Integrated Analysis of LncRNA-Mediated ceRNA Network in Calcific Aortic Valve Disease
Abstract
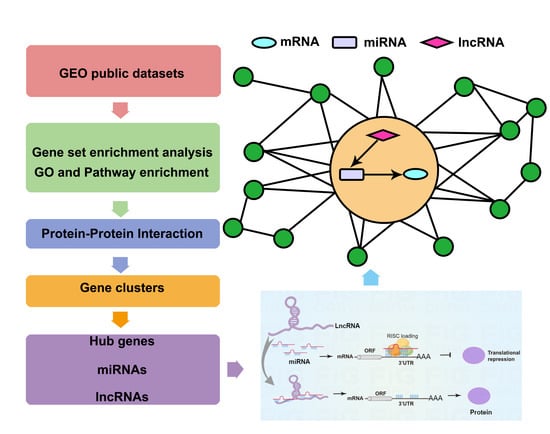
:1. Introduction
2. Materials and Methods
2.1. Data Collection and Process
2.2. Enrichment Analysis
2.3. Protein-Protein Interaction (PPI) Network Analysis and Gene Cluster Identification
2.4. Prediction of Hub miRNAs and Investigation of mRNA-miRNA Interaction Network Analysis
2.5. MiRNAs-lncRNAs Interaction Network Analysis
2.6. Real-Time Polymerase Chain Reaction
2.7. Statistical Analysis
3. Results
3.1. Processing of Sample Data and Identification of Differentially Expressed Genes
3.2. Functional Enrichment Analysis
3.3. Construction of Protein-Protein Interaction (PPI) Network and Investigation of Gene Clusters Participating in ECM-Related Biological Pathway
3.4. Further miRNA Mining and mRNA-miRNA Interaction Network Analysis
3.5. Construction of lncRNA-miRNA-mRNA ceRNA Network in CAVD
3.6. Verification of the Potential lncRNAs Expression in ceRNA Network
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yadgir, S.; Johnson, C.O.; Aboyans, V.; Adebayo, O.M.; Adedoyin, R.A.; Afarideh, M.; Alahdab, F.; Alashi, A.; Alipour, V.; Arabloo, J.; et al. Global, Regional, and National Burden of Calcific Aortic Valve and Degenerative Mitral Valve Diseases, 1990–2017. Circulation 2020, 141, 1670–1680. [Google Scholar] [CrossRef] [Green Version]
- Thaden, J.J.; Nkomo, V.T.; Enriquez-Sarano, M. The global burden of aortic stenosis. Prog. Cardiovasc. Dis. 2014, 56, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Goody, P.R.; Hosen, M.R.; Christmann, D.; Niepmann, S.T.; Zietzer, A.; Adam, M.; Bonner, F.; Zimmer, S.; Nickenig, G.; Jansen, F. Aortic Valve Stenosis: From Basic Mechanisms to Novel Therapeutic Targets. Arter. Thromb. Vasc. Biol. 2020, 40, 885–900. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barwari, T.; Joshi, A.; Mayr, M. MicroRNAs in Cardiovascular Disease. J. Am. Coll. Cardiol. 2016, 68, 2577–2584. [Google Scholar] [CrossRef] [Green Version]
- Hadji, F.; Boulanger, M.C.; Guay, S.P.; Gaudreault, N.; Amellah, S.; Mkannez, G.; Bouchareb, R.; Marchand, J.T.; Nsaibia, M.J.; Guauque-Olarte, S.; et al. Altered DNA Methylation of Long Noncoding RNA H19 in Calcific Aortic Valve Disease Promotes Mineralization by Silencing NOTCH1. Circulation 2016, 134, 1848–1862. [Google Scholar] [CrossRef]
- Yu, C.; Li, L.; Xie, F.; Guo, S.; Liu, F.; Dong, N.; Wang, Y. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc. Res. 2018, 114, 168–179. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Zhou, T.; Guo, S.; Guo, C.; Zhang, Q.; Dong, N.; Wang, Y. LncRNA MALAT1 sponges miR-204 to promote osteoblast differentiation of human aortic valve interstitial cells through up-regulating Smad4. Int. J. Cardiol. 2017, 243, 404–412. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Chen, Y.; Hao, S.; Qian, J. Uncovering novel landscape of cardiovascular diseases and therapeutic targets for cardioprotection via long noncoding RNA-miRNA-mRNA axes. Epigenomics 2018, 10, 661–671. [Google Scholar] [CrossRef]
- Yu, X.H.; Deng, W.Y.; Chen, J.J.; Xu, X.D.; Liu, X.X.; Chen, L.; Shi, M.W.; Liu, Q.X.; Tao, M.; Ren, K. LncRNA kcnq1ot1 promotes lipid accumulation and accelerates atherosclerosis via functioning as a ceRNA through the miR-452-3p/HDAC3/ABCA1 axis. Cell Death Dis. 2020, 11, 1043. [Google Scholar] [CrossRef] [PubMed]
- Bian, W.; Jiang, X.X.; Wang, Z.; Zhu, Y.R.; Zhang, H.; Li, X.; Liu, Z.; Xiong, J.; Zhang, D.M. Comprehensive analysis of the ceRNA network in coronary artery disease. Sci. Rep. 2021, 11, 24279. [Google Scholar] [CrossRef] [PubMed]
- Song, C.; Zhang, J.; Liu, Y.; Hu, Y.; Feng, C.; Shi, P.; Zhang, Y.; Wang, L.; Xie, Y.; Zhang, M.; et al. Characterization and Validation of ceRNA-Mediated Pathway-Pathway Crosstalk Networks Across Eight Major Cardiovascular Diseases. Front. Cell Dev. Biol. 2022, 10, 762129. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids. Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Soufan, O.; Ewald, J.; Hancock, R.E.W.; Basu, N.; Xia, J. NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res. 2019, 47, W234–W241. [Google Scholar] [CrossRef] [Green Version]
- Sticht, C.; De La Torre, C.; Parveen, A.; Gretz, N. MiRWalk: An online resource for prediction of microRNA binding sites. PLoS ONE 2018, 13, e0206239. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Li, J.H.; Liu, S.; Zhou, H.; Qu, L.H.; Yang, J.H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef] [Green Version]
- Long, C.; Hanning, L.; Cheng, S.; Jianqiu, P.; Jun, L.; Yue, L.; Ke, W.; Xiaoyi, W.; Peng, W.; Fangzhou, L. A Novel Long Noncoding RNA SNHG3 Promotes Osteoblast Differentiation through BMP2 Upregulation in Aortic Valve Calcification. JACC Basic Transl. Sci. 2022, in press.
- Chen, J.H.; Simmons, C.A. Cell-matrix interactions in the pathobiology of calcific aortic valve disease: Critical roles for matricellular, matricrine, and matrix mechanics cues. Circ. Res. 2011, 108, 1510–1524. [Google Scholar] [CrossRef] [Green Version]
- Di Vito, A.; Donato, A.; Presta, I.; Mancuso, T.; Brunetti, F.S.; Mastroroberto, P.; Amorosi, A.; Malara, N.; Donato, G. Extracellular Matrix in Calcific Aortic Valve Disease: Architecture, Dynamic and Perspectives. Int. J. Mol. Sci. 2021, 22, 913. [Google Scholar] [CrossRef]
- En, Q.; Zeping, H.; Yuetang, W.; Xu, W.; Wei, W. Metformin alleviates the calcification of aortic valve interstitial cells through activating the PI3K/AKT pathway in an AMPK dependent way. Mol. Med. 2021, 27, 156. [Google Scholar] [CrossRef]
- Eriksen, H.A.; Satta, J.; Risteli, J.; Veijola, M.; Vare, P.; Soini, Y. Type I and type III collagen synthesis and composition in the valve matrix in aortic valve stenosis. Atherosclerosis 2006, 189, 91–98. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Werkmeister, J.A.; Hilbert, S.L.; Ramshaw, J.A. Heart valve collagens: Cross-species comparison using immunohistological methods. J. Heart Valve Dis. 2010, 19, 766–771. [Google Scholar] [PubMed]
- Mourino-Alvarez, L.; Iloro, I.; de la Cuesta, F.; Azkargorta, M.; Sastre-Oliva, T.; Escobes, I.; Lopez-Almodovar, L.F.; Sanchez, P.L.; Urreta, H.; Fernandez-Aviles, F.; et al. MALDI-Imaging Mass Spectrometry: A step forward in the anatomopathological characterization of stenotic aortic valve tissue. Sci. Rep. 2016, 6, 27106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durand, A.L.; Dufour, A.; Aubert-Foucher, E.; Oger-Desfeux, C.; Pasdeloup, M.; Lustig, S.; Servien, E.; Vaz, G.; Perrier-Groult, E.; Mallein-Gerin, F.; et al. The Lysine Specific Demethylase-1 Negatively Regulates the COL9A1 Gene in Human Articular Chondrocytes. Int. J. Mol. Sci. 2020, 21, 6322. [Google Scholar] [CrossRef]
- Liu, C.Y.; Olsen, B.R.; Kao, W.W. Developmental patterns of two α 1(IX) collagen mRNA isoforms in mouse. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 1993, 198, 150–157. [Google Scholar] [CrossRef]
- Yip, C.Y.; Chen, J.H.; Zhao, R.; Simmons, C.A. Calcification by valve interstitial cells is regulated by the stiffness of the extracellular matrix. Arter. Thromb. Vasc. Biol. 2009, 29, 936–942. [Google Scholar] [CrossRef] [Green Version]
- Baldinger, A.; Brehm, B.R.; Richter, P.; Bossert, T.; Gruen, K.; Hekmat, K.; Kosmehl, H.; Neri, D.; Figulla, H.R.; Berndt, A.; et al. Comparative analysis of oncofetal fibronectin and tenascin-C expression in right atrial auricular and left ventricular human cardiac tissue from patients with coronary artery disease and aortic valve stenosis. Histochem. Cell Biol. 2011, 135, 427–441. [Google Scholar] [CrossRef]
- Satta, J.; Melkko, J.; Pöllänen, R.; Tuukkanen, J.; Pääkkö, P.; Ohtonen, P.; Mennander, A.; Soini, Y. Progression of human aortic valve stenosis is associated with tenascin-C expression. J. Am. Coll Cardiol. 2002, 39, 96–101. [Google Scholar] [CrossRef] [Green Version]
- Kaden, J.J.; Dempfle, C.E.; Grobholz, R.; Fischer, C.S.; Vocke, D.C.; Kiliç, R.; Sarikoç, A.; Piñol, R.; Hagl, S.; Lang, S.; et al. Inflammatory regulation of extracellular matrix remodeling in calcific aortic valve stenosis. Cardiovasc. Pathol. 2005, 14, 80–87. [Google Scholar] [CrossRef]
- Mohler, E.R., 3rd; Gannon, F.; Reynolds, C.; Zimmerman, R.; Keane, M.G.; Kaplan, F.S. Bone formation and inflammation in cardiac valves. Circulation 2001, 103, 1522–1528. [Google Scholar] [CrossRef]
- Liberman, M.; Bassi, E.; Martinatti, M.K.; Lario, F.C.; Wosniak, J., Jr.; Pomerantzeff, P.M.; Laurindo, F.R. Oxidant generation predominates around calcifying foci and enhances progression of aortic valve calcification. Arter. Thromb. Vasc. Biol. 2008, 28, 463–470. [Google Scholar] [CrossRef] [Green Version]
- Midwood, K.S.; Valenick, L.V.; Hsia, H.C.; Schwarzbauer, J.E. Coregulation of fibronectin signaling and matrix contraction by tenascin-C and syndecan-4. Mol. Biol. Cell. 2004, 15, 5670–5677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orend, G.; Huang, W.; Olayioye, M.A.; Hynes, N.E.; Chiquet-Ehrismann, R. Tenascin-C blocks cell-cycle progression of anchorage-dependent fibroblasts on fibronectin through inhibition of syndecan-4. Oncogene 2003, 22, 3917–3926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, X.; Miyamoto, S.; Mekada, E. Integrin α2 β1-dependent EGF receptor activation at cell-cell contact sites. J. Cell Sci. 2000, 113, 2139–2147. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Masters, K.S. Regulation of valvular interstitial cell calcification by adhesive peptide sequences. J. Biomed. Mater. Res. A 2010, 93, 1620–1630. [Google Scholar] [CrossRef] [Green Version]
- Butcher, J.T.; Norris, R.A.; Hoffman, S.; Mjaatvedt, C.H.; Markwald, R.R. Periostin promotes atrioventricular mesenchyme matrix invasion and remodeling mediated by integrin signaling through Rho/PI 3-kinase. Dev. Biol. 2007, 302, 256–266. [Google Scholar] [CrossRef] [Green Version]
- Carrion, K.; Dyo, J.; Patel, V.; Sasik, R.; Mohamed, S.A.; Hardiman, G.; Nigam, V. The long non-coding HOTAIR is modulated by cyclic stretch and WNT/β-CATENIN in human aortic valve cells and is a novel repressor of calcification genes. PLoS ONE 2014, 9, e96577. [Google Scholar] [CrossRef] [Green Version]
- Zheng, D.; Wang, B.; Zhu, X.; Hu, J.; Sun, J.; Xuan, J.; Ge, Z. LncRNA OIP5-AS1 inhibits osteoblast differentiation of valve interstitial cells via miR-137/TWIST11 axis. Biochem. Biophys. Res. Commun. 2019, 511, 826–832. [Google Scholar] [CrossRef]
- Zhang, L.; Li, G.; Wang, X.; Zhang, Y.; Huang, X.; Wu, H. lncRNA SNHG3 acts as oncogene in ovarian cancer through miR-139-5p and Notch1. Oncol. Lett. 2021, 21, 122. [Google Scholar] [CrossRef]
- Dacheng, W.; Songhe, L.; Weidong, J.; Shutao, Z.; Jingjing, L.; Jiaming, Z. LncRNA SNHG3 promotes the growth and metastasis of colorectal cancer by regulating miR-539/RUNX2 axis. Biomed. Pharm. 2020, 125, 110039. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Y.; Wu, G.; Chen, X.; Liu, Y.; Wang, X.; Yu, J.; Li, C.; Chen, X.; Jose, P.A.; et al. Plasma long non-coding RNA, CoroMarker, a novel biomarker for diagnosis of coronary artery disease. Clin. Sci. 2015, 129, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Zhang, Y.; Zhang, W.; Deng, S.Q.; Ge, Z.R. LncRNA-NRF is a Potential Biomarker of Heart Failure After Acute Myocardial Infarction. J. Cardiovasc. Transl. Res. 2020, 13, 1008–1015. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Wei, K.; Li, J.; Li, Y.; Cao, H.; Zheng, Z. Integrated Analysis of LncRNA-Mediated ceRNA Network in Calcific Aortic Valve Disease. Cells 2022, 11, 2204. https://doi.org/10.3390/cells11142204
Chen L, Wei K, Li J, Li Y, Cao H, Zheng Z. Integrated Analysis of LncRNA-Mediated ceRNA Network in Calcific Aortic Valve Disease. Cells. 2022; 11(14):2204. https://doi.org/10.3390/cells11142204
Chicago/Turabian StyleChen, Long, Ke Wei, Jun Li, Yue Li, Huiqing Cao, and Zhe Zheng. 2022. "Integrated Analysis of LncRNA-Mediated ceRNA Network in Calcific Aortic Valve Disease" Cells 11, no. 14: 2204. https://doi.org/10.3390/cells11142204
APA StyleChen, L., Wei, K., Li, J., Li, Y., Cao, H., & Zheng, Z. (2022). Integrated Analysis of LncRNA-Mediated ceRNA Network in Calcific Aortic Valve Disease. Cells, 11(14), 2204. https://doi.org/10.3390/cells11142204