [CrossRef]
- Sharma, P.; Ludwig, S.; Muller, L.; Hong, C.S.; Kirkwood, J.M.; Ferrone, S.; Whiteside, T.L. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J. Extracell. Vesicles 2018, 7, 1435138. [Google Scholar] [CrossRef]
- Konoshenko, M.Y.; Lekchnov, E.A.; Vlassov, A.V.; Laktionov, P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed. Res. Int. 2018, 2018, 8545347. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, Q.; Cheng, L.; Wang, Y.; Li, M.; Yang, Q.; Hu, L.; Lou, D.; Li, J.; Dong, X.; et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods 2021, 18, 212–218. [Google Scholar] [CrossRef]
- Gao, F.; Jiao, F.; Xia, C.; Zhao, Y.; Ying, W.; Xie, Y.; Guan, X.; Tao, M.; Zhang, Y.; Qin, W.; et al. A novel strategy for facile serum exosome isolation based on specific interactions between phospholipid bilayers and TiO2. Chem. Sci. 2019, 10, 1579–1588. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Li, L.; Iliuk, A.; Tao, W.A. Highly Efficient Phosphoproteome Capture and Analysis from Urinary Extracellular Vesicles. J. Proteome Res. 2018, 17, 3308–3316. [Google Scholar] [CrossRef]
- Iliuk, A.; Wu, X.; Li, L.; Sun, J.; Hadisurya, M.; Boris, R.S.; Tao, W.A. Plasma-Derived Extracellular Vesicle Phosphoproteomics through Chemical Affinity Purification. J. Proteome Res. 2020, 19, 2563–2574. [Google Scholar] [CrossRef]
- DeBlois, R.W.; Bean, C.P. Counting and Sizing of Submicron Particles by the Resistive Pulse Technique. Rev. Sci. Instrum. 1970, 41, 909–916. [Google Scholar] [CrossRef]
- DeBlois, R.W.; Wesley, R.K. Sizes and concentrations of several type C oncornaviruses and bacteriophage T2 by the resistive-pulse technique. J. Virol. 1977, 23, 227–233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Pan, C.; Kuhn, A.; Nievergelt, A.P.; Fantner, G.E.; Milenkovic, O.; Radenovic, A. Detecting topological variations of DNA at single-molecule level. Nat. Commun. 2019, 10, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [Green Version]
- Krichevsky, A.M.; Gabriely, G. miR-21: A small multi-faceted RNA. J. Cell Mol. Med. 2009, 13, 39–53. [Google Scholar] [CrossRef]
- Huang, L.; Luo, J.; Cai, Q.; Pan, Q.; Zeng, H.; Guo, Z.; Dong, W.; Huang, J.; Lin, T. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int. J. Cancer 2011, 128, 1758–1769. [Google Scholar] [CrossRef]
- Di Martino, M.T.; Rossi, M.; Caracciolo, D.; Gulla, A.; Tagliaferri, P.; Tassone, P. Mir-221/222 are promising targets for innovative anticancer therapy. Expert Opin. Ther. Targets 2016, 20, 1099–1108. [Google Scholar] [CrossRef]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.P.; Pan, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in extracellular vesicles as candidate markers for breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef] [Green Version]
- Latest Global Cancer Data: Cancer Burden Rises to 19.3 Million New Cases and 10.0 Million Cancer Deaths in 2020. Available online: https://www.iarc.who.int/fr/news-events/latest-global-cancer-data-cancer-burden-rises-to-19-3-million-new-cases-and-10-0-million-cancer-deaths-in-2020/ (accessed on 15 December 2020).
- Masuda, T.; Tomita, M.; Ishihama, Y. Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 2008, 7, 731–740. [Google Scholar] [CrossRef]
- Iliuk, A.B.; Martin, V.A.; Alicie, B.M.; Geahlen, R.L.; Tao, W.A. In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion-functionalized soluble nanopolymers. Mol. Cell Proteom. 2010, 9, 2162–2172. [Google Scholar] [CrossRef] [Green Version]
- Kelstrup, C.D.; Bekker-Jensen, D.B.; Arrey, T.N.; Hogrebe, A.; Harder, A.; Olsen, J.V. Performance Evaluation of the Q Exactive HF-X for Shotgun Proteomics. J. Proteome Res. 2018, 17, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Collins, B.C.; Hunter, C.L.; Liu, Y.; Schilling, B.; Rosenberger, G.; Bader, S.L.; Chan, D.W.; Gibson, B.W.; Gingras, A.C.; Held, J.M.; et al. Multi-laboratory assessment of reproducibility, qualitative and quantitative performance of SWATH-mass spectrometry. Nat. Commun. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Shankar, E.; Weis, M.C.; Avva, J.; Shukla, S.; Shukla, M.; Sreenath, S.N.; Gupta, S. Complex Systems Biology Approach in Connecting PI3K-Akt and NF-kappaB Pathways in Prostate Cancer. Cells 2019, 8, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelaez, R.; Ochoa, R.; Pariente, A.; Villanueva-Martinez, A.; Perez-Sala, A.; Larrayoz, I.M. Sterculic Acid Alters Adhesion Molecules Expression and Extracellular Matrix Compounds to Regulate Migration of Lung Cancer Cells. Cancers 2021, 13, 4370. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wu, C.J.; Chu, G.C.; Xiao, Y.; Ho, D.; Zhang, J.; Perry, S.R.; Labrot, E.S.; Wu, X.; Lis, R.; et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 2011, 470, 269–273. [Google Scholar] [CrossRef] [Green Version]
- Pang, X.; Xie, R.; Zhang, Z.; Liu, Q.; Wu, S.; Cui, Y. Identification of SPP1 as an Extracellular Matrix Signature for Metastatic Castration-Resistant Prostate Cancer. Front. Oncol. 2019, 9, 924. [Google Scholar] [CrossRef]
- Kariya, Y.; Oyama, M.; Kariya, Y.; Hashimoto, Y. Phosphorylated Osteopontin Secreted from Cancer Cells Induces Cancer Cell Motility. Biomolecules 2021, 11, 1323. [Google Scholar] [CrossRef]
- Vanhara, P.; Hampl, A.; Kozubik, A.; Soucek, K. Growth/differentiation factor-15: Prostate cancer suppressor or promoter? Prostate Cancer Prostatic Dis. 2012, 15, 320–328. [Google Scholar] [CrossRef] [Green Version]
- Potla, L.; Boghaert, E.R.; Armellino, D.; Frost, P.; Damle, N.K. Reduced expression of EphrinA1 (EFNA1) inhibits three-dimensional growth of HT29 colon carcinoma cells. Cancer Lett. 2002, 175, 187–195. [Google Scholar] [CrossRef]
- Xu, W.; Chang, J.; Du, X.; Hou, J. Long non-coding RNA PCAT-1 contributes to tumorigenesis by regulating FSCN1 via miR-145-5p in prostate cancer. Biomed. Pharm. 2017, 95, 1112–1118. [Google Scholar] [CrossRef]
- Chiyomaru, T.; Tatarano, S.; Kawakami, K.; Enokida, H.; Yoshino, H.; Nohata, N.; Fuse, M.; Seki, N.; Nakagawa, M. SWAP70, actin-binding protein, function as an oncogene targeting tumor-suppressive miR-145 in prostate cancer. Prostate 2011, 71, 1559–1567. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Ren, D.; Chen, X.; Tu, X.; Huang, S.; Wang, M.; Song, L.; Zou, X.; Peng, X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J. Cell Biochem. 2013, 114, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Zedan, A.H.; Hansen, T.F.; Assenholt, J.; Pleckaitis, M.; Madsen, J.S.; Osther, P.J.S. microRNA expression in tumour tissue and plasma in patients with newly diagnosed metastatic prostate cancer. Tumour Biol. 2018, 40, 1010428318775864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurul, N.O.; Ates, F.; Yilmaz, I.; Narli, G.; Yesildal, C.; Senkul, T. The association of let-7c, miR-21, miR-145, miR-182, and miR-221 with clinicopathologic parameters of prostate cancer in patients diagnosed with low-risk disease. Prostate 2019, 79, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Abramovic, I.; Ulamec, M.; Katusic Bojanac, A.; Bulic-Jakus, F.; Jezek, D.; Sincic, N. miRNA in prostate cancer: Challenges toward translation. Epigenomics 2020, 12, 543–558. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, L.; Liu, F.; Xiang, G.; Jiang, D.; Pu, X. Identification of endogenous controls for analyzing serum exosomal miRNA in patients with hepatitis B or hepatocellular carcinoma. Dis. Markers 2015, 2015, 893594. [Google Scholar] [CrossRef]
- Song, J.; Bai, Z.; Han, W.; Zhang, J.; Meng, H.; Bi, J.; Ma, X.; Han, S.; Zhang, Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012, 57, 897–904. [Google Scholar] [CrossRef]
- Donovan, M.J.; Noerholm, M.; Bentink, S.; Belzer, S.; Skog, J.; O’Neill, V.; Cochran, J.S.; Brown, G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Cai, Y.-H.; Ding, Y.; Zhang, G.; Liu, Y.; Sun, J.; Yang, Y.; Zhan, Z.; Iliuk, A.; Gu, Z.; et al. Proteomics, Phosphoproteomics and Mirna Analysis of Circulating Extracellular Vesicles through Automated and High-Throughput Isolation. Cells 2022, 11, 2070. https://doi.org/10.3390/cells11132070
Zhang H, Cai Y-H, Ding Y, Zhang G, Liu Y, Sun J, Yang Y, Zhan Z, Iliuk A, Gu Z, et al. Proteomics, Phosphoproteomics and Mirna Analysis of Circulating Extracellular Vesicles through Automated and High-Throughput Isolation. Cells. 2022; 11(13):2070. https://doi.org/10.3390/cells11132070
Chicago/Turabian StyleZhang, Hao, Yu-Han Cai, Yajie Ding, Guiyuan Zhang, Yufeng Liu, Jie Sun, Yuchen Yang, Zhen Zhan, Anton Iliuk, Zhongze Gu, and et al. 2022. "Proteomics, Phosphoproteomics and Mirna Analysis of Circulating Extracellular Vesicles through Automated and High-Throughput Isolation" Cells 11, no. 13: 2070. https://doi.org/10.3390/cells11132070
APA StyleZhang, H., Cai, Y. -H., Ding, Y., Zhang, G., Liu, Y., Sun, J., Yang, Y., Zhan, Z., Iliuk, A., Gu, Z., Gu, Y., & Tao, W. A. (2022). Proteomics, Phosphoproteomics and Mirna Analysis of Circulating Extracellular Vesicles through Automated and High-Throughput Isolation. Cells, 11(13), 2070. https://doi.org/10.3390/cells11132070






