HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress
Abstract
:1. Introduction
2. Methodology
2.1. Identification and Characterization of the HSP70 Gene Family
2.2. Phylogenetic Tree and Synteny Analysis of B.rapaHSP70 Family Proteins
2.3. B.rapaHSP70 Duplicated Gene Identification and Purity Selection Analysis
2.4. Cis-Element Analysis of B.rapaHSP70 Gene Promoters and Protein Structure
2.5. Targeted miRNA Prediction and Functional Analysis
2.6. Digital Expression Analysis of B.rapaHSP70
2.7. Plant Material and Stress Conditions
2.8. RNA Extraction and qRT-PCR Analysis
3. Results
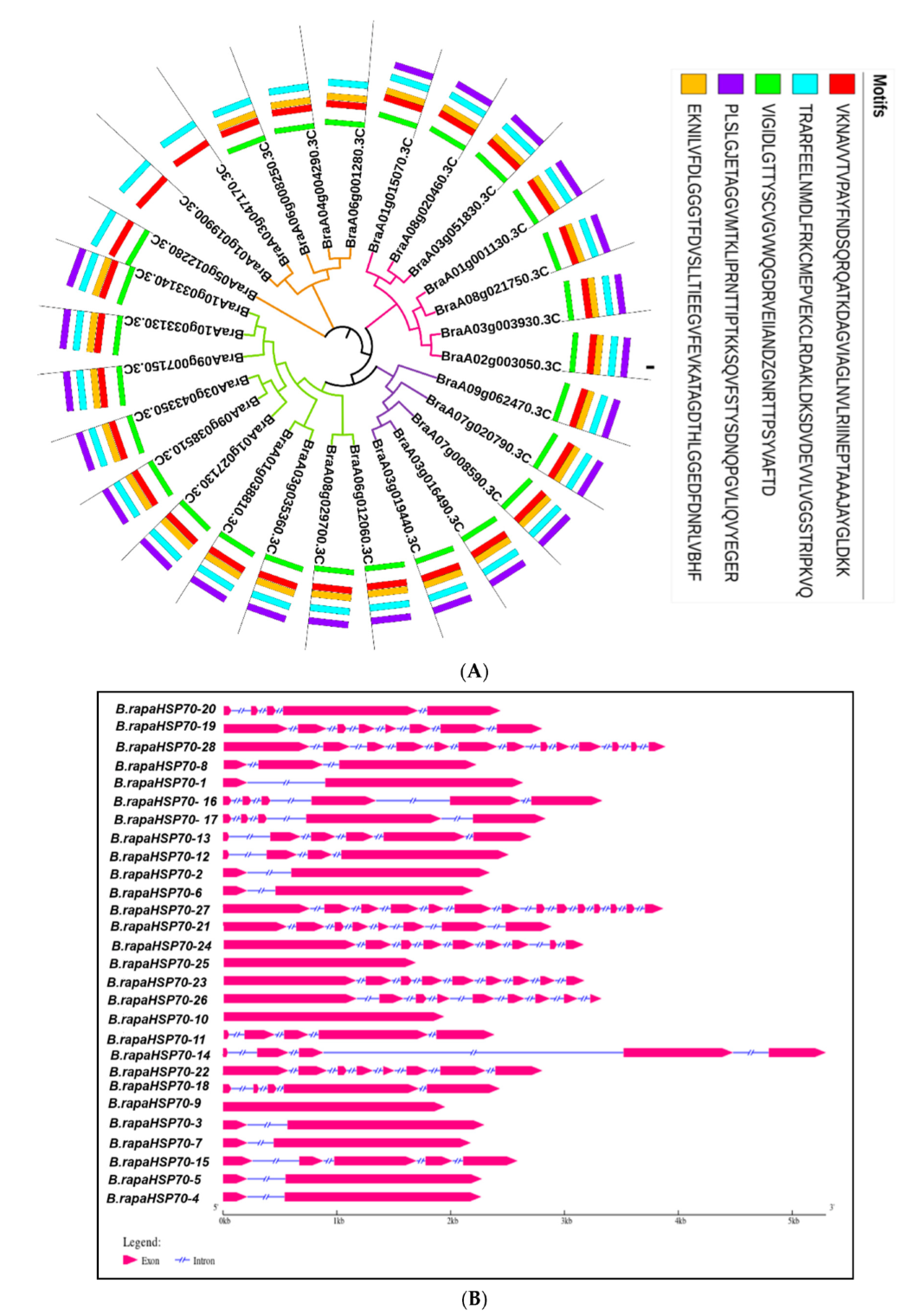
3.1. Identification and Characteristics of HSP70 Gene Family Members in B. rapa
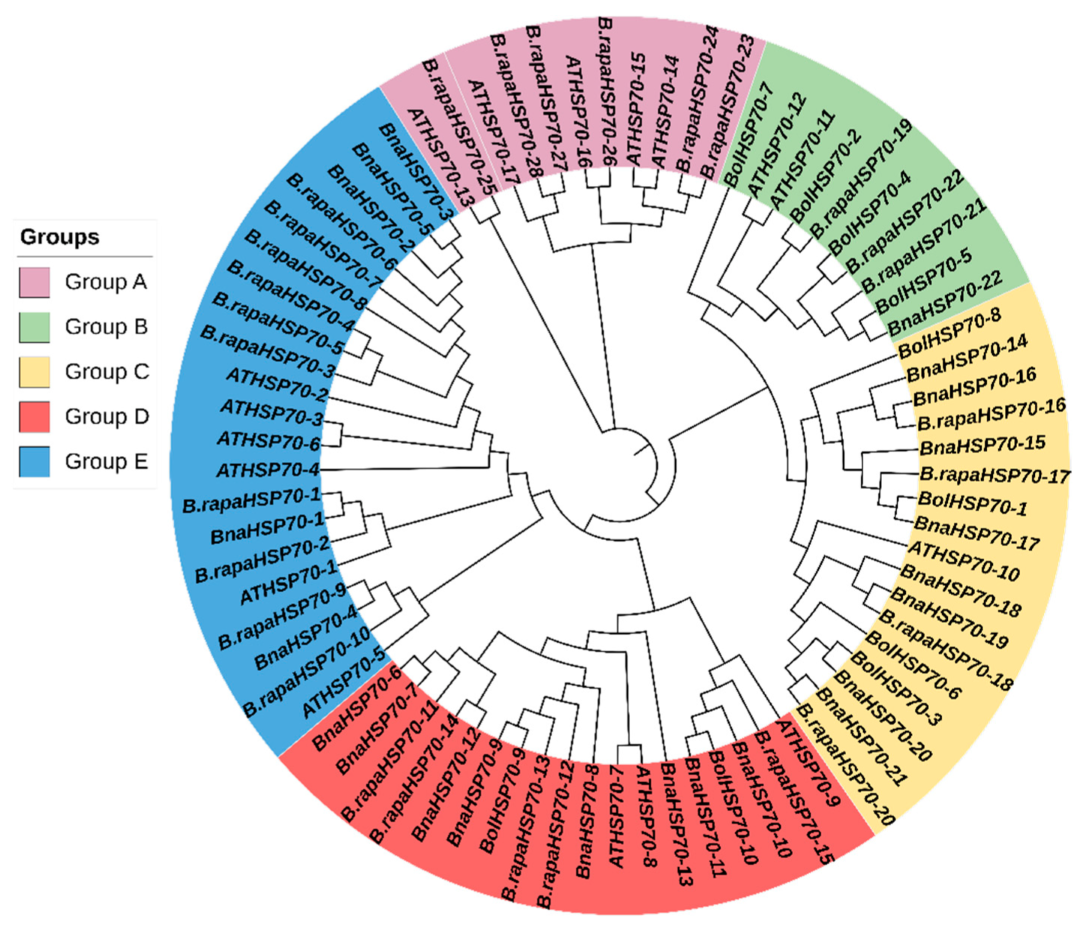
3.2. Evolutionary Relationship of B.rapaHSP70 Gene
3.3. B.rapaHSP70 Gene Duplication and Ka/Ks Analysis
3.4. B.rapaHSP70 Gene Duplication and Synteny Analysis of the B.rapaHSP70 Gene Family among Closely Related Species
3.5. Cis-Element Analysis in Promoter Regions of B.rapaHSP70 and Their Distribution
3.6. Prediction of the 3D Structures of B.rapaHSP70s
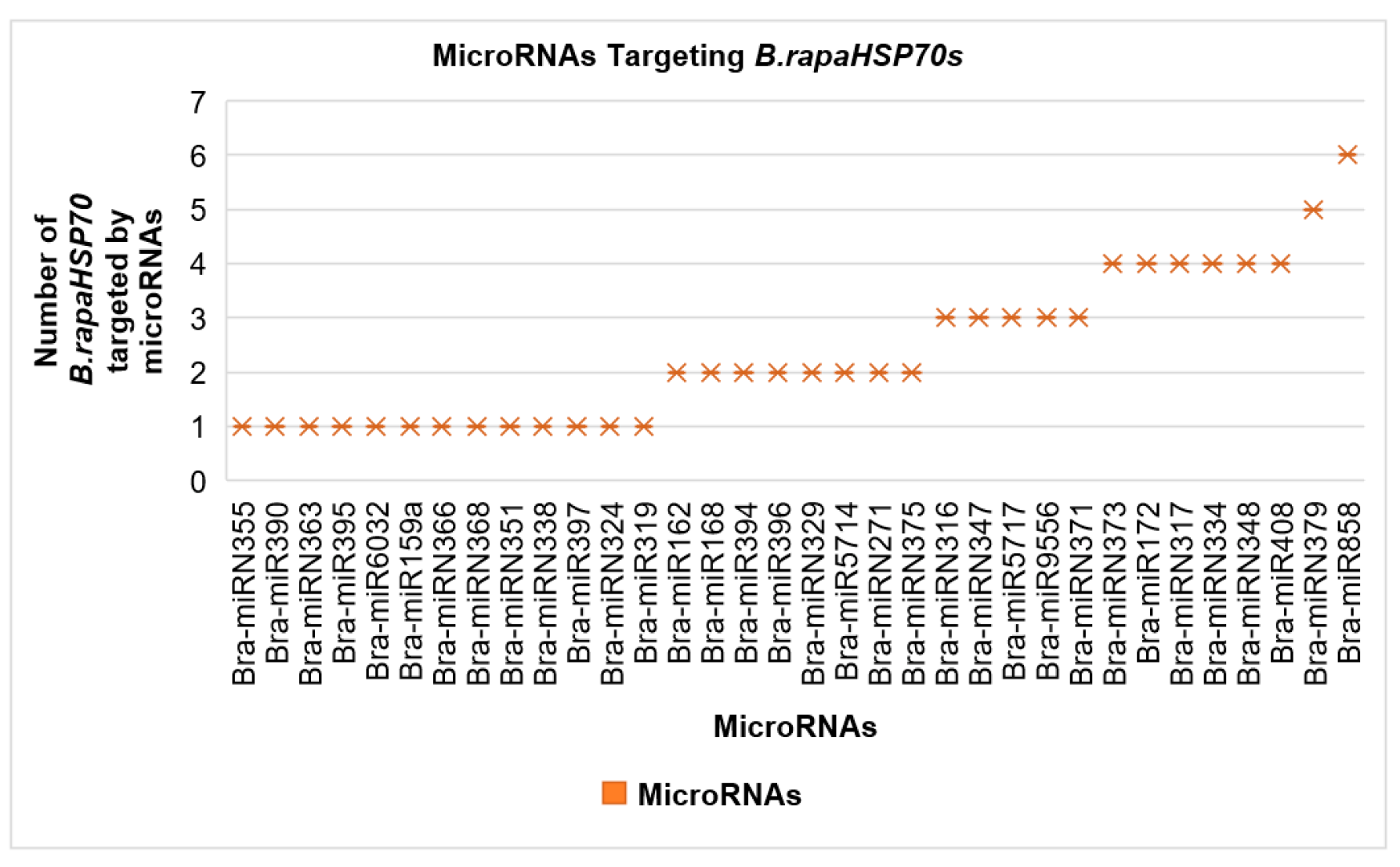
3.7. Genome-Wide Analysis of miRNA-Associated B.rapaHSP70 Genes
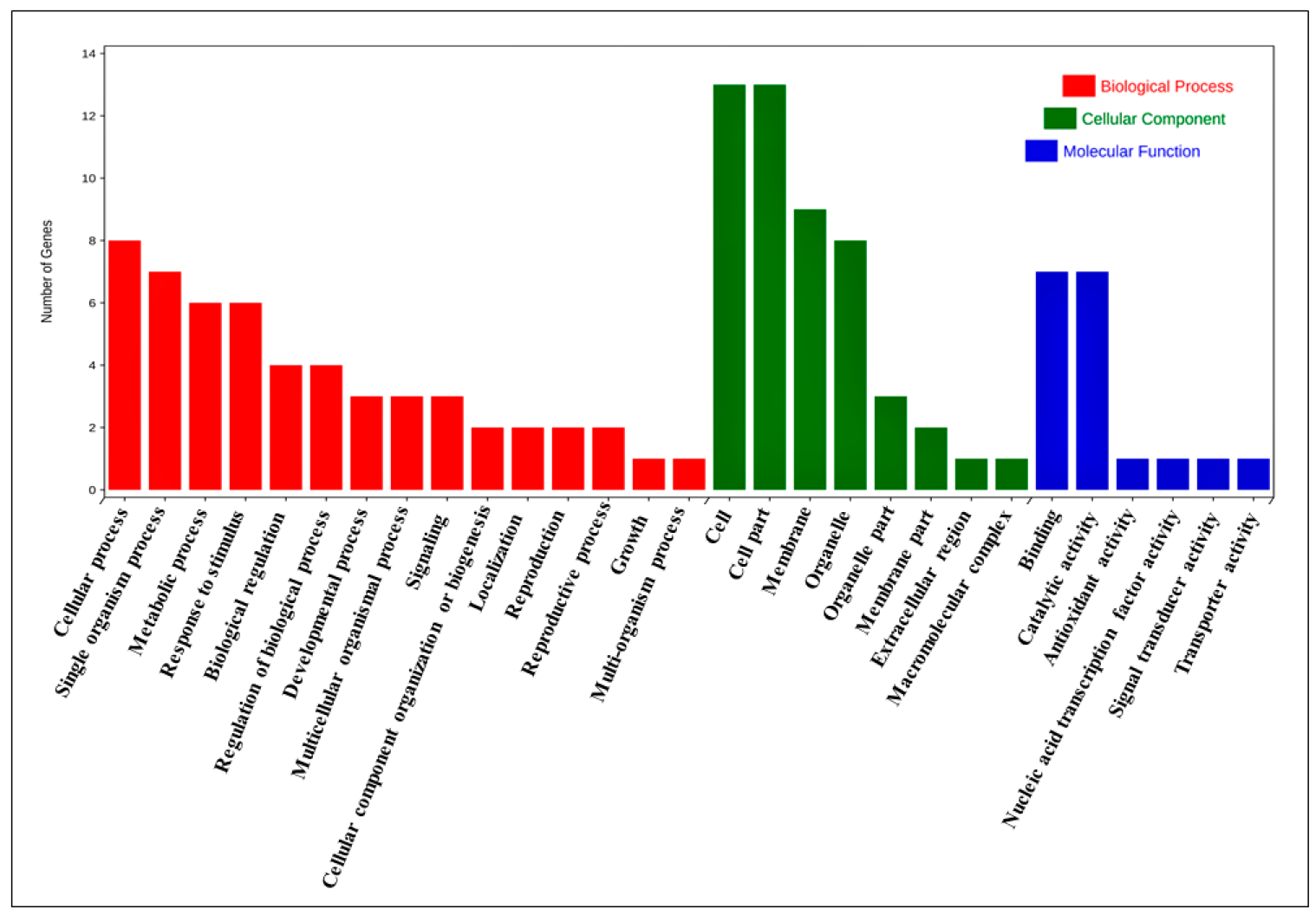
3.8. Functional Annotation Analysis of B.rapaHSP70 Genes
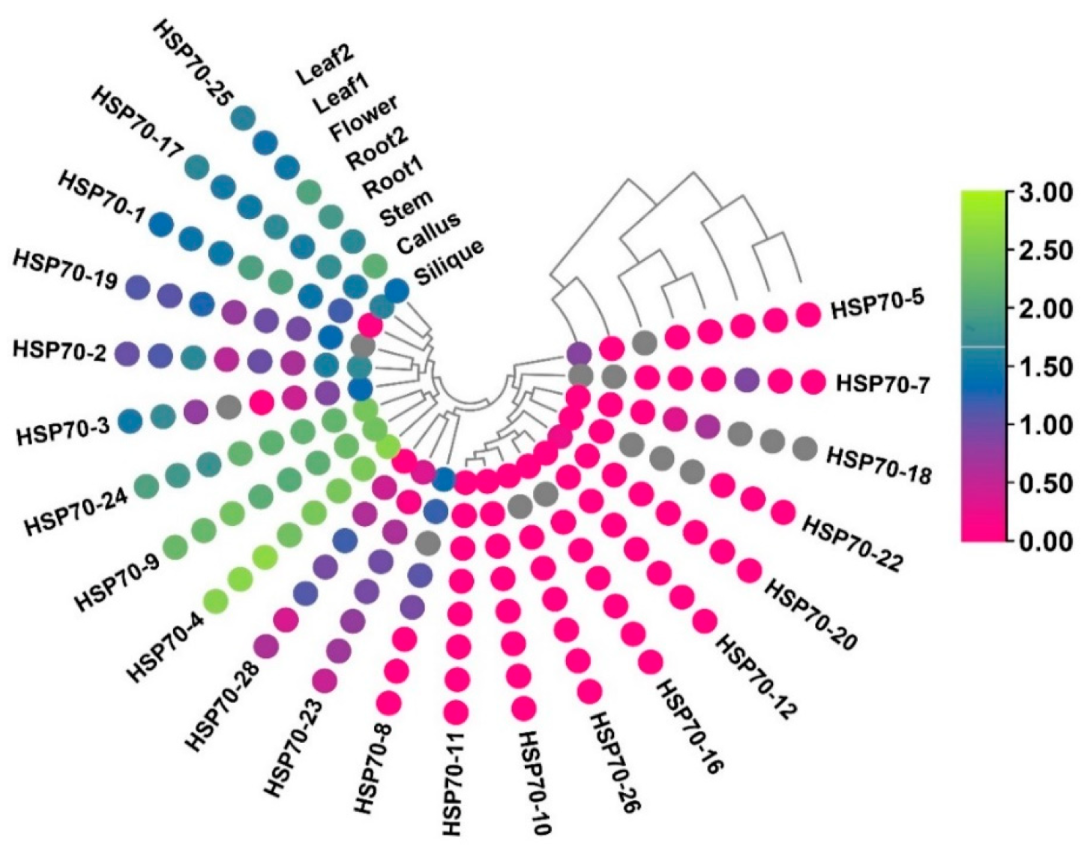
3.9. Expression Profiling of B.rapaHSP70 Genes in Various Tissues
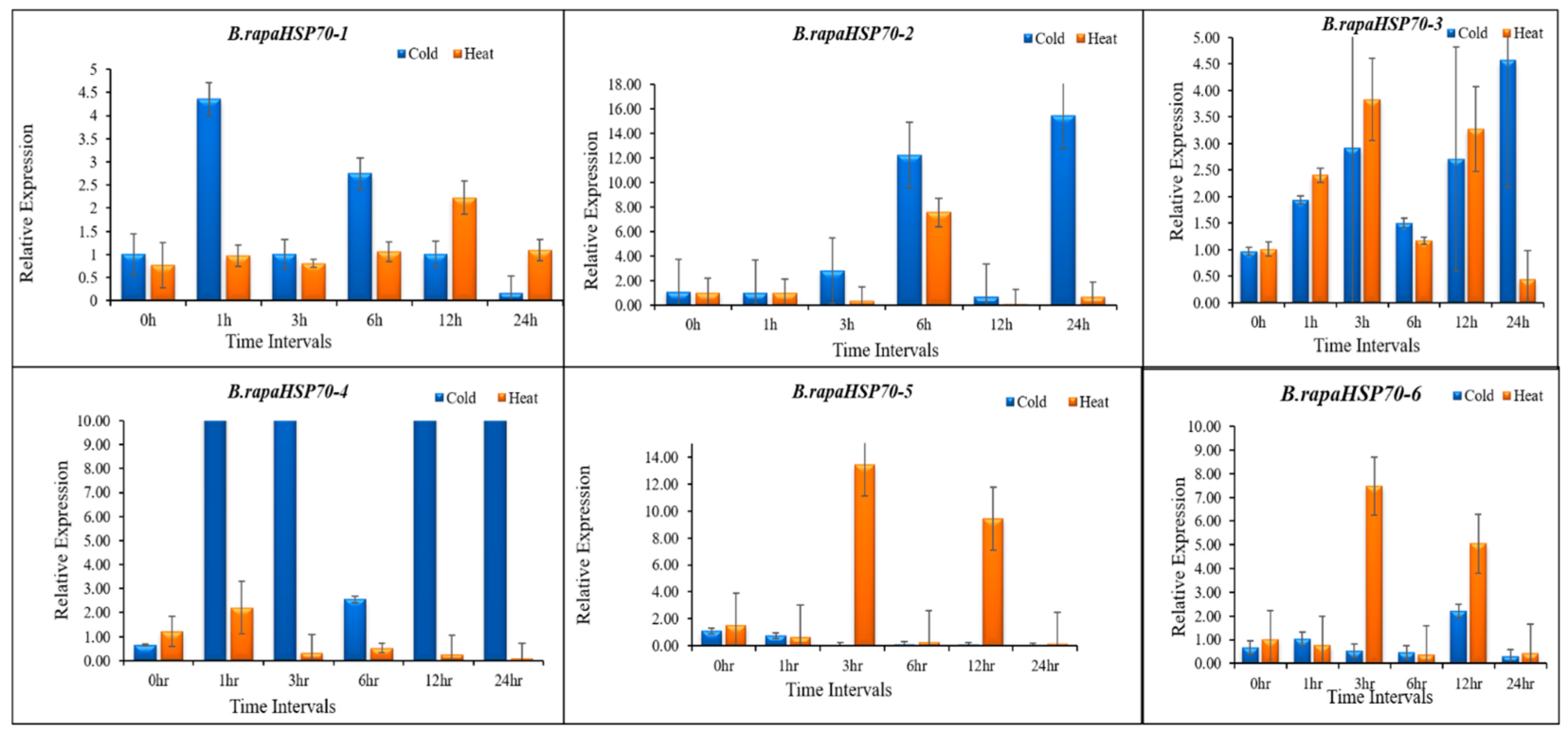
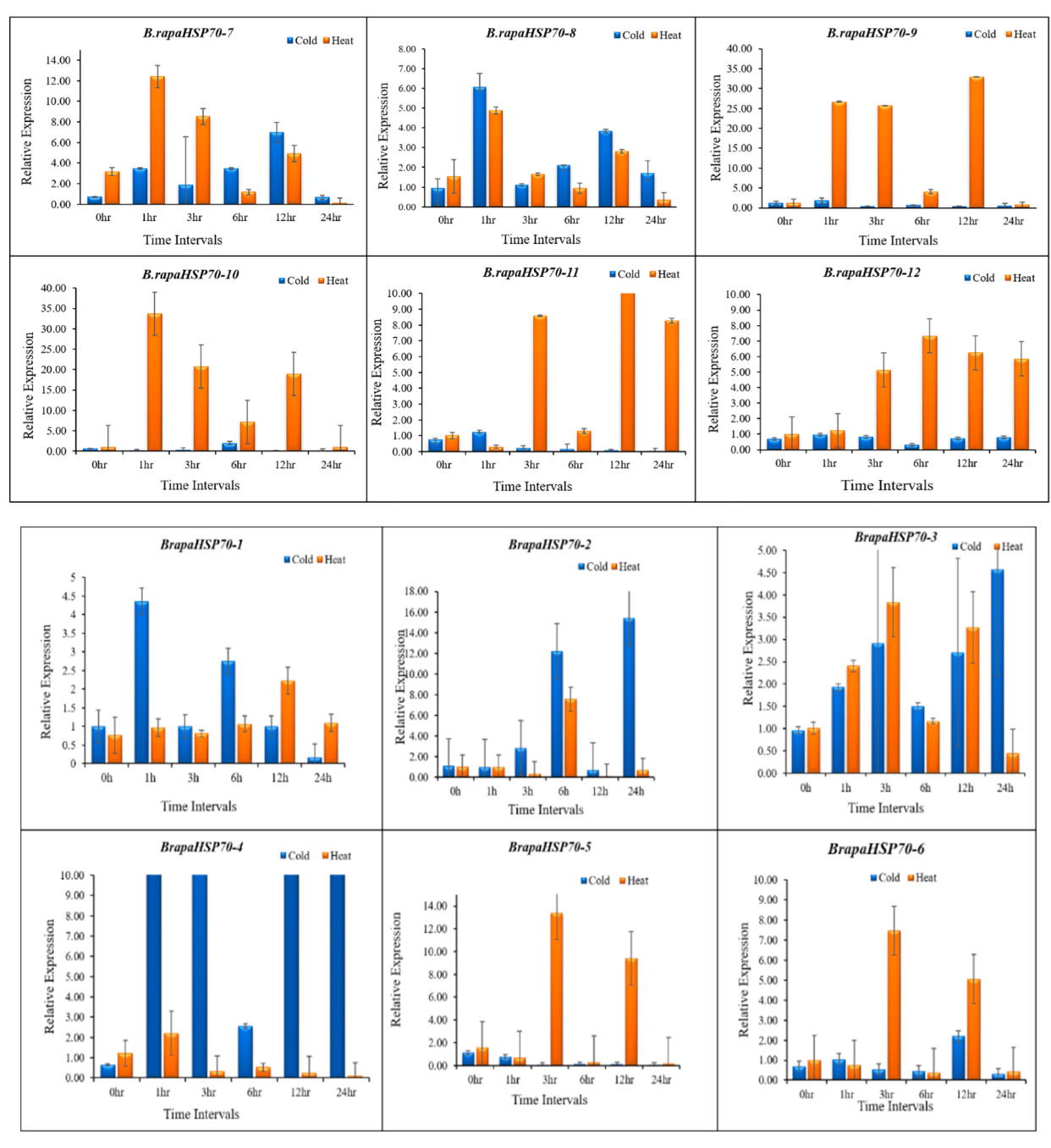
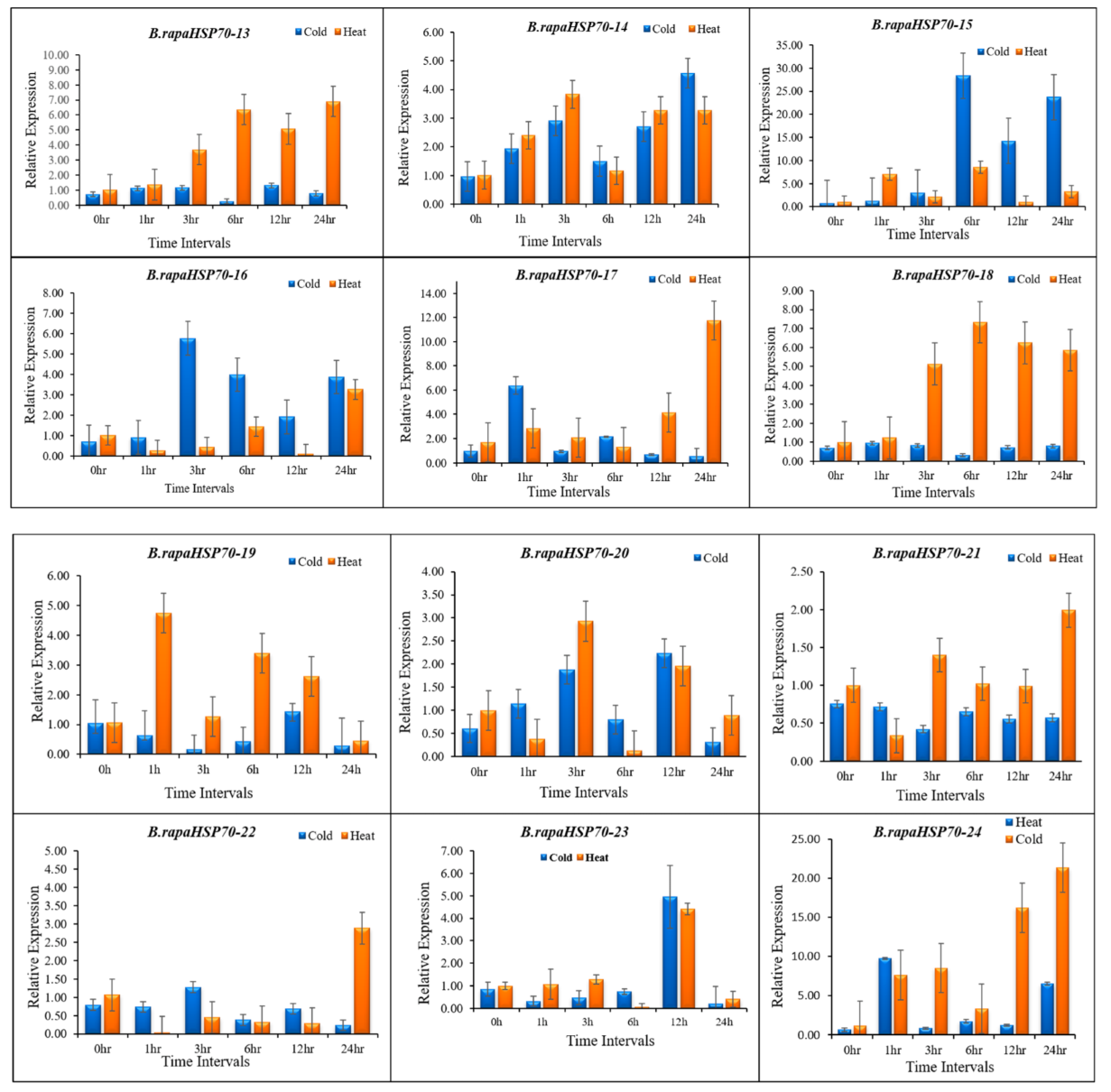
3.10. B.rapaHSP70 Gene Expression in Response to Heat and Cold Stress
4. Discussion
4.1. Characterization of the B.rapaHSP70 Gene Family in B. rapa
4.2. Promoter Analysis and Gene Ontology
4.3. miRNA: Key Players in the Regulation of Stress Responses
5. Differential Expression Analysis of B.rapaHSP70 Genes in Chinese Cabbage Leaves under Heat and Cold Stress
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Wu, P.; Zhang, B.; Hou, X. Retention, Molecular Evolution, and Expression Divergence of the Auxin/Indole Acetic Acid and Auxin Response Factor Gene Families in Brassica Rapa Shed Light on Their Evolution Patterns in Plants. Genome. Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef] [Green Version]
- Chinnusamy, V.; Zhu, J.; Zhu, J.K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Kotak, S.; Larkindale, J.; Lee, U.; von Koskull-Döring, P.; Vierling, E.; Scharf, K.-D. Complexity of the Heat Stress Response in Plants. Curr. Opin. Plant Biol. 2007, 10, 310–316. [Google Scholar] [CrossRef] [PubMed]
- Maimbo, M.; Ohnishi, K.; Hikichi, Y.; Yoshioka, H.; Kiba, A. Induction of a Small Heat Shock Protein and Its Functional Roles in Nicotiana Plants in the Defense Response against Ralstonia solanacearum. Plant Physiol. 2007, 145, 1588–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miemyk, J. The 70 kDa Stress-Related Proteins as Molecular Chaperones. Trends Plant Sci. 1997, 2, 180–187. [Google Scholar] [CrossRef]
- Radons, J. The Human HSP70 Family of Chaperones: Where Do We Stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef] [Green Version]
- Seguí-Simarro, J.; Testillano, P.; Risueño, M. Hsp70 and Hsp90 Change Their Expression and Subcellular Localization After Microspore Embryogenesis Induction in Brassica napus L. J. Struct. Biol. 2003, 142, 379–391. [Google Scholar] [CrossRef]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive Expression Profile Analysis of the Arabidopsis Hsp70 Gene Family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef] [Green Version]
- Lindquist, S.; Craig, E.A. The Heat-Shock Proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Jungkunz, I.; Link, K.; Vogel, F.; Voll, L.M.; Sonnewald, S.; Sonnewald, U. AtHsp70-15-Deficient Arabidopsis Plants are Characterized by Reduced Growth, a Constitutive Cytosolic Protein Response and Enhanced Resistance to TuMV. Plant J. 2011, 66, 983–995. [Google Scholar] [CrossRef]
- Su, P.-H.; Li, H.-M. Arabidopsis Stromal 70-kD Heat Shock Proteins Are Essential for Plant Development and Important for Thermotolerance of Germinating Seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, L.; Liang, Q.; Jiang, J.; Zhang, C.; Hao, Y.; Wang, X.; Su, W. A Subclass of HSP70s Regulate Development and Abiotic Stress Responses in Arabidopsis Thaliana. J. Plant Res. 2016, 130, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Wang, H.; Lu, Y.; De Ruiter, M.; Cariaso, M.; Prins, M.; Van Tunen, A.; He, Y. Identification of Conserved and Novel MicroRNAs that are Responsive to Heat Stress in Brassica Rapa. J. Exp. Bot. 2011, 63, 1025–1038. [Google Scholar] [CrossRef]
- Lee, S.-C.; Lim, M.-H.; Kim, J.A.; Lee, S.-I.; Kim, J.S.; Jin, M.; Kwon, S.-J.; Mun, J.-H.; Kim, Y.-K.; Kim, H.U. Tran-Scriptome Analysis in Brassica Rapa Under the Abiotic Stresses Using Brassica 24K Oligo Microarray. Mol. Cells 2008, 24, 595–605. [Google Scholar] [CrossRef]
- Wang, F.; Dong, Q.; Jiang, H.; Zhu, S.; Chen, B.; Xiang, Y. Genome-Wide Analysis of the Heat Shock Transcription Factors in Populus Trichocarpa and Medicago Truncatula. Mol. Biol. Rep. 2011, 39, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, R.; Jiang, T.; Ding, P.; Gao, Y.; Tan, X.; Zhu, K. Genome-Wide Analysis and Functional Characterization of the DELLA Gene Family Associated with Stress Tolerance in B. Napus. BMC Plant Biol. 2021, 21, 1–19. [Google Scholar] [CrossRef]
- He, L.; Zhang, F.; Wu, X.; Hu, Y.; Dong, L.; Dewitte, W.; Wen, B. Genome-Wide Characterization and Expression of Two-Component System Genes in Cytokinin-Regulated Gall Formation in Zizania latifolia. Plants 2020, 9, 1409. [Google Scholar] [CrossRef]
- Mun, J.-H.; Kwon, S.-J.; Yang, T.-J.; Seol, Y.-J.; Jin, M.; Kim, J.-A.; Lim, M.-H.; Kim, J.S.; Baek, S.; Choi, B.-S.; et al. Genome-Wide Comparative Analysis of the Brassica Rapa Gene Space Reveals Genome Shrinkage and Differential Loss of Duplicated Genes after Whole Genome Triplication. Genome Biol. 2009, 10, R111. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Bowers, J.E.; Wang, X.; Ming, R.; Alam, M.; Paterson, A.H. Synteny and Collinearity in Plant Genomes. Science 2008, 320, 486–488. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.; Kushwaha, H.R.; Kumar, K.; Verma, P.K. Comparative Structural Modeling of a Monothiol GRX from Chickpea: Insight in Iron–Sulfur Cluster Assembly. Int. J. Biol. Macromol. 2012, 51, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive Analysis of RNA-Seq Data Reveals the Complexity of the Transcriptome in Brassica Rapa. BMC Genom. 2013, 14, 689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, X.; Liu, G.; Duan, W.; Liu, T.; Huang, Z.; Ren, J.; Li, Y.; Hou, X. Genome-Wide Identification, Classification and Expression Analysis of the Heat Shock Transcription Factor Family in Chinese Cabbage. Mol. Genet. Genom. 2014, 289, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Guéguen, L.; Duret, L. Unbiased Estimate of Synonymous and Nonsynonymous Substitution Rates with Nonstationary Base Composition. Mol. Biol. Evol. 2017, 35, 734–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbas, M.; Li, Y.; Elbaiomy, R.G.; Yan, K.; Ragauskas, A.J.; Yadav, V.; Soaud, S.A.; Islam, M.; Saleem, N.; Noor, Z.; et al. Genome-Wide Analysis and Expression Profiling of SlHsp70 Gene Family in Solanum lycopersicum Revealed Higher Expression of SlHsp70-11 in Roots under Cd2+ Stress. Front. Biosci. 2022, 27, 186. [Google Scholar] [CrossRef]
- Chaudhary, R.; Baranwal, V.K.; Kumar, R.; Sircar, D.; Chauhan, H. Genome-Wide Identification and Expression Analysis of Hsp70, Hsp90, and Hsp100 Heat Shock Protein Genes in Barley Under Stress Conditions and Reproductive Development. Funct. Integr. Genom. 2019, 19, 1007–1022. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, T.; Liu, Y.; Li, Y.; Wang, M.; Zhu, B.; Liao, D.; Yun, T.; Huang, W.; Zhang, W.; et al. Pumpkin (Cucurbita moschata) HSP20 Gene Family Identification and Expression Under Heat Stress. Front. Genet. 2021, 12, 753953. [Google Scholar] [CrossRef]
- Liang, Z.; Li, M.; Liu, Z.; Wang, J. Genome-Wide Identification and Characterization of the Hsp70 Gene Family in Allo-polyploid Rapeseed (Brassica napus L.) Compared with Its Diploid Progenitors. PeerJ. 2019, 7, e7511. [Google Scholar] [CrossRef] [Green Version]
- Song, Z.; Pan, F.; Lou, X.; Wang, D.; Yang, C.; Zhang, B.; Zhang, H. Genome-Wide Identification and Characterization of Hsp70 Gene Family in Nicotiana Tabacum. Mol. Biol. Rep. 2019, 46, 1941–1954. [Google Scholar] [CrossRef]
- Su, H.; Xing, M.; Liu, X.; Fang, Z.; Yang, L.; Zhuang, M.; Zhang, Y.; Wang, Y.; Lv, H. Genome-Wide Analysis of HSP70 Family Genes in Cabbage (Brassica oleracea var. capitata) Reveals Their Involvement in Floral Development. BMC Genom. 2019, 20, 369. [Google Scholar] [CrossRef] [PubMed]
- Jeffares, D.C.; Penkett, C.J.; Bähler, J. Rapidly Regulated Genes are Intron Poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, Y.; Chen, D.; Liu, D.; Hu, M.; Dong, J.; Zhang, X.; Song, L.; Shen, F. The Catalase Gene Family in Cotton: Genome-Wide Characterization and Bioinformatics Analysis. Cells 2019, 8, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.P.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica Oleracea Genome Reveals the Asymmetrical Evolution of Polyploid Genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The Roles of Segmental and Tandem Gene Duplication in the Evolution of Large Gene Families in Arabidopsis Thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Hu, J.; Gao, C.; Chen, G.; Wang, B.; Lin, C.; Song, L.; Ding, Y.; Zhou, G. Genome-Wide Analysis of Long Non-Coding RNAs Unveils the Regulatory Roles in the Heat Tolerance of Chinese Cabbage (Brassica rapa ssp. Chinensis). Sci. Rep. 2019, 9, 5002. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yu, X.; Wang, H.; Lu, Y.-Z.; De Ruiter, M.; Prins, M.; He, Y.-K. A Novel Class of Heat-Responsive Small RNAs Derived from the Chloroplast Genome of Chinese Cabbage (Brassica rapa). BMC Genom. 2011, 12, 289. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhu, X.; Chen, X.; Song, C.; Zou, Z.; Wang, Y.; Wang, M.; Fang, W.; Li, X. Identification and Characterization of Cold-Responsive MicroRNAs in Tea Plant (Camellia sinensis) and Their Targets Using High-Throughput Sequencing and Degradome Analysis. BMC Plant Biol. 2014, 14, 271. [Google Scholar] [CrossRef] [Green Version]
- Neta-Sharir, I.; Isaacson, T.; Lurie, S.; Weiss, D. Dual Role for Tomato Heat Shock Protein 21: Protecting Photosystem II from Oxidative Stress and Promoting Color Changes during Fruit Maturation. Plant Cell 2005, 17, 1829–1838. [Google Scholar] [CrossRef] [Green Version]
- Waters, E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013, 64, 391–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Transcript ID | Gene Name | Chr Position | Location Start–End | Strand | CDS(BP) | Instability Index | Protein Length (A.A) | Protein MW (KDa) | PI | GRAVY | Intron, Exon | Sub-Cellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BraA01g038810.3C | B.rapaHSP70-1 | A01 | 26269513–26272150 | Positive | 1953 | 32.26 | 650 | 71.27 | 5.13 | −0.429 | 1,2 | Cytoplasm |
| BraA03g035360.3C | B.rapaHSP70-2 | A03 | 17438369–17440713 | Negative | 1958 | 33.17 | 652 | 71.38 | 5.13 | −0.443 | 1,2 | Cytoplasm |
| BraA09g007150.3C | B.rapaHSP70-3 | A09 | 4091084–4093380 | Negative | 1944 | 36.12 | 647 | 70.94 | 5.04 | −0.401 | 1,2 | Cytoplasm |
| BraA10g033140.3C | B.rapaHSP70-4 | A10 | 20445814–20448082 | positive | 1941 | 36.2 | 646 | 70.87 | 5.07 | −0.396 | 1,2 | Cytoplasm |
| BraA10g033130.3C | B.rapaHSP70-5 | A10 | 20434824–20437099 | Negative | 1941 | 36.2 | 646 | 70.87 | 5.07 | −0.396 | 1,2 | Cytoplasm |
| BraA03g043350.3C | B.rapaHSP70-6 | A03 | 21842760–21844958 | Positive | 1953 | 34.3 | 650 | 71.18 | 5.07 | −0.419 | 1,2 | Cytoplasm |
| BraA09g038510.3C | B.rapaHSP70-7 | A09 | 30333133–30335310 | Positive | 1950 | 35.5 | 649 | 71.05 | 5.02 | −0.416 | 1,2 | Cytoplasm |
| BraA01g027130.3C | B.rapaHSP70-8 | A01 | 17439040–17441266 | Negative | 1986 | 36.31 | 661 | 72.34 | 5.11 | −0.407 | 2,3 | Cytoplasm |
| BraA08g029700.3C | B.rapaHSP70-9 | A08 | 20365638–20367590 | Positive | 1953 | 35.47 | 650 | 71.34 | 5.28 | −0.434 | 0,1 | Cytoplasm |
| BraA06g012060.3C | B.rapaHSP70-10 | A06 | 6488327–6490270 | Negative | 1944 | 35.51 | 647 | 71.04 | 5.48 | −0.408 | 0,1 | Cytoplasm |
| BraA07g008590.3C | B.rapaHSP70-11 | A07 | 8612041–8614425 | Negative | 2010 | 30 | 669 | 73.84 | 5.08 | −0.477 | 4,5 | Endoplasmic Reticulum |
| BraA03g019440.3C | B.rapaHSP70-12 | A03 | 9196468–9198979 | Positive | 2010 | 27.96 | 669 | 73.62 | 5.08 | −0.464 | 3,4 | Endoplasmic Reticulum |
| BraA03g016490.3C | B.rapaHSP70-13 | A03 | 7627325–7630035 | Negative | 2010 | 27.79 | 669 | 73.63 | 5.08 | −0.468 | 5,6 | Endoplasmic Reticulum |
| BraA07g020790.3C | B.rapaHSP70-14 | A07 | 17199507–17204802 | Negative | 1998 | 30.99 | 665 | 73.49 | 5.11 | −0.466 | 4,5 | Endoplasmic Reticulum |
| BraA09g062470.3C | B.rapaHSP70-15 | A09 | 43345497–43348084 | Positive | 1911 | 32.8 | 636 | 70.55 | 4.94 | −0.48 | 4,5 | Endoplasmic Reticulum |
| BraA02g003050.3C | B.rapaHSP70-16 | A02 | 1492088–1495420 | Positive | 2043 | 34.42 | 680 | 72.87 | 5.82 | −0.335 | 5,6 | Mitochondrial |
| BraA03g003930.3C | B.rapaHSP70-17 | A03 | 1680387–1683221 | Positive | 2046 | 33.94 | 681 | 72.67 | 5.55 | −0.336 | 4,5 | Mitochondrial |
| BraA08g021750.3C | B.rapaHSP70-18 | A08 | 16460538–16462970 | Negative | 2031 | 36.79 | 676 | 72.48 | 5.79 | −0.281 | 4,5 | Mitochondrial |
| BraA01g015070.3C | B.rapaHSP70-19 | A01 | 7988766–7991570 | Positive | 2136 | 31.48 | 711 | 76.1 | 5.09 | −0.316 | 4,5 | Chloroplast |
| BraA01g001130.3C | B.rapaHSP70-20 | A01 | 523816–526255 | Positive | 2049 | 38.26 | 682 | 73.01 | 5.56 | −0.306 | 7,8 | Mitochondrial |
| BraA03g051830.3C | B.rapaHSP70-21 | A03 | 26797918–26800806 | Positive | 2133 | 27.71 | 710 | 75.91 | 5.27 | −0.316 | 7,8 | Chloroplast |
| BraA08g020460.3C | B.rapaHSP70-22 | A08 | 15677635-15680441 | Negative | 2154 | 28.52 | 717 | 76.42 | 5.14 | −0.313 | 7,8 | Chloroplast |
| BraA06g001280.3C | B.rapaHSP70-23 | A06 | 802745–805920 | Positive | 2457 | 39.54 | 818 | 90.29 | 5.17 | −0.402 | 8,9 | Cytoplasmic |
| BraA04g004290.3C | B.rapaHSP70-24 | A04 | 2645163–2648333 | Negative | 2373 | 40.91 | 790 | 87.16 | 5.23 | −0.4 | 8,9 | Cytoplasmic |
| BraA05g012280.3C | B.rapaHSP70-25 | A05 | 6664594–6666288 | Negative | 1695 | 41.55 | 564 | 60.81 | 5.39 | 0.051 | 0,1 | Chloroplast |
| BraA06g008250.3C | B.rapaHSP70-26 | A06 | 4497297–4500623 | Positive | 2286 | 47.68 | 761 | 84.54 | 5.73 | −0.493 | 8,9 | Nuclear |
| BraA03g047170.3C | B.rapaHSP70-27 | A03 | 23836812-23840680 | Positive | 2631 | 42.89 | 876 | 97.59 | 5.78 | −0.483 | 13,14 | Nuclear |
| BraA01g019900.3C | B.rapaHSP70-28 | A01 | 10687460–10691347 | Negative | 2685 | 44.47 | 894 | 99.97 | 5.89 | −0.499 | 12,13 | Nuclear |
| Seq_1 | Seq_2 | Ka | Ks | Ka_Ks | T(MYA) |
|---|---|---|---|---|---|
| BraA10g033140.3C | BraA10g033130.3C | 0 | 0.0983403 | 0 | 3278010.814 |
| BraA09g038510.3C | BraA01g027130.3C | 0.0056856 | 0.2035601 | 0.027931049 | 6.785335295 |
| BraA01g038810.3C | BraA03g035360.3C | 0.0086499 | 0.9733226 | 0.008887026 | 32.44408789 |
| BraA08g029700.3C | BraA06g012060.3C | 0.0273781 | 0.7299868 | 0.037504912 | 24.33289231 |
| BraA03g019440.3C | BraA03g016490.3C | 0.0035436 | 0.2455705 | 0.014430195 | 8.185684806 |
| BraA03g051830.3C | BraA08g020460.3C | 0.0218032 | 0.5165388 | 0.042210144 | 17.21796007 |
| BraA02g003050.3C | BraA03g003930.3C | 0.0206864 | 0.3771192 | 0.054853846 | 12.57063953 |
| BraA08g021750.3C | BraA01g001130.3C | 0.0253008 | 0.4383311 | 0.057720824 | 14.61103702 |
| BraA03g047170.3C | BraA01g019900.3C | 0.0506028 | 0.315182 | 0.160551192 | 10.50606656 |
| BraA06g001280.3C | BraA04g004290.3C | 0.0160146 | 0.3383936 | 0.047325361 | 11.27978551 |
| MicroRNAs | MicroRNA Targeting B.rapaHSP70s | |||||
|---|---|---|---|---|---|---|
| Bra-miRN355 | B.rapaHSP70-3 | |||||
| Bra-miR390 | B.rapaHSP70-8 | |||||
| Bra-miRN363 | B.rapaHSP70-10 | |||||
| Bra-miR395 | B.rapaHSP70-13 | |||||
| Bra-miR6032 | B.rapaHSP70-14 | |||||
| Bra-miR159a | B.rapaHSP70-15 | |||||
| Bra-miRN366 | B.rapaHSP70-15 | |||||
| Bra-miRN368 | B.rapaHSP70-15 | |||||
| Bra-miRN351 | B.rapaHSP70-16 | |||||
| Bra-miRN338 | B.rapaHSP70-19 | |||||
| Bra-miR397 | B.rapaHSP70-23 | |||||
| Bra-miRN324 | B.rapaHSP70-25 | |||||
| Bra-miR319 | B.rapaHSP70-26 | |||||
| Bra-miR162 | B.rapaHSP70-21 | B.rapaHSP70-22 | ||||
| Bra-miR168 | B.rapaHSP70-2 | B.rapaHSP70-13 | ||||
| Bra-miR394 | B.rapaHSP70-14 | B.rapaHSP70-27 | ||||
| Bra-miR396 | B.rapaHSP70-23 | B.rapaHSP70-24 | ||||
| Bra-miRN329 | B.rapaHSP70-4 | B.rapaHSP70-5 | ||||
| Bra-miR5714 | B.rapaHSP70-18 | B.rapaHSP70-22 | ||||
| Bra-miRN271 | B.rapaHSP70-16 | B.rapaHSP70-17 | ||||
| Bra-miRN375 | B.rapaHSP70-21 | B.rapaHSP70-23 | ||||
| Bra-miRN316 | B.rapaHSP70-15 | B.rapaHSP70-23 | B.rapaHSP70-24 | |||
| Bra-miRN347 | B.rapaHSP70-6 | B.rapaHSP70-12 | B.rapaHSP70-13 | |||
| Bra-miR5717 | B.rapaHSP70-20 | B.rapaHSP70-21 | B.rapaHSP70-22 | |||
| Bra-miR9556 | B.rapaHSP70-9 | B.rapaHSP70-13 | B.rapaHSP70-15 | |||
| Bra-miRN371 | B.rapaHSP70-19 | B.rapaHSP70-21 | B.rapaHSP70-22 | |||
| Bra-miRN373 | B.rapaHSP70-1 | B.rapaHSP70-22 | B.rapaHSP70-27 | B.rapaHSP70-28 | ||
| Bra-miR172 | B.rapaHSP70-2 | B.rapaHSP70-9 | B.rapaHSP70-14 | B.rapaHSP70-23 | ||
| Bra-miRN317 | B.rapaHSP70-15 | B.rapaHSP70-19 | B.rapaHSP70-27 | B.rapaHSP70-28 | ||
| Bra-miRN334 | B.rapaHSP70-7 | B.rapaHSP70-8 | B.rapaHSP70-16 | B.rapaHSP70-17 | ||
| Bra-miRN348 | B.rapaHSP70-3 | B.rapaHSP70-4 | B.rapaHSP70-5 | B.rapaHSP70-8 | ||
| Bra-miR408 | B.rapaHSP70-6 | B.rapaHSP70-11 | B.rapaHSP70-21 | B.rapaHSP70-22 | ||
| Bra-miRN379 | B.rapaHSP70-1 | B.rapaHSP70-2 | B.rapaHSP70-25 | B.rapaHSP70-27 | B.rapaHSP70-28 | |
| Bra-miR858 | B.rapaHSP70-12 | B.rapaHSP70-13 | B.rapaHSP70-18 | B.rapaHSP70-19 | B.rapaHSP70-20 | B.rapaHSP70-22 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabusam, J.; Shi, Q.; Feng, D.; Zulfiqar, S.; Shen, S.; Ma, W.; Zhao, J. HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress. Cells 2022, 11, 2316. https://doi.org/10.3390/cells11152316
Tabusam J, Shi Q, Feng D, Zulfiqar S, Shen S, Ma W, Zhao J. HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress. Cells. 2022; 11(15):2316. https://doi.org/10.3390/cells11152316
Chicago/Turabian StyleTabusam, Javaria, Qiling Shi, Daling Feng, Sumer Zulfiqar, Shuxing Shen, Wei Ma, and Jianjun Zhao. 2022. "HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress" Cells 11, no. 15: 2316. https://doi.org/10.3390/cells11152316
APA StyleTabusam, J., Shi, Q., Feng, D., Zulfiqar, S., Shen, S., Ma, W., & Zhao, J. (2022). HSP70 Gene Family in Brassica rapa: Genome-Wide Identification, Characterization, and Expression Patterns in Response to Heat and Cold Stress. Cells, 11(15), 2316. https://doi.org/10.3390/cells11152316





