Vertebrate Cell Differentiation, Evolution, and Diseases: The Vertebrate-Specific Developmental Potential Guardians VENTX/NANOG and POU5/OCT4 Enter the Stage
Abstract
:1. Introduction
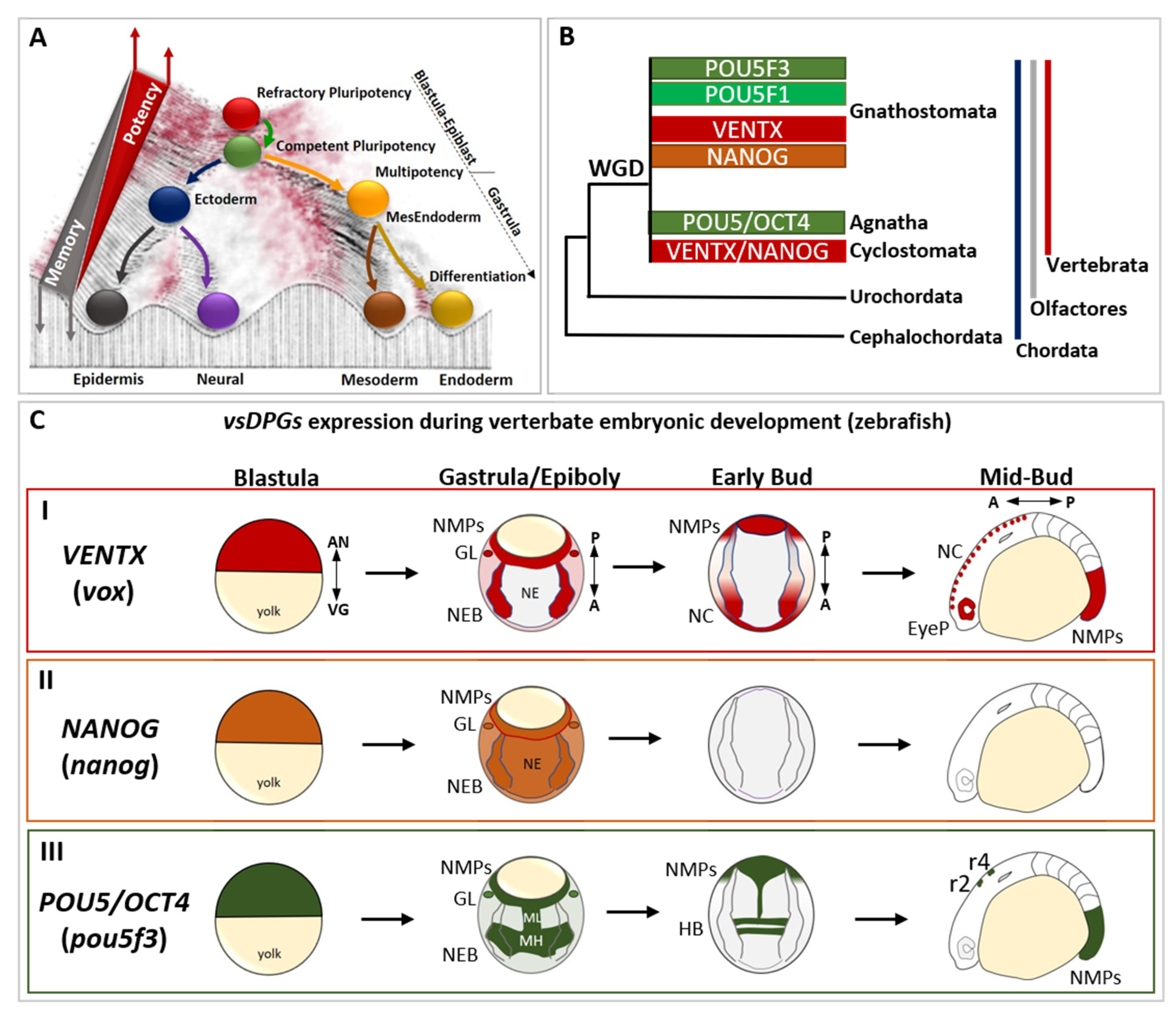
2. Role of Developmental Potential Guardians in Stem Cells Pluripotency
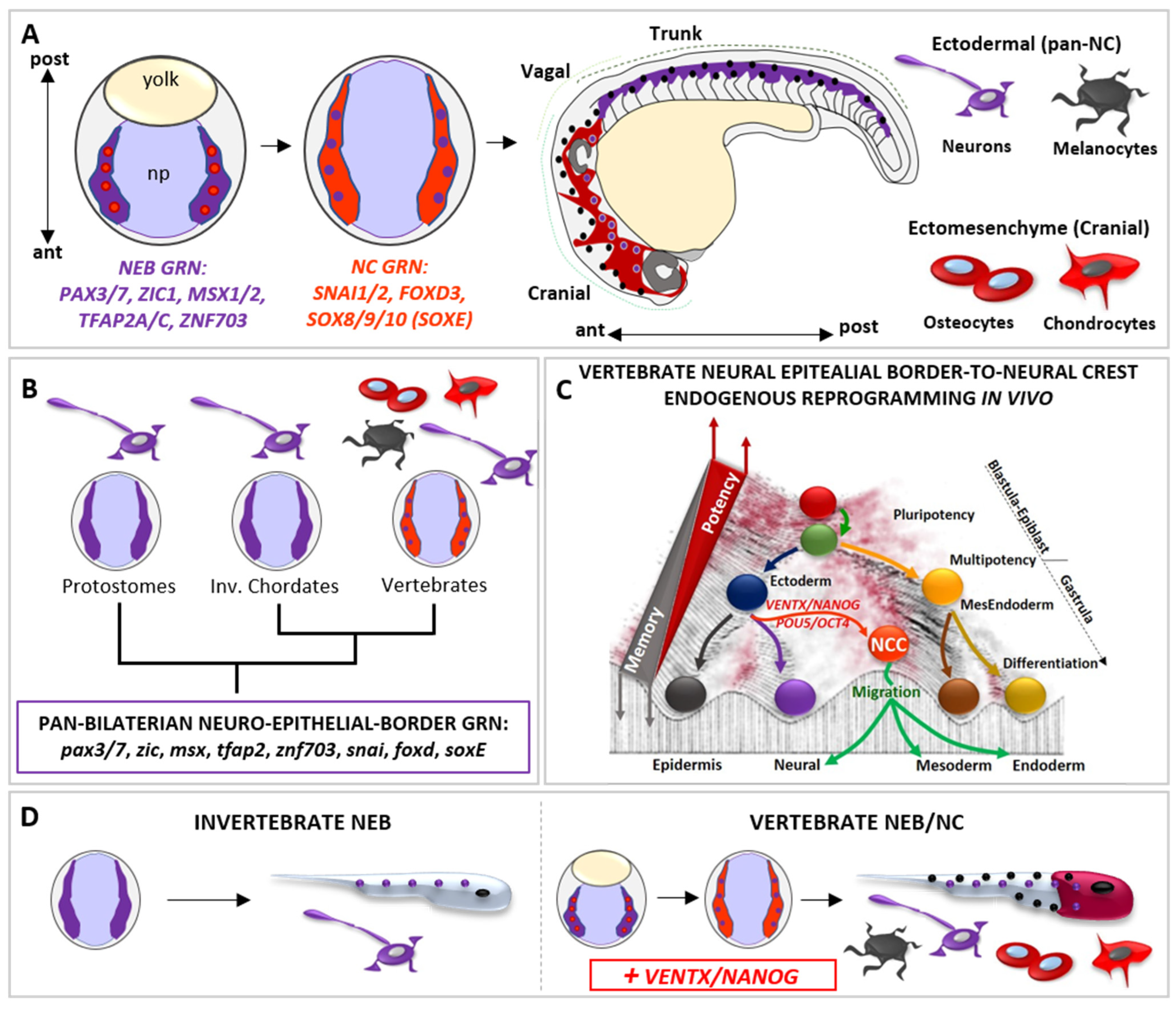
3. Reprogramming Capacity of Developmental Potential Guardians Shapes Neural Crest Multipotency and Vertebrate Evolution
4. Developmental Potential Guardians Role in Neuro-Mesodermal Progenitors and Vertebrate Axial Length
5. Developmental Potential Guardians in Human Diseases
6. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boroviak, T.; Nichols, J. The birth of embryonic pluripotency. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snape, A.; Wylie, C.C.; Smith, J.C.; Heasman, J. Changes in states of commitment of single animal pole blastomeres of Xenopus laevis. Dev. Biol. 1987, 119, 503–510. [Google Scholar] [CrossRef]
- Hong, Y.; Winkler, C.; Schartl, M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes). Mech. Dev. 1996, 60, 33–44. [Google Scholar] [CrossRef]
- Lavial, F.; Acloque, H.; Bertocchini, F.; Macleod, D.J.; Boast, S.; Bachelard, E.; Montillet, G.; Thenot, S.; Sang, H.M.; Stern, C.D.; et al. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development 2007, 134, 3549–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014, 15, 471–487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, K.; Sato, K.; Michiue, T.; Okabayashi, K.; Ohnuma, K.; Danno, H.; Asashima, M. Developmental potential for morphogenesis in vivo and in vitro. J. Exp. Zool. B Mol. Dev. Evol. 2008, 310, 492–503. [Google Scholar] [CrossRef]
- Simunovic, M.; Brivanlou, A.H. Embryoids, organoids and gastruloids: New approaches to understanding embryogenesis. Development 2017, 144, 976–985. [Google Scholar] [CrossRef] [Green Version]
- Martinez Arias, A.; Nichols, J.; Schröter, C. A molecular basis for developmental plasticity in early mammalian embryos. Development 2013, 140, 3499–3510. [Google Scholar] [CrossRef] [Green Version]
- Nichols, J.; Silva, J.; Roode, M.; Smith, A. Suppression of Erk signalling promotes ground state pluripotency in the mouse embryo. Development 2009, 136, 3215–3222. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.; Stirparo, G.G.; Strawbridge, S.E.; Spindlow, D.; Yang, J.; Clarke, J.; Dattani, A.; Yanagida, A.; Li, M.A.; Myers, S.; et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 2021, 28, 1040–1056.e6. [Google Scholar] [CrossRef]
- Kirschner, M.; Gerhart, J. Evolvability. Proc. Natl. Acad. Sci. USA 1998, 95, 8420–8427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirschner, M.; Gerhart, J. The Plausibility of Life: Resolving Darwin’s Dilemma; Yale University Press: New Haven, CN, USA, 2006. [Google Scholar]
- Smith, A. Formative pluripotency: The executive phase in a developmental continuum. Development 2017, 144, 365–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scerbo, P.; Marchal, L.; Kodjabachian, L. Lineage commitment of embryonic cells involves MEK1-dependent clearance of pluripotency regulator Ventx2. eLife 2017, 6, e21526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scerbo, P.; Monsoro-Burq, A.H. The vertebrate-specific VENTX/NANOG gene empowers neural crest with ectomesenchyme potential. Sci. Adv. 2020, 6, eaaz1469. [Google Scholar] [CrossRef]
- Zhao, J.; Lambert, G.; Meijer, A.H.; Rosa, F.M. The transcription factor Vox represses endoderm development by interacting with Casanova and Pou2. Development 2013, 140, 1090–1099. [Google Scholar] [CrossRef] [Green Version]
- Morrison, G.M.; Brickman, J.M. Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development 2006, 133, 2011–2022. [Google Scholar] [CrossRef] [Green Version]
- Theunissen, T.W.; Costa, Y.; Radzisheuskaya, A.; van Oosten, A.L.; Lavial, F.; Pain, B.; Castro, L.F.; Silva, J.C. Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. Development 2011, 138, 4853–4865. [Google Scholar] [CrossRef] [Green Version]
- Tapia, N.; Reinhardt, P.; Duemmler, A.; Wu, G.; Araúzo-Bravo, M.J.; Esch, D.; Greber, B.; Cojocaru, V.; Rascon, C.A.; Tazaki, A.; et al. Reprogramming to pluripotency is an ancient trait of vertebrate Oct4 and Pou2 proteins. Nat. Commun. 2012, 3, 1279. [Google Scholar] [CrossRef] [Green Version]
- Swaidan, N.T.; Salloum-Asfar, S.; Palangi, F.; Errafii, K.; Soliman, N.H.; Aboughalia, A.T.; Wali, A.H.S.; Abdulla, S.A.; Emara, M.M. Identification of potential transcription factors that enhance human iPSC generation. Sci. Rep. 2020, 10, 21950. [Google Scholar] [CrossRef]
- Livigni, A.; Peradziryi, H.; Sharov, A.A.; Chia, G.; Hammachi, F.; Migueles, R.P.; Sukparangsi, W.; Pernagallo, S.; Bradley, M.; Nichols, J.; et al. A conserved Oct4/POUV-dependent network links adhesion and migration to progenitor maintenance. Curr. Biol. 2013, 23, 2233–2244. [Google Scholar] [CrossRef] [Green Version]
- Scerbo, P.; Girardot, F.; Vivien, C.; Markov, G.V.; Luxardi, G.; Demeneix, B.; Kodjabachian, L.; Coen, L. Ventx factors function as Nanog-like guardians of developmental potential in Xenopus. PLoS ONE 2012, 7, e36855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scerbo, P.; Markov, G.V.; Vivien, C.; Kodjabachian, L.; Demeneix, B.; Coen, L.; Girardot, F. On the origin and evolutionary history of NANOG. PLoS ONE 2014, 9, e85104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boroviak, T.; Stirparo, G.G.; Dietmann, S.; Hernando-Herraez, I.; Mohammed, H.; Reik, W.; Smith, A.; Sasaki, E.; Nichols, J.; Bertone, P. Single cell transcriptome analysis of human, marmoset and mouse embryos reveals common and divergent features of preimplantation development. Development 2018, 145, dev167833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, L.; Yin, Y.; Zheng, Y.; Ma, Y.; Li, Y.; Zhao, Z.; Guo, J.; Ai, Z.; Niu, Y.; Duan, K.; et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature 2020, 577, 537–542. [Google Scholar] [CrossRef]
- Perez-Camps, M.; Tian, J.; Chng, S.C.; Sem, K.P.; Sudhaharan, T.; Teh, C.; Wachsmuth, M.; Korzh, V.; Ahmed, S.; Reversade, B. Quantitative imaging reveals real-time Pou5f3-Nanog complexes driving dorsoventral mesendoderm patterning in zebrafish. eLife 2016, 5, e11475. [Google Scholar] [CrossRef]
- He, M.; Zhang, R.; Jiao, S.; Zhang, F.; Ye, D.; Wang, H.; Sun, Y. Nanog safeguards early embryogenesis against global activation of maternal beta-catenin activity by interfering with TCF factors. PLoS Biol. 2020, 18, e3000561. [Google Scholar] [CrossRef]
- Kushwaha, R.; Jagadish, N.; Kustagi, M.; Tomishima, M.J.; Mendiratta, G.; Bansal, M.; Kim, H.R.; Sumazin, P.; Alvarez, M.J.; Lefebvre, C.; et al. Interrogation of a context-specific transcription factor network identifies novel regulators of pluripotency. Stem Cells 2015, 33, 367–377. [Google Scholar] [CrossRef] [Green Version]
- Cevallos, R.R.; Edwards, Y.J.K.; Parant, J.M.; Yoder, B.K.; Hu, K. Human transcription factors responsive to initial reprogramming predominantly undergo legitimate reprogramming during fibroblast conversion to iPSCs. Sci. Rep. 2020, 10, 19710. [Google Scholar] [CrossRef]
- Zhou, H.; Morales, M.G.; Hashimoto, H.; Dickson, M.E.; Song, K.; Ye, W.; Kim, M.S.; Niederstrasser, H.; Wang, Z.; Chen, B.; et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017, 31, 1770–1783. [Google Scholar] [CrossRef] [Green Version]
- Vivien, C.; Scerbo, P.; Girardot, F.; Le Blay, K.; Demeneix, B.A.; Coen, L. Non-viral expression of mouse Oct4, Sox2, and Klf4 transcription factors efficiently reprograms tadpole muscle fibers in vivo. J. Biol. Chem. 2012, 287, 7427–7435. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Knöchel, S.; Donow, C.; Miethe, J.; Kaufmann, E.; Knöchel, W. The POU factor Oct-25 regulates the Xvent-2B gene and counteracts terminal differentiation in Xenopus embryos. J. Biol. Chem. 2004, 279, 43735–43743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Siegel, D.; Oswald, F.; Knöchel, W. Oct25 represses transcription of nodal/activin target genes by interaction with signal transducers during Xenopus gastrulation. J. Biol. Chem. 2008, 283, 34168–34177. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Siegel, D.; Donow, C.; Knöchel, S.; Yuan, L.; Knöchel, W. POU-V factors antagonize maternal VegT activity and beta-Catenin signaling in Xenopus embryos. EMBO J. 2007, 26, 2942–2954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Wu, B.; Giese, R.; Zhu, Z. Xom interacts with and stimulates transcriptional activity of LEF1/TCFs: Implications for ventral cell fate determination during vertebrate embryogenesis. Cell Res. 2007, 17, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.; Weinreb, C.; Wagner, D.E.; Megason, S.; Peshkin, L.; Kirschner, M.W.; Klein, A.M. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 2018, 360, eaar5780. [Google Scholar] [CrossRef] [Green Version]
- Bright, A.R.; van Genesen, S.; Li, Q.; Grasso, A.; Frölich, S.; van der Sande, M.; van Heeringen, S.J.; Veenstra, G.J.C. Combinatorial transcription factor activities on open chromatin induce embryonic heterogeneity in vertebrates. EMBO J. 2021, 40, e104913. [Google Scholar] [CrossRef]
- Wagner, D.E.; Weinreb, C.; Collins, Z.M.; Briggs, J.A.; Megason, S.G.; Klein, A.M. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 2018, 360, 981–987. [Google Scholar] [CrossRef] [Green Version]
- Fang, F.; Angulo, B.; Xia, N.; Sukhwani, M.; Wang, Z.; Carey, C.C.; Mazurie, A.; Cui, J.; Wilkinson, R.; Wiedenheft, B.; et al. A PAX5-OCT4-PRDM1 developmental switch specifies human primordial germ cells. Nat. Cell Biol. 2018, 20, 655–665. [Google Scholar] [CrossRef]
- Sybirna, A.; Tang, W.W.C.; Pierson Smela, M.; Dietmann, S.; Gruhn, W.H.; Brosh, R.; Surani, M.A. A critical role of PRDM14 in human primordial germ cell fate revealed by inducible degrons. Nat. Commun. 2020, 11, 1282. [Google Scholar] [CrossRef]
- Murakami, K.; Günesdogan, U.; Zylicz, J.J.; Tang, W.W.C.; Sengupta, R.; Kobayashi, T.; Kim, S.; Butler, R.; Dietmann, S.; Surani, M.A. NANOG alone induces germ cells in primed epiblast in vitro by activation of enhancers. Nature 2016, 529, 403–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatarama, T.; Lai, F.; Luo, X.; Zhou, Y.; Newman, K.; King, M.L. Repression of zygotic gene expression in the Xenopus germline. Development 2010, 137, 651–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Sánchez, A.V.; Camp, E.; Leal-Tassias, A.; Atkinson, S.P.; Armstrong, L.; Díaz-Llopis, M.; Mullor, J.L. Nanog regulates primordial germ cell migration through Cxcr4b. Stem Cells 2010, 28, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Lignell, A.; Kerosuo, L.; Streichan, S.J.; Cai, L.; Bronner, M.E. Identification of a neural crest stem cell niche by Spatial Genomic Analysis. Nat. Commun. 2017, 8, 1830. [Google Scholar] [CrossRef] [Green Version]
- Zalc, A.; Sinha, R.; Gulati, G.S.; Wesche, D.J.; Daszczuk, P.; Swigut, T.; Weissman, I.L.; Wysocka, J. Reactivation of the pluripotency program precedes formation of the cranial neural crest. Science 2021, 371, eabb4776. [Google Scholar] [CrossRef]
- Lukoseviciute, M.; Gavriouchkina, D.; Williams, R.M.; Hochgreb-Hagele, T.; Senanayake, U.; Chong-Morrison, V.; Thongjuea, S.; Repapi, E.; Mead, A.; Sauka-Spengler, T. From Pioneer to Repressor: Bimodal foxd3 Activity Dynamically Remodels Neural Crest Regulatory Landscape In Vivo. Dev. Cell. 2018, 47, 608–628.e6. [Google Scholar] [CrossRef] [Green Version]
- Lukoseviciute, M.; Mayes, S.; Sauka-Spengler, T. Neuromesodermal progenitor origin of trunk neural crest in vivo. bioRxiv 2021. [Google Scholar] [CrossRef]
- Boroviak, T.; Loos, R.; Bertone, P.; Smith, A.; Nichols, J. The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nat. Cell Biol. 2014, 16, 516–528. [Google Scholar] [CrossRef] [Green Version]
- Le Petillon, Y.; Luxardi, G.; Scerbo, P.; Cibois, M.; Leon, A.; Subirana, L.; Irimia, M.; Kodjabachian, L.; Escriva, H.; Bertrand, S. Nodal/Activin Pathway is a Conserved Neural Induction Signal in Chordates. Nat. Ecol. Evol. 2017, 1, 1192–1200. [Google Scholar] [CrossRef] [Green Version]
- Hudson, C.; Lemaire, P. Induction of anterior neural fates in the ascidian Ciona intestinalis. Mech. Dev. 2001, 100, 189–203. [Google Scholar] [CrossRef]
- Green, S.A.; Norris, R.P.; Terasaki, M.; Lowe, C.J. FGF signaling induces mesoderm in the hemichordate Saccoglossus kowalevskii. Development 2013, 140, 1024–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blakeley, P.; Fogarty, N.M.; del Valle, I.; Wamaitha, S.E.; Hu, T.X.; Elder, K.; Snell, P.; Christie, L.; Robson, P.; Niakan, K.K. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development 2015, 142, 3151–3165. [Google Scholar] [CrossRef] [Green Version]
- Faunes, F.; Hayward, P.; Descalzo, S.M.; Chatterjee, S.S.; Balayo, T.; Trott, J.; Christoforou, A.; Ferrer-Vaquer, A.; Hadjantonakis, A.K.; Dasgupta, R.; et al. A membrane-associated β-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 2013, 140, 1171–1183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogarty, N.M.E.; McCarthy, A.; Snijders, K.E.; Powell, B.E.; Kubikova, N.; Blakeley, P.; Lea, R.; Elder, K.; Wamaitha, S.E.; Kim, D.; et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature 2017, 550, 67–73. [Google Scholar] [CrossRef]
- Loh, Y.H.; Wu, Q.; Chew, J.L.; Vega, V.B.; Zhang, W.; Chen, X.; Bourque, G.; George, J.; Leong, B.; Liu, J.; et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006, 38, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Sharov, A.A.; Masui, S.; Sharova, L.V.; Piao, Y.; Aiba, K.; Matoba, R.; Xin, L.; Niwa, H.; Ko, M.S. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC Genom. 2008, 9, 269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Oron, E.; Nelson, B.; Razis, S.; Ivanova, N. Distinct lineage specification roles for NANOG, OCT4, and SOX2 in human embryonic stem cells. Cell Stem Cell 2012, 10, 440–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Liu, J.; Chen, C.; Liu, Z.; Jiang, C.; Chu, H.; Pan, W.; Wang, X.; Zhang, L.; Li, B.; et al. The deubiquitinase USP21 maintains the stemness of mouse embryonic stem cells via stabilization of Nanog. Nat. Commun. 2016, 7, 13594. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ni, P.; Mou, C.; Zhang, Y.; Guo, H.; Zhao, T.; Loh, Y.H.; Chen, L. Cops2 promotes pluripotency maintenance by Stabilizing Nanog Protein and Repressing Transcription. Sci. Rep. 2016, 6, 26804. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishna, S.; Suresh, B.; Lim, K.H.; Cha, B.H.; Lee, S.H.; Kim, K.S.; Baek, K.H. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011, 20, 1511–1519. [Google Scholar] [CrossRef]
- Zhu, Z.; Kirschner, M. Regulated proteolysis of Xom mediates dorsoventral pattern formation during early Xenopus development. Dev. Cell. 2002, 3, 557–568. [Google Scholar] [CrossRef] [Green Version]
- Brown, K.; Loh, K.M.; Nusse, R. Live Imaging Reveals that the First Division of Differentiating Human Embryonic Stem Cells Often Yields Asymmetric Fates. Cell. Rep. 2017, 21, 301–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habib, S.J.; Chen, B.C.; Tsai, F.C.; Anastassiadis, K.; Meyer, T.; Betzig, E.; Nusse, R. A localized Wnt signal orients asymmetric stem cell division in vitro. Science 2013, 339, 1445–1448. [Google Scholar] [CrossRef] [Green Version]
- Han, D.; Wu, G.; Chen, R.; Drexler, H.C.A.; MacCarthy, C.M.; Kim, K.P.; Adachi, K.; Gerovska, D.; Mavrommatis, L.; Bedzhov, I.; et al. A balanced Oct4 interactome is crucial for maintaining pluripotency. Sci. Adv. 2022, 8, eabe4375. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Park, K.M.; Gontarz, P.; Zhang, B.; Pan, J.; McKenzie, Z.; Fischer, L.A.; Dong, C.; Dietmann, S.; Xing, X.; et al. OCT4 cooperates with distinct ATP-dependent chromatin remodelers in naive and primed pluripotent states in human. Nat. Commun. 2021, 12, 5123. [Google Scholar] [CrossRef]
- Tien, C.L.; Mohammadparast, S.; Chang, C. Heterochromatin protein 1 beta regulates neural and neural crest development by repressing pluripotency-associated gene pou5f3.2/oct25 in Xenopus. Dev. Dyn. 2021, 250, 1113–1124. [Google Scholar] [CrossRef]
- Peng, J.C.; Valouev, A.; Swigut, T.; Zhang, J.; Zhao, Y.; Sidow, A.; Wysocka, J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell 2009, 139, 1290–1302. [Google Scholar] [CrossRef] [Green Version]
- Kuznetsov, J.N.; Aguero, T.H.; Owens, D.A.; Kurtenbach, S.; Field, M.G.; Durante, M.A.; Rodriguez, D.A.; King, M.L.; Harbour, J.W. BAP1 regulates epigenetic switch from pluripotency to differentiation in developmental lineages giving rise to BAP1-mutant cancers. Sci. Adv. 2019, 5, eaax1738. [Google Scholar] [CrossRef] [Green Version]
- Nicetto, D.; Hahn, M.; Jung, J.; Schneider, T.D.; Straub, T.; David, R.; Schotta, G.; Rupp, R.A. Suv4-20h histone methyltransferases promote neuroectodermal differentiation by silencing the pluripotency-associated Oct-25 gene. PLoS Genet. 2013, 9, e1003188. [Google Scholar] [CrossRef] [Green Version]
- Galvagni, F.; Lentucci, C.; Neri, F.; Dettori, D.; De Clemente, C.; Orlandini, M.; Anselmi, F.; Rapelli, S.; Grillo, M.; Borghi, S.; et al. Snai1 promotes ESC exit from the pluripotency by direct repression of self-renewal genes. Stem Cells 2015, 33, 742–750. [Google Scholar] [CrossRef] [Green Version]
- Yamamizu, K.; Schlessinger, D.; Ko, M.S. SOX9 accelerates ESC differentiation to three germ layer lineages by repressing SOX2 expression through P21 (WAF1/CIP1). Development 2014, 141, 4254–4266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rousso, S.Z.; Schyr, R.B.; Gur, M.; Zouela, N.; Kot-Leibovich, H.; Shabtai, Y.; Koutsi-Urshanski, N.; Baldessari, D.; Pillemer, G.; Niehrs, C.; et al. Negative autoregulation of Oct3/4 through Cdx1 promotes the onset of gastrulation. Dev. Dyn. 2011, 240, 796–807. [Google Scholar] [CrossRef] [PubMed]
- Steventon, B.; Busby, L.; Arias, A.M. Establishment of the vertebrate body plan: Rethinking gastrulation through stem cell models of early embryogenesis. Dev. Cell. 2021, 56, 2405–2418. [Google Scholar] [CrossRef] [PubMed]
- Martinez Arias, A.; Steventon, B. On the nature and function of organizers. Development 2018, 145, dev159525. [Google Scholar] [CrossRef] [Green Version]
- Moris, N.; Martinez Arias, A.; Steventon, B. Experimental embryology of gastrulation: Pluripotent stem cells as a new model system. Curr. Opin. Genet. Dev. 2020, 64, 78–83. [Google Scholar] [CrossRef]
- Blauwkamp, T.A.; Nigam, S.; Ardehali, R.; Weissman, I.L.; Nusse, R. Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun. 2012, 3, 1070. [Google Scholar] [CrossRef]
- Funa, N.S.; Schachter, K.A.; Lerdrup, M.; Ekberg, J.; Hess, K.; Dietrich, N.; Honoré, C.; Hansen, K.; Semb, H. β-Catenin Regulates Primitive Streak Induction through Collaborative Interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell 2015, 16, 639–652. [Google Scholar] [CrossRef] [Green Version]
- Menendez, L.; Yatskievych, T.A.; Antin, P.B.; Dalton, S. Wnt signaling and a Smad pathway blockade direct the differentiation of human pluripotent stem cells to multipotent neural crest cells. Proc. Natl. Acad. Sci. USA 2011, 108, 19240–19245. [Google Scholar] [CrossRef] [Green Version]
- Thier, M.C.; Hommerding, O.; Panten, J.; Pinna, R.; García-González, D.; Berger, T.; Wörsdörfer, P.; Assenov, Y.; Scognamiglio, R.; Przybylla, A.; et al. Identification of Embryonic Neural Plate Border Stem Cells and Their Generation by Direct Reprogramming from Adult Human Blood Cells. Cell Stem Cell 2019, 24, 166–182.e13. [Google Scholar] [CrossRef] [Green Version]
- Britton, G.; Heemskerk, I.; Hodge, R.; Qutub, A.A.; Warmflash, A. A novel self-organizing embryonic stem cell system reveals signaling logic underlying the patterning of human ectoderm. Development 2019, 146, dev179093. [Google Scholar] [CrossRef]
- Blythe, S.A.; Cha, S.W.; Tadjuidje, E.; Heasman, J.; Klein, P.S. Beta-Catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Dev. Cell 2010, 19, 220–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charney, R.M.; Forouzmand, E.; Cho, J.S.; Cheung, J.; Paraiso, K.D.; Yasuoka, Y.; Takahashi, S.; Taira, M.; Blitz, I.L.; Xie, X.; et al. Foxh1 Occupies cis-Regulatory Modules Prior to Dynamic Transcription Factor Interactions Controlling the Mesendoderm Gene Program. Dev. Cell 2017, 40, 595–607.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, W.T.; Charney Le, R.; Blitz, I.L.; Fish, M.B.; Li, Y.; Biesinger, J.; Xie, X.; Cho, K.W. Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program. Development 2014, 141, 4537–4547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schohl, A.; Fagotto, F. Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development 2002, 129, 37–52. [Google Scholar] [CrossRef]
- Xu, P.F.; Houssin, N.; Ferri-Lagneau, K.F.; Thisse, B.; Thisse, C. Construction of a vertebrate embryo from two opposing morphogen gradients. Science 2014, 344, 87–89. [Google Scholar] [CrossRef]
- Fauny, J.D.; Thisse, B.; Thisse, C. The entire zebrafish blastula-gastrula margin acts as an organizer dependent on the ratio of Nodal to BMP activity. Development 2009, 136, 3811–3819. [Google Scholar] [CrossRef] [Green Version]
- Soh, G.H.; Pomreinke, A.P.; Müller, P. Integration of Nodal and BMP Signaling by Mutual Signaling Effector Antagonism. Cell Rep. 2020, 31, 107487. [Google Scholar] [CrossRef]
- Kunath, T.; Saba-El-Leil, M.K.; Almousailleakh, M.; Wray, J.; Meloche, S.; Smith, A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 2007, 134, 2895–2902. [Google Scholar] [CrossRef] [Green Version]
- Nakanoh, S.; Okazaki, K.; Agata, K. Inhibition of MEK and GSK3 supports ES cell-like domed colony formation from avian and reptile embryos. Zoolog. Sci. 2013, 30, 543–552. [Google Scholar] [CrossRef]
- Nakanoh, S.; Agata, K. Evolutionary view of pluripotency seen from early development of non-mammalian amniotes. Dev. Biol. 2019, 452, 95–103. [Google Scholar] [CrossRef]
- Crispatzu, G.; Rehimi, R.; Pachano, T.; Bleckwehl, T.; Cruz-Molina, S.; Xiao, C.; Mahabir, E.; Bazzi, H.; Rada-Iglesias, A. The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. Nat. Commun. 2021, 12, 4344. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.H.; van Genesen, S.; Perino, M.; Bark, M.R.; Veenstra, G.J.C. Loss of PRC2 subunits primes lineage choice during exit of pluripotency. Nat. Commun. 2021, 12, 6985. [Google Scholar] [CrossRef] [PubMed]
- Jansen, C.; Paraiso, K.D.; Zhou, J.J.; Blitz, I.L.; Fish, M.B.; Charney, R.M.; Cho, J.S.; Yasuoka, Y.; Sudou, N.; Bright, A.R.; et al. Uncovering the mesendoderm gene regulatory network through multi-omic data integration. Cell Rep. 2022, 38, 110364. [Google Scholar] [CrossRef]
- Eames, B.A.; Meulemans Madeiros, D.; Adameyko, I. Evolving Neural Crest Cells; CRC Press: Boca Raton, FL, USA, 2020. [Google Scholar]
- Kotov, A.; Alkobtawi, M.; Seal, S.; Kappès, V.; Medina Ruiz, S.; Arbès, H.; Harland, R.; Peshkin, L.; Monsoro-Burq, A.H. From neural border to migratory stage: A comprehensive single cell roadmap of the timing and regulatory logic driving cranial and vagal neural crest emergence. bioRxiv 2022. [Google Scholar] [CrossRef]
- Williams, R.M.; Lukoseviciute, M.; Sauka-Spengler, T.; Bronner, M.E. Single-cell atlas of early chick development reveals gradual segregation of neural crest lineage from the neural plate border during neurulation. eLife 2022, 11, e74464. [Google Scholar] [CrossRef]
- Rossi, C.C.; Hernandez-Lagunas, L.; Zhang, C.; Choi, I.F.; Kwok, L.; Klymkowsky, M.; Artinger, K.B. Rohon-Beard sensory neurons are induced by BMP4 expressing non-neural ectoderm in Xenopus laevis. Dev. Biol. 2008, 314, 351–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoes-Costa, M.; Bronner, M.E. Insights into neural crest development and evolution from genomic analysis. Genome Res. 2013, 23, 1069–1080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, D.; Rothstein, M.; Azambuja, A.P.; Simoes-Costa, M. Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. eLife 2018, 7, e40556. [Google Scholar] [CrossRef] [PubMed]
- Gillis, J.A.; Alsema, E.C.; Criswell, K.E. Trunk neural crest origin of dermal denticles in a cartilaginous fish. Proc. Natl. Acad. Sci. USA 2017, 114, 13200–13205. [Google Scholar] [CrossRef] [Green Version]
- Calloni, G.W.; Glavieux-Pardanaud, C.; Le Douarin, N.M.; Dupin, E. Sonic Hedgehog promotes the development of multipotent neural crest progenitors endowed with both mesenchymal and neural potentials. Proc. Natl. Acad. Sci. USA 2007, 104, 19879–19884. [Google Scholar] [CrossRef] [Green Version]
- Coelho-Aguiar, J.M.; Le Douarin, N.M.; Dupin, E. Environmental factors unveil dormant developmental capacities in multipotent progenitors of the trunk neural crest. Dev. Biol. 2013, 384, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Martik, M.L.; Gandhi, S.; Uy, B.R.; Gillis, J.A.; Green, S.A.; Simoes-Costa, M.; Bronner, M.E. Evolution of the new head by gradual acquisition of neural crest regulatory circuits. Nature 2019, 574, 675–678. [Google Scholar] [CrossRef] [Green Version]
- Stundl, J.; Bertucci, P.Y.; Lauri, A.; Arendt, D.; Bronner, M.E. Evolution of new cell types at the lateral neural border. Curr. Top. Dev. Biol. 2021, 141, 173–205. [Google Scholar] [PubMed]
- Stolfi, A.; Ryan, K.; Meinertzhagen, I.A.; Christiaen, L. Migratory neuronal progenitors arise from the neural plate borders in tunicates. Nature 2015, 527, 371–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, R.; Hazbun, A.; Chen, K.; Cao, C.; Levine, M.; Horie, T. Shared evolutionary origin of vertebrate neural crest and cranial placodes. Nature 2018, 560, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Abitua, P.B.; Wagner, E.; Navarrete, I.A.; Levine, M. Identification of a rudimentary neural crest in a non-vertebrate chordate. Nature 2012, 492, 104–107. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhao, D.; Horie, T.; Chen, G.; Bao, H.; Chen, S.; Liu, W.; Horie, R.; Liang, T.; Dong, B.; et al. Conserved gene regulatory module specifies lateral neural borders across bilaterians. Proc. Natl. Acad. Sci. USA 2017, 114, E6352–E6360. [Google Scholar] [CrossRef] [Green Version]
- Williams, R.M.; Candido-Ferreira, I.; Repapi, E.; Gavriouchkina, D.; Senanayake, U.; Ling, I.T.C.; Telenius, J.; Taylor, S.; Hughes, J.; Sauka-Spengler, T. Reconstruction of the Global Neural Crest Gene Regulatory Network In Vivo. Dev. Cell. 2019, 51, 255–276.e7. [Google Scholar] [CrossRef] [Green Version]
- Hockman, D.; Chong-Morrison, V.; Green, S.A.; Gavriouchkina, D.; Candido-Ferreira, I.; Ling, I.T.C.; Williams, R.M.; Amemiya, C.T.; Smith, J.J.; Bronner, M.E.; et al. A genome-wide assessment of the ancestral neural crest gene regulatory network. Nat. Commun. 2019, 10, 4689. [Google Scholar] [CrossRef] [Green Version]
- Green, S.A.; Uy, B.R.; Bronner, M.E. Ancient evolutionary origin of vertebrate enteric neurons from trunk-derived neural crest. Nature 2017, 544, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simoes-Costa, M.; Bronner, M.E. Reprogramming of avian neural crest axial identity and cell fate. Science 2016, 352, 1570–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandhi, S.; Ezin, M.; Bronner, M.E. Reprogramming Axial Level Identity to Rescue Neural-Crest-Related Congenital Heart Defects. Dev. Cell. 2020, 53, 300–315.e4. [Google Scholar] [CrossRef] [PubMed]
- Square, T.A.; Jandzik, D.; Massey, J.L.; Romášek, M.; Stein, H.P.; Hansen, A.W.; Purkayastha, A.; Cattell, M.V.; Medeiros, D.M. Evolution of the endothelin pathway drove neural crest cell diversification. Nature 2020, 585, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Jandzik, D.; Garnett, A.T.; Square, T.A.; Cattell, M.V.; Yu, J.K.; Medeiros, D.M. Evolution of the new vertebrate head by co-option of an ancient chordate skeletal tissue. Nature 2015, 518, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Albertson, R.C.; Streelman, J.T.; Kocher, T.D.; Yelick, P.C. Integration and evolution of the cichlid mandible: The molecular basis of alternate feeding strategies. Proc. Natl. Acad. Sci. USA 2005, 102, 16287–16292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parsons, K.J.; Trent Taylor, A.; Powder, K.E.; Albertson, R.C. Wnt signalling underlies the evolution of new phenotypes and craniofacial variability in Lake Malawi cichlids. Nat. Commun. 2014, 5, 3629. [Google Scholar] [CrossRef] [Green Version]
- Navon, D.; Male, I.; Tetrault, E.R.; Aaronson, B.; Karlstrom, R.O.; Albertson, R.C. Hedgehog signaling is necessary and sufficient to mediate craniofacial plasticity in teleosts. Proc. Natl. Acad. Sci. USA 2020, 117, 19321–19327. [Google Scholar] [CrossRef]
- Abzhanov, A.; Protas, M.; Grant, B.R.; Grant, P.R.; Tabin, C.J. Bmp4 and morphological variation of beaks in Darwin’s finches. Science 2004, 305, 1462–1465. [Google Scholar] [CrossRef] [Green Version]
- Abzhanov, A.; Kuo, W.P.; Hartmann, C.; Grant, B.R.; Grant, P.R.; Tabin, C.J. The calmodulin pathway and evolution of elongated beak morphology in Darwin’s finches. Nature 2006, 442, 563–567. [Google Scholar] [CrossRef]
- Conith, A.J.; Albertson, R.C. The cichlid oral and pharyngeal jaws are evolutionarily and genetically coupled. Nat. Commun. 2021, 12, 5477. [Google Scholar] [CrossRef]
- Xie, M.; Kamenev, D.; Kaucka, M.; Kastriti, M.E.; Zhou, B.; Artemov, A.V.; Storer, M.; Fried, K.; Adameyko, I.; Dyachuk, V.; et al. Schwann cell precursors contribute to skeletal formation during embryonic development in mice and zebrafish. Proc. Natl. Acad. Sci. USA 2019, 116, 15068–15073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaucka, M.; Ivashkin, E.; Gyllborg, D.; Zikmund, T.; Tesarova, M.; Kaiser, J.; Xie, M.; Petersen, J.; Pachnis, V.; Nicolis, S.K.; et al. Analysis of neural crest-derived clones reveals novel aspects of facial development. Sci. Adv. 2016, 2, e1600060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamouri, F.; Zhang, W.; Aujard, I.; Le Saux, T.; Ducos, B.; Vriz, S.; Jullien, L.; Bensimon, D. Optical control of protein activity and gene expression by photoactivation of caged cyclofen. Methods Enzymol. 2019, 624, 1–23. [Google Scholar] [PubMed]
- Zhang, W.; Hamouri, F.; Feng, Z.; Aujard, I.; Ducos, B.; Ye, S.; Weiss, S.; Volovitch, M.; Vriz, S.; Jullien, L.; et al. Control of Protein Activity and Gene Expression by Cyclofen-OH Uncaging. ChemBioChem 2018, 19, 1232–1238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Nam, S.; Hamouri, F.; Aujard, I.; Ducos, B.; Vriz, S.; Volovitch, M.; Jullien, L.; Lin, S.; Weiss, S.; et al. Optical Control of Tumor Induction in the Zebrafish. Sci. Rep. 2017, 7, 9195. [Google Scholar] [CrossRef] [Green Version]
- Sinha, D.K.; Neveu, P.; Gagey, N.; Aujard, I.; Le Saux, T.; Rampon, C.; Gauron, C.; Kawakami, K.; Leucht, C.; Bally-Cuif, L.; et al. Photoactivation of the CreER T2 recombinase for conditional site-specific recombination with high spatiotemporal resolution. Zebrafish 2010, 7, 199–204. [Google Scholar] [CrossRef]
- Xu, L.; Feng, Z.; Sinha, D.; Ducos, B.; Ebenstein, Y.; Tadmor, A.D.; Gauron, C.; Le Saux, T.; Lin, S.; Weiss, S.; et al. Spatiotemporal manipulation of retinoic acid activity in zebrafish hindbrain development via photo-isomerization. Development 2012, 139, 3355–3362. [Google Scholar] [CrossRef] [Green Version]
- Sinha, D.K.; Neveu, P.; Gagey, N.; Aujard, I.; Benbrahim-Bouzidi, C.; Le Saux, T.; Rampon, C.; Gauron, C.; Goetz, B.; Dubruille, S.; et al. Photocontrol of protein activity in cultured cells and zebrafish with one- and two-photon illumination. ChemBioChem 2010, 11, 653–663. [Google Scholar] [CrossRef]
- Steventon, B.; Duarte, F.; Lagadec, R.; Mazan, S.; Nicolas, J.F.; Hirsinger, E. Species-specific contribution of volumetric growth and tissue convergence to posterior body elongation in vertebrates. Development 2016, 143, 1732–1741. [Google Scholar]
- Olivera-Martinez, I.; Harada, H.; Halley, P.A.; Storey, K.G. Loss of FGF-dependent mesoderm identity and rise of endogenous retinoid signalling determine cessation of body axis elongation. PLoS Biol. 2012, 10, e1001415. [Google Scholar] [CrossRef] [Green Version]
- Attardi, A.; Fulton, T.; Florescu, M.; Shah, G.; Muresan, L.; Lenz, M.O.; Lancaster, C.; Huisken, J.; van Oudenaarden, A.; Steventon, B. Neuromesodermal progenitors are a conserved source of spinal cord with divergent growth dynamics. Development 2018, 145, dev166728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuikawa, T.; Ikeda, M.; Tsuda, S.; Saito, S.; Yamasu, K. Involvement of Oct4-type transcription factor Pou5f3 in posterior spinal cord formation in zebrafish embryos. Dev. Growth Differ. 2021, 63, 306–322. [Google Scholar] [CrossRef]
- Cambray, N.; Wilson, V. Axial progenitors with extensive potency are localised to the mouse chordoneural hinge. Development 2002, 129, 4855–4866. [Google Scholar] [CrossRef]
- Tzouanacou, E.; Wegener, A.; Wymeersch, F.J.; Wilson, V.; Nicolas, J.F. Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Dev. Cell. 2009, 17, 365–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, B.L.; Steventon, B. A fishy tail: Insights into the cell and molecular biology of neuromesodermal cells from zebrafish embryos. Dev. Biol. 2022, 487, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Aires, R.; Jurberg, A.D.; Leal, F.; Nóvoa, A.; Cohn, M.J.; Mallo, M. Oct4 Is a Key Regulator of Vertebrate Trunk Length Diversity. Dev. Cell 2016, 38, 262–274. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Jimenez, E.; de Aja, J.S.; Badia-Careaga, C.; Barral, A.; Rollan, I.; Ruco, R.; Santos, E.; Tiana, M.; Victorino, J.; Sanches-Iranzo, H.; et al. Pluripotency factors regulate the onset of Hox cluster activation in the early embryo. Sci Adv. 2022, 8, eabo3583. [Google Scholar]
- Arthur, W. D’Arcy Thompson and the theory of transformations. Nat. Rev. Genet. 2006, 7, 401–406. [Google Scholar] [CrossRef]
- Thompson, D.’A.W. On Growth and Form; Cambridge University Press: Cambridge, MA, USA, 1917. [Google Scholar]
- Banavar, S.P.; Carn, E.K.; Rowghanian, P.; Stooke-Vaughan, G.; Kim, S.; Campàs, O. Mechanical control of tissue shape and morphogenetic flows during vertebrate body axis elongation. Sci. Rep. 2021, 11, 8591. [Google Scholar] [CrossRef]
- Mongera, A.; Rowghanian, P.; Gustafson, H.J.; Shelton, E.; Kealhofer, D.A.; Carn, E.K.; Serwane, F.; Lucio, A.A.; Giammona, J.; Campàs, O. A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 2018, 561, 401–405. [Google Scholar] [CrossRef]
- Zhang, W.; Scerbo, P.; Delagrange, M.; Candat, V.; Mayr, V.; Vriz, S.; Distel, M.; Ducos, B.; Bensimon, D. Fgf8 dynamics and critical slowing down may account for the temperature independence of somitogenesis. Commun. Biol. 2022, 5, 113. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz LAJr Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Friedmann-Morvinski, D.; Verma, I.M. Dedifferentiation and reprogramming: Origins of cancer stem cells. EMBO Rep. 2014, 15, 244–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Park, J.; Ahn, J.W.; Sim, J.M.; Kang, S.J.; Kim, S.; Hwang, S.J.; Han, S.H.; Sung, K.S.; Lim, J. Transcriptomic Landscape of Lower Grade Glioma Based on Age-Related Non-Silent Somatic Mutations. Curr. Oncol. 2021, 28, 2281–2295. [Google Scholar] [CrossRef]
- Polano, M.; Fabbiani, E.; Adreuzzi, E.; Cintio, F.D.; Bedon, L.; Gentilini, D.; Mongiat, M.; Ius, T.; Arcicasa, M.; Skrap, M.; et al. A New Epigenetic Model to Stratify Glioma Patients According to Their Immunosuppressive State. Cells 2021, 10, 576. [Google Scholar] [CrossRef]
- Zbinden, M.; Duquet, A.; Lorente-Trigos, A.; Ngwabyt, S.N.; Borges, I.; Ruiz, I.A.A. NANOG regulates glioma stem cells and is essential in vivo acting in a cross-functional network with GLI1 and p53. EMBO J. 2010, 29, 2659–2674. [Google Scholar] [CrossRef] [Green Version]
- He, Q.L.; Jiang, H.X.; Zhang, X.L.; Qin, S.Y. Relationship between a 7-mRNA signature of the pancreatic adenocarcinoma microenvironment and patient prognosis (a STROBE-compliant article). Medicine 2020, 99, e21287. [Google Scholar] [CrossRef]
- Tew, B.Y.; Durand, J.K.; Bryant, K.L.; Hayes, T.K.; Peng, S.; Tran, N.L.; Gooden, G.C.; Buckley, D.N.; Der, C.J.; Baldwin, A.S.; et al. Genome-wide DNA methylation analysis of KRAS mutant cell lines. Sci. Rep. 2020, 10, 10149. [Google Scholar] [CrossRef]
- Tan, P.; Xu, Y.; Du, Y.; Wu, L.; Guo, B.; Huang, S.; Zhu, J.; Li, B.; Lin, F.; Yao, L. SPOP suppresses pancreatic cancer progression by promoting the degradation of NANOG. Cell Death Dis. 2019, 10, 794. [Google Scholar] [CrossRef] [Green Version]
- Tsuyukubo, T.; Ishida, K.; Osakabe, M.; Shiomi, E.; Kato, R.; Takata, R.; Obara, W.; Sugai, T. Comprehensive analysis of somatic copy number alterations in clear cell renal cell carcinoma. Mol. Carcinog. 2020, 59, 412–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasti, A.; Mehrazma, M.; Madjd, Z.; Abolhasani, M.; Saeednejad Zanjani, L.; Asgari, M. Co-expression of Cancer Stem Cell Markers OCT4 and NANOG Predicts Poor Prognosis in Renal Cell Carcinomas. Sci. Rep. 2018, 8, 11739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Assarnia, S.; Ardalan Khales, S.; Forghanifard, M.M. Correlation between SALL4 stemness marker and bone morphogenetic protein signaling genes in esophageal squamous cell carcinoma. J. Biochem. Mol. Toxicol. 2019, 33, e22262. [Google Scholar] [CrossRef]
- Piazzolla, D.; Palla, A.R.; Pantoja, C.; Cañamero, M.; de Castro, I.P.; Ortega, S.; Gómez-López, G.; Dominguez, O.; Megías, D.; Roncador, G.; et al. Lineage-restricted function of the pluripotency factor NANOG in stratified epithelia. Nat. Commun. 2014, 5, 4226. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-García, M.P.; Lucena-Cacace, A.; Robles-Frías, M.J.; Narlik-Grassow, M.; Blanco-Aparicio, C.; Carnero, A. The role of PIM1/PIM2 kinases in tumors of the male reproductive system. Sci. Rep. 2016, 6, 38079. [Google Scholar] [CrossRef]
- Korkola, J.E.; Houldsworth, J.; Chadalavada, R.S.; Olshen, A.B.; Dobrzynski, D.; Reuter, V.E.; Bosl, G.J.; Chaganti, R.S.K. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006, 66, 820–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sainz de Aja, J.; Menchero, S.; Rollan, I.; Barral, A.; Tiana, M.; Jawaid, W.; Cossio, I.; Alvarez, A.; Carreño-Tarragona, G.; Badia-Careaga, C.; et al. The pluripotency factor NANOG controls primitive hematopoiesis and directly regulates Tal1. EMBO J. 2019, 38, e99122. [Google Scholar] [CrossRef]
- Gentner, E.; Vegi, N.M.; Mulaw, M.A.; Mandal, T.; Bamezai, S.; Claus, R.; Tasdogan, A.; Quintanilla-Martinez, L.; Grunenberg, A.; Döhner, K.; et al. VENTX induces expansion of primitive erythroid cells and contributes to the development of acute myeloid leukemia in mice. Oncotarget 2016, 7, 86889–86901. [Google Scholar] [CrossRef] [Green Version]
- Rawat, V.P.; Arseni, N.; Ahmed, F.; Mulaw, M.A.; Thoene, S.; Heilmeier, B.; Sadlon, T.; D’Andrea, R.J.; Hiddemann, W.; Bohlander, S.K.; et al. The vent-like homeobox gene VENTX promotes human myeloid differentiation and is highly expressed in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2010, 107, 16946–16951. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.D.; Wang, Y.; Zhou, P.J.; Qin, S.R.; Zhang, R.; Zhang, Y.; Xue, X.; Wang, J.; Wang, X.; Chen, H.C.; et al. The IGF2/IGF1R/Nanog Signaling Pathway Regulates the Proliferation of Acute Myeloid Leukemia Stem Cells. Front. Pharmacol. 2018, 9, 687. [Google Scholar] [CrossRef]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef]
- Hochedlinger, K.; Yamada, Y.; Beard, C.; Jaenisch, R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell 2005, 121, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villodre, E.S.; Kipper, F.C.; Pereira, M.B.; Lenz, G. Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat. Rev. 2016, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Raj Rajan, R.; Mohan, G.; Kollenchery Puthenveettil, P.; Maliekal, T.T. Markers and Reporters to Reveal the Hierarchy in Heterogeneous Cancer Stem Cells. Cell Dev. Biol. 2021, 9, 668851. [Google Scholar] [CrossRef] [PubMed]
- Jeter, C.R.; Yang, T.; Wang, J.; Chao, H.P.; Tang, D.G. Concise Review: NANOG in Cancer Stem Cells and Tumor Development: An Update and Outstanding Questions. Stem Cells 2015, 33, 2381–2390. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.M.; Liu, S.; Lu, H.; Zhang, H.; Zhang, P.J.; Gimotty, P.A.; Guerra, M.; Guo, W.; Xu, X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene 2012, 31, 4898–4911. [Google Scholar] [CrossRef] [Green Version]
- Irie, N.; Weinberger, L.; Tang, W.W.; Kobayashi, T.; Viukov, S.; Manor, Y.S.; Dietmann, S.; Hanna, J.H.; Surani, M.A. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef] [Green Version]
- Leitch, H.G.; Tang, W.W.; Surani, M.A. Primordial germ-cell development and epigenetic reprogramming in mammals. Curr. Top. Dev. Biol. 2013, 104, 149–187. [Google Scholar]
- Sabour, D.; Schöler, H.R. Reprogramming and the mammalian germline: The Weismann barrier revisited. Curr. Opin. Cell Biol. 2012, 24, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Visvader, J.E. Cells of origin in cancer. Nature 2011, 469, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Gautier, A.; Gauron, C.; Volovitch, M.; Bensimon, D.; Jullien, L.; Vriz, S. How to control proteins with light in living systems. Nat. Chem. Biol. 2014, 10, 533–541. [Google Scholar] [CrossRef] [PubMed]


| Cell Type | Cell Line | Phenotype | Author (Year) | PMID (NCBI) |
|---|---|---|---|---|
| Glioma | HS-683 | cell proliferation | Meyers RM (2017) | 29083409 |
| Glioblastoma | G549NS (patient-derived) | cell proliferation | MacLeod G (2019) | 30995489 |
| Neural Stem Cell | HF7450 (primary-derived) | cell proliferation | MacLeod G (2019) | 30995489 |
| Pancreatic Cancer | PANC-1 | response to chemicals | Ramaker RC (2021) | 34049503 |
| Pancreatic Adenocarcinoma | HPAF-2 | cell proliferation | Steinhart Z (2017) | 27869803 |
| Chronic Myeloid Leukemia | K-562 | cell proliferation | Liu J (2019) | 31316073 |
| Renal Cell Carcinoma | RCC 786-O | response to chemicals | Zou Y (2019) | 30962421 |
| Non-Small Cell Lung Adenocarcinoma | A549 | response to chemicals | Gobbi G (2019) | 31406246 |
| Non-Small Cell Lung Adenocarcinoma | A549 | cell proliferation | Gobbi G (2019) | 31406246 |
| Ovarian Cancer | TOV-21G | cell proliferation | Meyers RM (2017) | 29083409 |
| Ovarian Cancer | PEO1 | cell proliferation | Wheeler LJ (2019) | 31437751 |
| Urinary Bladder Cancer | MGH-U4 | response to chemicals | Goodspeed A (2019) | 30414698 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ducos, B.; Bensimon, D.; Scerbo, P. Vertebrate Cell Differentiation, Evolution, and Diseases: The Vertebrate-Specific Developmental Potential Guardians VENTX/NANOG and POU5/OCT4 Enter the Stage. Cells 2022, 11, 2299. https://doi.org/10.3390/cells11152299
Ducos B, Bensimon D, Scerbo P. Vertebrate Cell Differentiation, Evolution, and Diseases: The Vertebrate-Specific Developmental Potential Guardians VENTX/NANOG and POU5/OCT4 Enter the Stage. Cells. 2022; 11(15):2299. https://doi.org/10.3390/cells11152299
Chicago/Turabian StyleDucos, Bertrand, David Bensimon, and Pierluigi Scerbo. 2022. "Vertebrate Cell Differentiation, Evolution, and Diseases: The Vertebrate-Specific Developmental Potential Guardians VENTX/NANOG and POU5/OCT4 Enter the Stage" Cells 11, no. 15: 2299. https://doi.org/10.3390/cells11152299
APA StyleDucos, B., Bensimon, D., & Scerbo, P. (2022). Vertebrate Cell Differentiation, Evolution, and Diseases: The Vertebrate-Specific Developmental Potential Guardians VENTX/NANOG and POU5/OCT4 Enter the Stage. Cells, 11(15), 2299. https://doi.org/10.3390/cells11152299






