RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function
Abstract
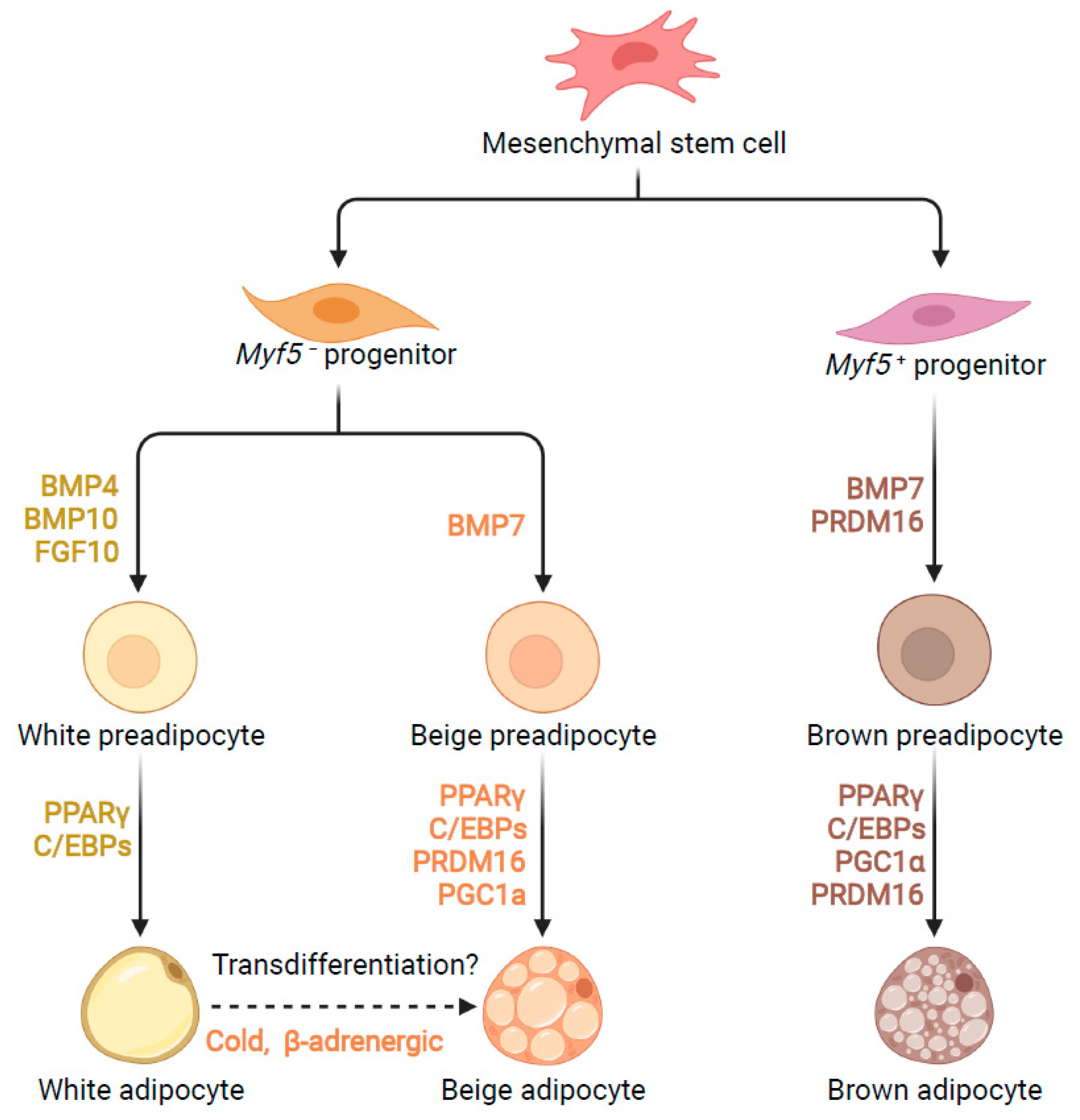
:1. Introduction
2. Transcriptional Control of Adipogenesis
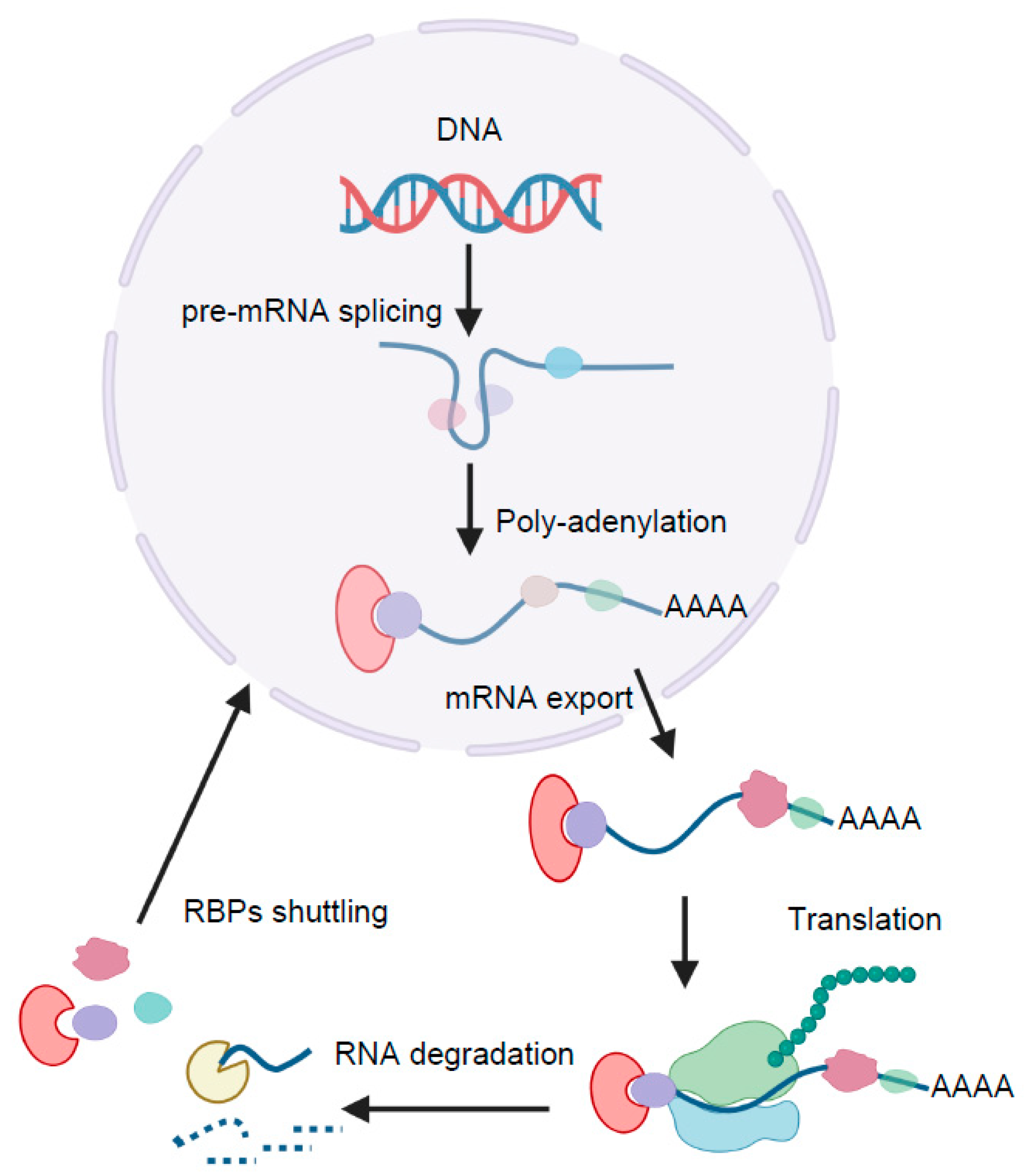
3. Post-Transcriptional Control of Adipogenesis
4. Regulation of Adipogenesis and Adipose Function by RBPs
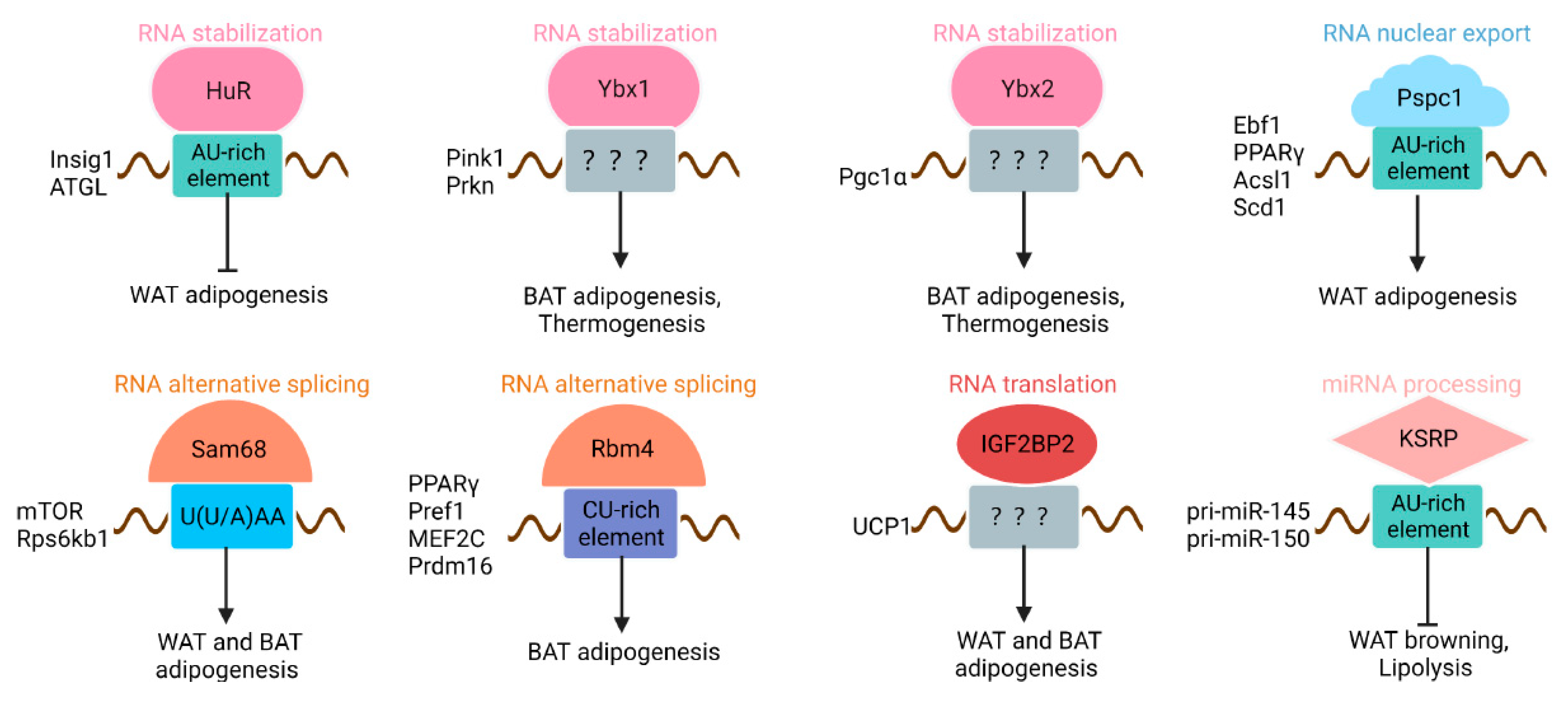
4.1. HuR
4.2. PSPC1
4.3. Sam68
4.4. RBM4
4.5. Y-Box Binding Proteins
4.6. IGF2BP2
4.7. KH-Type Splicing Regulatory Protein
5. Concluding Remarks and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luo, L.; Liu, M. Adipose Tissue in Control of Metabolism. J. Endocrinol. 2016, 231, R77–R99. [Google Scholar] [CrossRef] [Green Version]
- Scheja, L.; Heeren, J. The Endocrine Function of Adipose Tissues in Health and Cardiometabolic Disease. Nat. Rev. Endocrinol. 2019, 15, 507–524. [Google Scholar] [CrossRef] [PubMed]
- Wronska, A.; Kmiec, Z. Structural and Biochemical Characteristics of Various White Adipose Tissue Depots. Acta Physiol. Oxf. Engl. 2012, 205, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Sharma, A.K.; Wolfrum, C. Novel Insights into Adipose Tissue Heterogeneity. Rev. Endocr. Metab. Disord. 2022, 23, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Frontini, A.; Cinti, S. Distribution and Development of Brown Adipocytes in the Murine and Human Adipose Organ. Cell Metab. 2010, 11, 253–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Townsend, K.L.; Tseng, Y.-H. Brown Fat Fuel Utilization and Thermogenesis. Trends Endocrinol. Metab. 2014, 25, 168–177. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Boström, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.-H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Berry, D.C.; Graff, J.M. Distinct Cellular and Molecular Mechanisms for Β3 Adrenergic Receptor-Induced Beige Adipocyte Formation. eLife 2017, 6, e30329. [Google Scholar] [CrossRef]
- Wu, J.; Cohen, P.; Spiegelman, B.M. Adaptive Thermogenesis in Adipocytes: Is Beige the New Brown? Genes Dev. 2013, 27, 234–250. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Plutzky, J. Brown Fat and Browning for the Treatment of Obesity and Related Metabolic Disorders. Diabetes Metab. J. 2016, 40, 12–21. [Google Scholar] [CrossRef]
- Longo, M.; Zatterale, F.; Naderi, J.; Parrillo, L.; Formisano, P.; Raciti, G.A.; Beguinot, F.; Miele, C. Adipose Tissue Dysfunction as Determinant of Obesity-Associated Metabolic Complications. Int. J. Mol. Sci. 2019, 20, 2358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, A.; Kim, W.K.; Bae, K.-H. Distinction of White, Beige and Brown Adipocytes Derived from Mesenchymal Stem Cells. World J. Stem Cells 2014, 6, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Gulyaeva, O.; Dempersmier, J.; Sul, H.S. Genetic and Epigenetic Control of Adipose Development. Biochim. Biophys. Acta BBA - Mol. Cell Biol. Lipids 2019, 1864, 3–12. [Google Scholar] [CrossRef]
- Inagaki, T.; Sakai, J.; Kajimura, S. Transcriptional and Epigenetic Control of Brown and Beige Adipose Cell Fate and Function. Nat. Rev. Mol. Cell Biol. 2016, 17, 480–495. [Google Scholar] [CrossRef] [Green Version]
- Shapira, S.N.; Seale, P. Transcriptional Control of Brown and Beige Fat Development and Function. Obes. Silver Spring Md 2019, 27, 13–21. [Google Scholar] [CrossRef] [Green Version]
- Lefterova, M.I.; Lazar, M.A. New Developments in Adipogenesis. Trends Endocrinol. Metab. 2009, 20, 107–114. [Google Scholar] [CrossRef]
- Tanaka, T. Defective Adipocyte Differentiation in Mice Lacking the C/EBPbeta and/or C/EBPdelta Gene. EMBO J. 1997, 16, 7432–7443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hishida, T.; Nishizuka, M.; Osada, S.; Imagawa, M. The Role of C/EBPδ in the Early Stages of Adipogenesis. Biochimie 2009, 91, 654–657. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Scheller, E.L.; MacDougald, O.A. Adipose Tissue Stem Cells Meet Preadipocyte Commitment: Going Back to the Future. J. Lipid Res. 2012, 53, 227–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Otto, T.C.; Lane, M.D. Adipose Development: From Stem Cell to Adipocyte. Crit. Rev. Biochem. Mol. Biol. 2005, 40, 229–242. [Google Scholar] [CrossRef] [PubMed]
- Ghaben, A.L.; Scherer, P.E. Adipogenesis and Metabolic Health. Nat. Rev. Mol. Cell Biol. 2019, 20, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Sakaue, H.; Ogawa, W.; Matsumoto, M.; Kuroda, S.; Takata, M.; Sugimoto, T.; Spiegelman, B.M.; Kasuga, M. Posttranscriptional Control of Adipocyte Differentiation through Activation of Phosphoinositide 3-Kinase. J. Biol. Chem. 1998, 273, 28945–28952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.-J.; Belaghzal, H.; Hsiao, W.-Y.; Qi, J.; Bradner, J.E.; Guertin, D.A.; Sif, S.; Imbalzano, A.N. Transcriptional and Post-Transcriptional Control of Adipocyte Differentiation by Jumonji Domain-Containing Protein 6. Nucleic Acids Res. 2015, 43, 7790–7804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelaini, S.; Chan, C.; Cornelius, V.A.; Margariti, A. RNA-Binding Proteins Hold Key Roles in Function, Dysfunction, and Disease. Biology 2021, 10, 366. [Google Scholar] [CrossRef] [PubMed]
- Corley, M.; Burns, M.C.; Yeo, G.W. How RNA-Binding Proteins Interact with RNA: Molecules and Mechanisms. Mol. Cell 2020, 78, 9–29. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wu, J.; Li, Z.; Han, J.; Xia, P.; Shen, Y.; Ma, J.; Liu, X.; Zhang, J.; Yu, P. Advances in the Study of RNA-Binding Proteins in Diabetic Complications. Mol. Metab. 2022, 62, 101515. [Google Scholar] [CrossRef]
- Zhao, Y.; Mir, C.; Garcia-Mayea, Y.; Paciucci, R.; Kondoh, H.; LLeonart, M.E. RNA-Binding Proteins: Underestimated Contributors in Tumorigenesis. Semin. Cancer Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Jonas, K.; Calin, G.A.; Pichler, M. RNA-Binding Proteins as Important Regulators of Long Non-Coding RNAs in Cancer. Int. J. Mol. Sci. 2020, 21, 2969. [Google Scholar] [CrossRef] [Green Version]
- Weskamp, K.; Olwin, B.B.; Parker, R. Post-Transcriptional Regulation in Skeletal Muscle Development, Repair, and Disease. Trends Mol. Med. 2021, 27, 469–481. [Google Scholar] [CrossRef]
- Shi, D.-L.; Grifone, R. RNA-Binding Proteins in the Post-Transcriptional Control of Skeletal Muscle Development, Regeneration and Disease. Front. Cell Dev. Biol. 2021, 9, 738978. [Google Scholar] [CrossRef]
- Nussbacher, J.K.; Tabet, R.; Yeo, G.W.; Lagier-Tourenne, C. Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 2019, 102, 294–320. [Google Scholar] [CrossRef] [PubMed]
- Nutter, C.A.; Kuyumcu-Martinez, M.N. Emerging Roles of RNA-Binding Proteins in Diabetes and Their Therapeutic Potential in Diabetic Complications. WIREs RNA 2018, 9, e1459. [Google Scholar] [CrossRef] [PubMed]
- Hinman, M.N.; Lou, H. Diverse Molecular Functions of Hu Proteins. Cell. Mol. Life Sci. 2008, 65, 3168–3181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripin, N.; Boudet, J.; Duszczyk, M.M.; Hinniger, A.; Faller, M.; Krepl, M.; Gadi, A.; Schneider, R.J.; Šponer, J.; Meisner-Kober, N.C.; et al. Molecular Basis for AU-Rich Element Recognition and Dimerization by the HuR C-Terminal RRM. Proc. Natl. Acad. Sci. USA 2019, 116, 2935–2944. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.C.; Steitz, J.A. HNS, a Nuclear-Cytoplasmic Shuttling Sequence in HuR. Proc. Natl. Acad. Sci. USA 1998, 95, 15293–15298. [Google Scholar] [CrossRef] [Green Version]
- Schultz, C.W.; Preet, R.; Dhir, T.; Dixon, D.A.; Brody, J.R. Understanding and Targeting the Disease-Related RNA Binding Protein Human Antigen R (HuR). WIREs RNA 2020, 11, e1581. [Google Scholar] [CrossRef]
- Giles, K.M.; Daly, J.M.; Beveridge, D.J.; Thomson, A.M.; Voon, D.C.; Furneaux, H.M.; Jazayeri, J.A.; Leedman, P.J. The 3’-Untranslated Region of P21WAF1 MRNA Is a Composite Cis-Acting Sequence Bound by RNA-Binding Proteins from Breast Cancer Cells, Including HuR and Poly(C)-Binding Protein. J. Biol. Chem. 2003, 278, 2937–2946. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Caldwell, M.C.; Lin, S.; Furneaux, H.; Gorospe, M. HuR Regulates Cyclin A and Cyclin B1 MRNA Stability during Cell Proliferation. EMBO J. 2000, 19, 2340–2350. [Google Scholar] [CrossRef] [Green Version]
- Beauchamp, P.; Nassif, C.; Hillock, S.; van der Giessen, K.; von Roretz, C.; Jasmin, B.J.; Gallouzi, I.-E. The Cleavage of HuR Interferes with Its Transportin-2-Mediated Nuclear Import and Promotes Muscle Fiber Formation. Cell Death Differ. 2010, 17, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- von Roretz, C.; Beauchamp, P.; Di Marco, S.; Gallouzi, I.-E. HuR and Myogenesis: Being in the Right Place at the Right Time. Biochim. Biophys. Acta 2011, 1813, 1663–1667. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Gong, L.; Liu, S.; Zhang, Y.; Zhang, C.; Tian, M.; Lu, H.; Bu, P.; Yang, J.; Ouyang, C.; et al. Adipose HuR Protects against Diet-Induced Obesity and Insulin Resistance. Nat. Commun. 2019, 10, 2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siang, D.T.C.; Lim, Y.C.; Kyaw, A.M.M.; Win, K.N.; Chia, S.Y.; Degirmenci, U.; Hu, X.; Tan, B.C.; Walet, A.C.E.; Sun, L.; et al. The RNA-Binding Protein HuR Is a Negative Regulator in Adipogenesis. Nat. Commun. 2020, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Carobbio, S.; Hagen, R.M.; Lelliott, C.J.; Slawik, M.; Medina-Gomez, G.; Tan, C.-Y.; Sicard, A.; Atherton, H.J.; Barbarroja, N.; Bjursell, M.; et al. Adaptive Changes of the Insig1/SREBP1/SCD1 Set Point Help Adipose Tissue to Cope With Increased Storage Demands of Obesity. Diabetes 2013, 62, 3697–3708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akaike, Y.; Masuda, K.; Kuwano, Y.; Nishida, K.; Kajita, K.; Kurokawa, K.; Satake, Y.; Shoda, K.; Imoto, I.; Rokutan, K. HuR Regulates Alternative Splicing of the TRA2β Gene in Human Colon Cancer Cells under Oxidative Stress. Mol. Cell. Biol. 2014, 34, 2857–2873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantt, K.; Cherry, J.; Tenney, R.; Karschner, V.; Pekala, P.H. An Early Event in Adipogenesis, the Nuclear Selection of the CCAAT Enhancer-Binding Protein β (C/EBPβ) MRNA by HuR and Its Translocation to the Cytosol. J. Biol. Chem. 2005, 280, 24768–24774. [Google Scholar] [CrossRef] [Green Version]
- Dai, W.; Zhang, G.; Makeyev, E.V. RNA-Binding Protein HuR Autoregulates Its Expression by Promoting Alternative Polyadenylation Site Usage. Nucleic Acids Res. 2012, 40, 787–800. [Google Scholar] [CrossRef] [Green Version]
- Knott, G.J.; Chong, Y.S.; Passon, D.M.; Liang, X.; Deplazes, E.; Conte, M.R.; Marshall, A.C.; Lee, M.; Fox, A.H.; Bond, C.S. Structural Basis of Dimerization and Nucleic Acid Binding of Human DBHS Proteins NONO and PSPC1. Nucleic Acids Res. 2022, 50, 522–535. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Casas Garcia, G.P.; Perugini, M.A.; Fox, A.H.; Bond, C.S.; Lee, M. Crystal Structure of a SFPQ/PSPC1 Heterodimer Provides Insights into Preferential Heterodimerization of Human DBHS Family Proteins. J. Biol. Chem. 2018, 293, 6593–6602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuwahara, S.; Ikei, A.; Taguchi, Y.; Tabuchi, Y.; Fujimoto, N.; Obinata, M.; Uesugi, S.; Kurihara, Y. PSPC1, NONO, and SFPQ Are Expressed in Mouse Sertoli Cells and May Function as Coregulators of Androgen Receptor-Mediated Transcription1. Biol. Reprod. 2006, 75, 352–359. [Google Scholar] [CrossRef] [Green Version]
- Lang, Y.-D.; Jou, Y.-S. PSPC1: A Contextual Determinant of Tumor Progression. Mol. Cell. Oncol. 2020, 7, 1721253. [Google Scholar] [CrossRef]
- Lowery, L.A.; Rubin, J.; Sive, H. Whitesnake/Sfpq Is Required for Cell Survival and Neuronal Development in the Zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 1347–1357. [Google Scholar] [CrossRef]
- Li, S.; Li, Z.; Shu, F.-J.; Xiong, H.; Phillips, A.C.; Dynan, W.S. Double-Strand Break Repair Deficiency in NONO Knockout Murine Embryonic Fibroblasts and Compensation by Spontaneous Upregulation of the PSPC1 Paralog. Nucleic Acids Res. 2014, 42, 9771–9780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Rajbhandari, P.; Damianov, A.; Han, A.; Sallam, T.; Waki, H.; Villanueva, C.J.; Lee, S.D.; Nielsen, R.; Mandrup, S.; et al. RNA-Binding Protein PSPC1 Promotes the Differentiation-Dependent Nuclear Export of Adipocyte RNAs. J. Clin. Investig. 2017, 127, 987–1004. [Google Scholar] [CrossRef] [Green Version]
- Fumagalli, S.; Totty, N.F.; Hsuan, J.J.; Courtneidge, S.A. A Target for Src in Mitosis. Nature 1994, 368, 871–874. [Google Scholar] [CrossRef]
- Lin, Q.; Taylor, S.J.; Shalloway, D. Specificity and Determinants of Sam68 RNA Binding. J. Biol. Chem. 1997, 272, 27274–27280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vernet, C.; Artzt, K. STAR, a Gene Family Involved in Signal Transduction and Activation of RNA. Trends Genet. 1997, 13, 479–484. [Google Scholar] [CrossRef]
- Bielli, P.; Busà, R.; Paronetto, M.P.; Sette, C. The RNA-Binding Protein Sam68 Is a Multifunctional Player in Human Cancer. Endocr. Relat. Cancer 2011, 18, R91–R102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Jiménez, F.; Sánchez-Margalet, V. Role of Sam68 in Post-Transcriptional Gene Regulation. Int. J. Mol. Sci. 2013, 14, 23402–23419. [Google Scholar] [CrossRef] [Green Version]
- Messina, V.; Meikar, O.; Paronetto, M.P.; Calabretta, S.; Geremia, R.; Kotaja, N.; Sette, C. The RNA Binding Protein SAM68 Transiently Localizes in the Chromatoid Body of Male Germ Cells and Influences Expression of Select MicroRNAs. PLoS ONE 2012, 7, e39729. [Google Scholar] [CrossRef]
- Zhao, X.; Li, Z.; He, B.; Liu, J.; Li, S.; Zhou, L.; Pan, C.; Yu, Z.; Xu, Z. Sam68 Is a Novel Marker for Aggressive Neuroblastoma. OncoTargets Ther. 2013, 6, 1751–1760. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Li, Y.; Cheng, J.; Chen, L.; Xu, H.; Li, Q.; Pang, T. Sam68 Affects Cell Proliferation and Apoptosis of Human Adult T-Acute Lymphoblastic Leukemia Cells via AKT/MTOR Signal Pathway. Leuk. Res. 2016, 46, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Richard, S.; Torabi, N.; Franco, G.V.; Tremblay, G.A.; Chen, T.; Vogel, G.; Morel, M.; Cléroux, P.; Forget-Richard, A.; Komarova, S.; et al. Ablation of the Sam68 RNA Binding Protein Protects Mice from Age-Related Bone Loss. PLOS Genet. 2005, 1, e74. [Google Scholar] [CrossRef] [PubMed]
- Paronetto, M.P.; Messina, V.; Barchi, M.; Geremia, R.; Richard, S.; Sette, C. Sam68 Marks the Transcriptionally Active Stages of Spermatogenesis and Modulates Alternative Splicing in Male Germ Cells. Nucleic Acids Res. 2011, 39, 4961–4974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huot, M.-É.; Vogel, G.; Zabarauskas, A.; Ngo, C.T.-A.; Coulombe-Huntington, J.; Majewski, J.; Richard, S. The Sam68 STAR RNA-Binding Protein Regulates MTOR Alternative Splicing during Adipogenesis. Mol. Cell 2012, 46, 187–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, J.; Richard, S. Sam68 Regulates S6K1 Alternative Splicing during Adipogenesis. Mol. Cell. Biol. 2015, 35, 1926–1939. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Hébert, S.; Song, J.; Kleinman, C.L.; Richard, S. Transcriptome Profiling in Preadipocytes Identifies Long Noncoding RNAs as Sam68 Targets. Oncotarget 2017, 8, 81994–82005. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Yang, Y.; Wu, J. TNFα-Induced up-Regulation of MiR-155 Inhibits Adipogenesis by down-Regulating Early Adipogenic Transcription Factors. Biochem. Biophys. Res. Commun. 2011, 414, 618–624. [Google Scholar] [CrossRef]
- McNeil, G.P.; Zhang, X.; Genova, G.; Jackson, F.R. A Molecular Rhythm Mediating Circadian Clock Output in Drosophila. Neuron 1998, 20, 297–303. [Google Scholar] [CrossRef] [Green Version]
- Markus, M.A.; Morris, B.J. Lark Is the Splicing Factor RBM4 and Exhibits Unique Subnuclear Localization Properties. DNA Cell Biol. 2006, 25, 457–464. [Google Scholar] [CrossRef]
- Lai, M.-C.; Kuo, H.-W.; Chang, W.-C.; Tarn, W.-Y. A Novel Splicing Regulator Shares a Nuclear Import Pathway with SR Proteins. EMBO J. 2003, 22, 1359–1369. [Google Scholar] [CrossRef] [Green Version]
- Markus, M.A.; Morris, B.J. RBM4: A Multifunctional RNA-Binding Protein. Int. J. Biochem. Cell Biol. 2009, 41, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, D.; Qian, H.; Tsai, Y.S.; Shao, S.; Liu, Q.; Dominguez, D.; Wang, Z. The Splicing Factor RBM4 Controls Apoptosis, Proliferation, and Migration to Suppress Tumor Progression. Cancer Cell 2014, 26, 374–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, K.; Zhang, X.; Song, Q.; Feng, Q. G-Quadruplex Regulation of VEGFA MRNA Translation by RBM4. Int. J. Mol. Sci. 2022, 23, 743. [Google Scholar] [CrossRef] [PubMed]
- Kojima, S.; Matsumoto, K.; Hirose, M.; Shimada, M.; Nagano, M.; Shigeyoshi, Y.; Hoshino, S.; Ui-Tei, K.; Saigo, K.; Green, C.B.; et al. LARK Activates Posttranscriptional Expression of an Essential Mammalian Clock Protein, PERIOD1. Proc. Natl. Acad. Sci. USA 2007, 104, 1859–1864. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-C.; Tarn, W.-Y.; Hsieh, W.-K. Emerging Role for RNA Binding Motif Protein 4 in the Development of Brown Adipocytes. Biochim. Biophys. Acta BBA - Mol. Cell Res. 2014, 1843, 769–779. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Zhang, F.; Zhang, X.; Xue, C.; Namwanje, M.; Fan, L.; Reilly, M.P.; Hu, F.; Qiang, L. Distinct Functions of PPARγ Isoforms in Regulating Adipocyte Plasticity. Biochem. Biophys. Res. Commun. 2016, 481, 132–138. [Google Scholar] [CrossRef] [Green Version]
- Hudak, C.S.; Sul, H.S. Pref-1, a Gatekeeper of Adipogenesis. Front. Endocrinol. 2013, 4, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- da Silva, C.; Durandt, C.; Kallmeyer, K.; Ambele, M.A.; Pepper, M.S. The Role of Pref-1 during Adipogenic Differentiation: An Overview of Suggested Mechanisms. Int. J. Mol. Sci. 2020, 21, 4104. [Google Scholar] [CrossRef]
- Estrella, N.L.; Desjardins, C.A.; Nocco, S.E.; Clark, A.L.; Maksimenko, Y.; Naya, F.J. MEF2 Transcription Factors Regulate Distinct Gene Programs in Mammalian Skeletal Muscle Differentiation. J. Biol. Chem. 2015, 290, 1256–1268. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.-C. RBM4-MEF2C Network Constitutes a Feed-Forward Circuit That Facilitates the Differentiation of Brown Adipocytes. RNA Biol. 2015, 12, 208–220. [Google Scholar] [CrossRef] [Green Version]
- Harms, M.J.; Ishibashi, J.; Wang, W.; Lim, H.-W.; Goyama, S.; Sato, T.; Kurokawa, M.; Won, K.-J.; Seale, P. Prdm16 Is Required for the Maintenance of Brown Adipocyte Identity and Function in Adult Mice. Cell Metab. 2014, 19, 593–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mordovkina, D.; Lyabin, D.N.; Smolin, E.A.; Sogorina, E.M.; Ovchinnikov, L.P.; Eliseeva, I. Y-Box Binding Proteins in MRNP Assembly, Translation, and Stability Control. Biomolecules 2020, 10, 591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, M.T. Nucleic Acid-Binding Properties of the Xenopus Oocyte Y Box Protein MRNP3+4. Biochemistry 1994, 33, 13910–13917. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Imamura, T.; Nagatani, G.; Ise, T.; Murakami, T.; Uramoto, H.; Torigoe, T.; Ishiguchi, H.; Yoshida, Y.; Nomoto, M.; et al. Y Box-Binding Protein-1 Binds Preferentially to Single-Stranded Nucleic Acids and Exhibits 3’-->5’ Exonuclease Activity. Nucleic Acids Res. 2001, 29, 1200–1207. [Google Scholar] [CrossRef] [Green Version]
- Tafuri, S.R.; Wolffe, A.P. DNA Binding, Multimerization, and Transcription Stimulation by the Xenopus Y Box Proteins in Vitro. New Biol. 1992, 4, 349–359. [Google Scholar]
- Cooke, A.; Schwarzl, T.; Huppertz, I.; Kramer, G.; Mantas, P.; Alleaume, A.-M.; Huber, W.; Krijgsveld, J.; Hentze, M.W. The RNA-Binding Protein YBX3 Controls Amino Acid Levels by Regulating SLC MRNA Abundance. Cell Rep. 2019, 27, 3097–3106.e5. [Google Scholar] [CrossRef] [Green Version]
- Lyabin, D.N.; Eliseeva, I.A.; Smolin, E.A.; Doronin, A.N.; Budkina, K.S.; Kulakovskiy, I.V.; Ovchinnikov, L.P. YB-3 Substitutes YB-1 in Global MRNA Binding. RNA Biol. 2020, 17, 487–499. [Google Scholar] [CrossRef] [Green Version]
- Rabiee, A.; Plucińska, K.; Isidor, M.S.; Brown, E.L.; Tozzi, M.; Sidoli, S.; Petersen, P.S.S.; Agueda-Oyarzabal, M.; Torsetnes, S.B.; Chehabi, G.N.; et al. White Adipose Remodeling during Browning in Mice Involves YBX1 to Drive Thermogenic Commitment. Mol. Metab. 2021, 44, 101137. [Google Scholar] [CrossRef]
- Wu, R.; Cao, S.; Li, F.; Feng, S.; Shu, G.; Wang, L.; Gao, P.; Zhu, X.; Zhu, C.; Wang, S.; et al. RNA-Binding Protein YBX1 Promotes Brown Adipogenesis and Thermogenesis via PINK1/PRKN-Mediated Mitophagy. FASEB J. 2022, 36, e22219. [Google Scholar] [CrossRef]
- Xu, D.; Xu, S.; Kyaw, A.M.M.; Lim, Y.C.; Chia, S.Y.; Chee Siang, D.T.; Alvarez-Dominguez, J.R.; Chen, P.; Leow, M.K.-S.; Sun, L. RNA Binding Protein Ybx2 Regulates RNA Stability During Cold-Induced Brown Fat Activation. Diabetes 2017, 66, 2987–3000. [Google Scholar] [CrossRef] [Green Version]
- Snyder, E.; Soundararajan, R.; Sharma, M.; Dearth, A.; Smith, B.; Braun, R.E. Compound Heterozygosity for Y Box Proteins Causes Sterility Due to Loss of Translational Repression. PLOS Genet. 2015, 11, e1005690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleene, K.C. Position-Dependent Interactions of Y-Box Protein 2 (YBX2) with MRNA Enable MRNA Storage in Round Spermatids by Repressing MRNA Translation and Blocking Translation-Dependent MRNA Decay. Mol. Reprod. Dev. 2016, 83, 190–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergman, D.; Halje, M.; Nordin, M.; Engström, W. Insulin-Like Growth Factor 2 in Development and Disease: A Mini-Review. Gerontology 2013, 59, 240–249. [Google Scholar] [CrossRef]
- Sussenbach, J.S.; Rodenburg, R.J.T.; Scheper, W.; Holthuizen, P. Transcriptional and Post-Transcriptional Regulation of the Human IGF-II Gene Expression. In Current Directions in Insulin-Like Growth Factor Research; Le Roith, D., Raizada, M.K., Eds.; Advances in Experimental Medicine and Biology; Springer: Boston, MA, USA, 1993; pp. 63–71. ISBN 978-1-4615-2988-0. [Google Scholar]
- Nielsen, J.; Christiansen, J.; Lykke-Andersen, J.; Johnsen, A.H.; Wewer, U.M.; Nielsen, F.C. A Family of Insulin-Like Growth Factor II MRNA-Binding Proteins Represses Translation in Late Development. Mol. Cell. Biol. 1999, 19, 1262–1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudoukha, S.; Cuvellier, S.; Polesskaya, A. Role of the RNA-Binding Protein IMP-2 in Muscle Cell Motility. Mol. Cell. Biol. 2010, 30, 5710–5725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, N.; Zhao, L.; Wrighting, D.; Krämer, D.; Majithia, A.; Wang, Y.; Cracan, V.; Borges-Rivera, D.; Mootha, V.K.; Nahrendorf, M.; et al. IGF2BP2/IMP2-Deficient Mice Resist Obesity through Enhanced Translation of Ucp1 MRNA and Other MRNAs Encoding Mitochondrial Proteins. Cell Metab. 2015, 21, 609–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, N. The Diverse Functions of IMP2/IGF2BP2 in Metabolism. Trends Endocrinol. Metab. 2020, 31, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Votsi, C.; Toufexis, C.; Michailidou, K.; Antoniades, A.; Skordis, N.; Karaolis, M.; Pattichis, C.S.; Christodoulou, K. Type 2 Diabetes Susceptibility in the Greek-Cypriot Population: Replication of Associations with TCF7L2, FTO, HHEX, SLC30A8 and IGF2BP2 Polymorphisms. Genes 2017, 8, 16. [Google Scholar] [CrossRef] [Green Version]
- Phani, N.M.; Adhikari, P.; Nagri, S.K.; D’Souza, S.C.; Satyamoorthy, K.; Rai, P.S. Replication and Relevance of Multiple Susceptibility Loci Discovered from Genome Wide Association Studies for Type 2 Diabetes in an Indian Population. PLoS ONE 2016, 11, e0157364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, M.C.Y.; Shriner, D.; Chen, B.H.; Li, J.; Chen, W.-M.; Guo, X.; Liu, J.; Bielinski, S.J.; Yanek, L.R.; Nalls, M.A.; et al. Meta-Analysis of Genome-Wide Association Studies in African Americans Provides Insights into the Genetic Architecture of Type 2 Diabetes. PLOS Genet. 2014, 10, e1004517. [Google Scholar] [CrossRef]
- Han, L.; Li, Y.; Tang, L.; Chen, Z.; Zhang, T.; Chen, S.; Liu, S.; Peng, X.; Mai, Y.; Zhuo, R.; et al. IGF2BP2 Rs11705701 Polymorphisms Are Associated with Prediabetes in a Chinese Population: A Population-Based Case-Control Study. Exp. Ther. Med. 2016, 12, 1849–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regué, L.; Ji, F.; Flicker, D.; Kramer, D.; Pierce, W.; Davidoff, T.; Widrick, J.J.; Houstis, N.; Minichiello, L.; Dai, N.; et al. IMP2 Increases Mouse Skeletal Muscle Mass and Voluntary Activity by Enhancing Autocrine Insulin-Like Growth Factor 2 Production and Optimizing Muscle Metabolism. Mol. Cell. Biol. 2019, 39, e00528-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenwald, W.W.; Chiou, J.; Yan, J.; Qiu, Y.; Dai, N.; Wang, A.; Nariai, N.; Aylward, A.; Han, J.Y.; Kadakia, N.; et al. Pancreatic Islet Chromatin Accessibility and Conformation Reveals Distal Enhancer Networks of Type 2 Diabetes Risk. Nat. Commun. 2019, 10, 2078. [Google Scholar] [CrossRef] [PubMed]
- Regué, L.; Minichiello, L.; Avruch, J.; Dai, N. Liver-Specific Deletion of IGF2 MRNA Binding Protein-2/IMP2 Reduces Hepatic Fatty Acid Oxidation and Increases Hepatic Triglyceride Accumulation. J. Biol. Chem. 2019, 294, 11944–11951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briata, P.; Chen, C.-Y.; Ramos, A.; Gherzi, R. Functional and Molecular Insights into KSRP Function in MRNA Decay. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 2013, 1829, 689–694. [Google Scholar] [CrossRef]
- García-Mayoral, M.F.; Hollingworth, D.; Masino, L.; Díaz-Moreno, I.; Kelly, G.; Gherzi, R.; Chou, C.-F.; Chen, C.-Y.; Ramos, A. The Structure of the C-Terminal KH Domains of KSRP Reveals a Noncanonical Motif Important for MRNA Degradation. Structure 2007, 15, 485–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gherzi, R.; Lee, K.-Y.; Briata, P.; Wegmüller, D.; Moroni, C.; Karin, M.; Chen, C.-Y. A KH Domain RNA Binding Protein, KSRP, Promotes ARE-Directed MRNA Turnover by Recruiting the Degradation Machinery. Mol. Cell 2004, 14, 571–583. [Google Scholar] [CrossRef]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A New Regulatory Protein, KSRP, Mediates Exon Inclusion through an Intronic Splicing Enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [Green Version]
- Dhamija, S.; Kuehne, N.; Winzen, R.; Doerrie, A.; Dittrich-Breiholz, O.; Thakur, B.K.; Kracht, M.; Holtmann, H. Interleukin-1 Activates Synthesis of Interleukin-6 by Interfering with a KH-Type Splicing Regulatory Protein (KSRP)-Dependent Translational Silencing Mechanism. J. Biol. Chem. 2011, 286, 33279–33288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trabucchi, M.; Briata, P.; Garcia-Mayoral, M.; Haase, A.D.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. The RNA-Binding Protein KSRP Promotes the Biogenesis of a Subset of MicroRNAs. Nature 2009, 459, 1010–1014. [Google Scholar] [CrossRef] [Green Version]
- Gebert, L.F.R.; MacRae, I.J. Regulation of MicroRNA Function in Animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Gulei, D.; Raduly, L.; Broseghini, E.; Ferracin, M.; Berindan-Neagoe, I. The Extensive Role of MiR-155 in Malignant and Non-Malignant Diseases. Mol. Aspects Med. 2019, 70, 33–56. [Google Scholar] [CrossRef] [PubMed]
- Palzer, K.-A.; Bolduan, V.; Käfer, R.; Kleinert, H.; Bros, M.; Pautz, A. The Role of KH-Type Splicing Regulatory Protein (KSRP) for Immune Functions and Tumorigenesis. Cells 2022, 11, 1482. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.-F.; Lin, Y.-Y.; Wang, H.-K.; Zhu, X.; Giovarelli, M.; Briata, P.; Gherzi, R.; Garvey, W.T.; Chen, C.-Y. KSRP Ablation Enhances Brown Fat Gene Program in White Adipose Tissue through Reduced MiR-150 Expression. Diabetes 2014, 63, 2949–2961. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.-Y.; Chou, C.-F.; Giovarelli, M.; Briata, P.; Gherzi, R.; Chen, C.-Y. KSRP and MicroRNA 145 Are Negative Regulators of Lipolysis in White Adipose Tissue. Mol. Cell. Biol. 2014, 34, 2339–2349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lass, A.; Zimmermann, R.; Haemmerle, G.; Riederer, M.; Schoiswohl, G.; Schweiger, M.; Kienesberger, P.; Strauss, J.G.; Gorkiewicz, G.; Zechner, R. Adipose Triglyceride Lipase-Mediated Lipolysis of Cellular Fat Stores Is Activated by CGI-58 and Defective in Chanarin-Dorfman Syndrome. Cell Metab. 2006, 3, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, P.; Kandror, K.V. FoxO1 Controls Insulin-Dependent Adipose Triglyceride Lipase (ATGL) Expression and Lipolysis in Adipocytes. J. Biol. Chem. 2009, 284, 13296–13300. [Google Scholar] [CrossRef] [Green Version]
- Spangenberg, L.; Correa, A.; Dallagiovanna, B.; Naya, H. Role of Alternative Polyadenylation during Adipogenic Differentiation: An In Silico Approach. PLoS ONE 2013, 8, e75578. [Google Scholar] [CrossRef]
- Erson-Bensan, A.E. Alternative Polyadenylation and RNA-Binding Proteins. J. Mol. Endocrinol. 2016, 57, F29–F34. [Google Scholar] [CrossRef]
- Cui, J.; Li, C.; Cui, X.; Liu, X.; Meng, C.; Zhou, G. Shortening of HO1 3′UTRs by Alternative Polyadenylation Suppresses Adipogenesis in 3T3-L1. J. Agric. Food Chem. 2021, 69, 8038–8049. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potapov, V.; Fu, X.; Dai, N.; Corrêa, I.R.; Tanner, N.A.; Ong, J.L. Base Modifications Affecting RNA Polymerase and Reverse Transcriptase Fidelity. Nucleic Acids Res. 2018, 46, 5753–5763. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Yang, Y.; Jiang, S.; Peng, J. Novel Insights into Adipogenesis from the Perspective of Transcriptional and RNA N6-Methyladenosine-Mediated Post-Transcriptional Regulation. Adv. Sci. 2020, 7, 2001563. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Song, C.; Wang, N.; Li, S.; Liu, Q.; Sun, Z.; Wang, K.; Yu, S.-C.; Yang, Q. NADP Modulates RNA M6A Methylation and Adipogenesis via Enhancing FTO Activity. Nat. Chem. Biol. 2020, 16, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.J.T.; Pan, T.; Kalsotra, A. RNA Modifications and Structures Cooperate to Guide RNA–Protein Interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202–210. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Fang, X.; Zhong, P.; Song, Z.; Hu, X. N6-Methyladenosine Modifications: Interactions with Novel RNA-Binding Proteins and Roles in Signal Transduction. RNA Biol. 2019, 16, 991–1000. [Google Scholar] [CrossRef]
- Visvanathan, A.; Patil, V.; Arora, A.; Hegde, A.S.; Arivazhagan, A.; Santosh, V.; Somasundaram, K. Essential Role of METTL3-Mediated M6A Modification in Glioma Stem-like Cells Maintenance and Radioresistance. Oncogene 2018, 37, 522–533. [Google Scholar] [CrossRef]
- Li, L.; Zang, L.; Zhang, F.; Chen, J.; Shen, H.; Shu, L.; Liang, F.; Feng, C.; Chen, D.; Tao, H.; et al. Fat Mass and Obesity-Associated (FTO) Protein Regulates Adult Neurogenesis. Hum. Mol. Genet. 2017, 26, 2398–2411. [Google Scholar] [CrossRef]
- Ray, D.; Kazan, H.; Cook, K.B.; Weirauch, M.T.; Najafabadi, H.S.; Li, X.; Gueroussov, S.; Albu, M.; Zheng, H.; Yang, A.; et al. A Compendium of RNA-Binding Motifs for Decoding Gene Regulation. Nature 2013, 499, 172–177. [Google Scholar] [CrossRef] [Green Version]
- Jens, M.; Rajewsky, N. Competition between Target Sites of Regulators Shapes Post-Transcriptional Gene Regulation. Nat. Rev. Genet. 2015, 16, 113–126. [Google Scholar] [CrossRef]
- Hafner, M.; Landthaler, M.; Burger, L.; Khorshid, M.; Hausser, J.; Berninger, P.; Rothballer, A.; Ascano, M.; Jungkamp, A.-C.; Munschauer, M.; et al. Transcriptome-Wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP. Cell 2010, 141, 129–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| RBP | RNA-Binding Domain | Target Cis-Elements | RNA Targets | Effect on RNA Targets |
|---|---|---|---|---|
| HuR | RRMs | AU-rich element | Insig1, ATGL | Stabilization |
| PSPC1 | RRMs | AU-rich element | Ebf1, PPARγ, Acsl1, and Scd1 | Nuclear export |
| Sam68 | KH | U(U/A)AA | mTOR, Rps6kb1 | Alternative splicing |
| RBM4 | RRMs, ZF | CU-rich element | PPARγ, Pref1, MEF2C, Prdm16 | Alternative splicing |
| Ybx1 | CSD, C-terminal domain | Not determined | Pink1, Prkn | Stabilization |
| Ybx2 | CSD, C-terminal domain | Not determined | Pgc1α | Stabilization |
| IGF2BP2 | RRM, KH | Not determined | UCP1 | Inhibit translation |
| KSRP | KH | AU-rich element | pri-miR-145, pri-miR-150 | miRNA processing |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, P.; Wu, W.; Ma, C.; Du, C.; Huang, Y.; Xu, H.; Li, C.; Cheng, X.; Hao, R.; Xu, Y. RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function. Cells 2022, 11, 2357. https://doi.org/10.3390/cells11152357
Zhang P, Wu W, Ma C, Du C, Huang Y, Xu H, Li C, Cheng X, Hao R, Xu Y. RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function. Cells. 2022; 11(15):2357. https://doi.org/10.3390/cells11152357
Chicago/Turabian StyleZhang, Pengpeng, Wenyan Wu, Chaofeng Ma, Chunyu Du, Yueru Huang, Haixia Xu, Cencen Li, Xiaofang Cheng, Ruijie Hao, and Yongjie Xu. 2022. "RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function" Cells 11, no. 15: 2357. https://doi.org/10.3390/cells11152357
APA StyleZhang, P., Wu, W., Ma, C., Du, C., Huang, Y., Xu, H., Li, C., Cheng, X., Hao, R., & Xu, Y. (2022). RNA-Binding Proteins in the Regulation of Adipogenesis and Adipose Function. Cells, 11(15), 2357. https://doi.org/10.3390/cells11152357






