Whole Transcriptome Analysis Identifies Platycodin D-Mediated RNA Regulatory Network in Non–Small-Cell Lung Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Model and Treatment
2.2. RNA Extraction, Library Construction and RNA Sequencing
2.3. Screening of Differentially Expressed ncRNAs and Coding RNAs
2.4. PCR and Quantitative Real-Time qPCR Validation of Candidate DE-circRNAs and DE-IncRNAs
2.5. Weighted Gene Co-Expression Network Analysis, Gene Ontology, Pathway Enrichment Analysis, and Functional GSEA Analysis
2.6. Microarray Data Analysis for the Identification of Clinical Significance of DEGs in NSCLC Patients
2.7. Structurally Variation Analysis
2.8. Molecular Docking of PD
2.9. Cell Apoptosis Assay and Cell Cycle Analysis
2.10. Statistical Analysis
3. Results
3.1. Overview of RNA-Seq Data
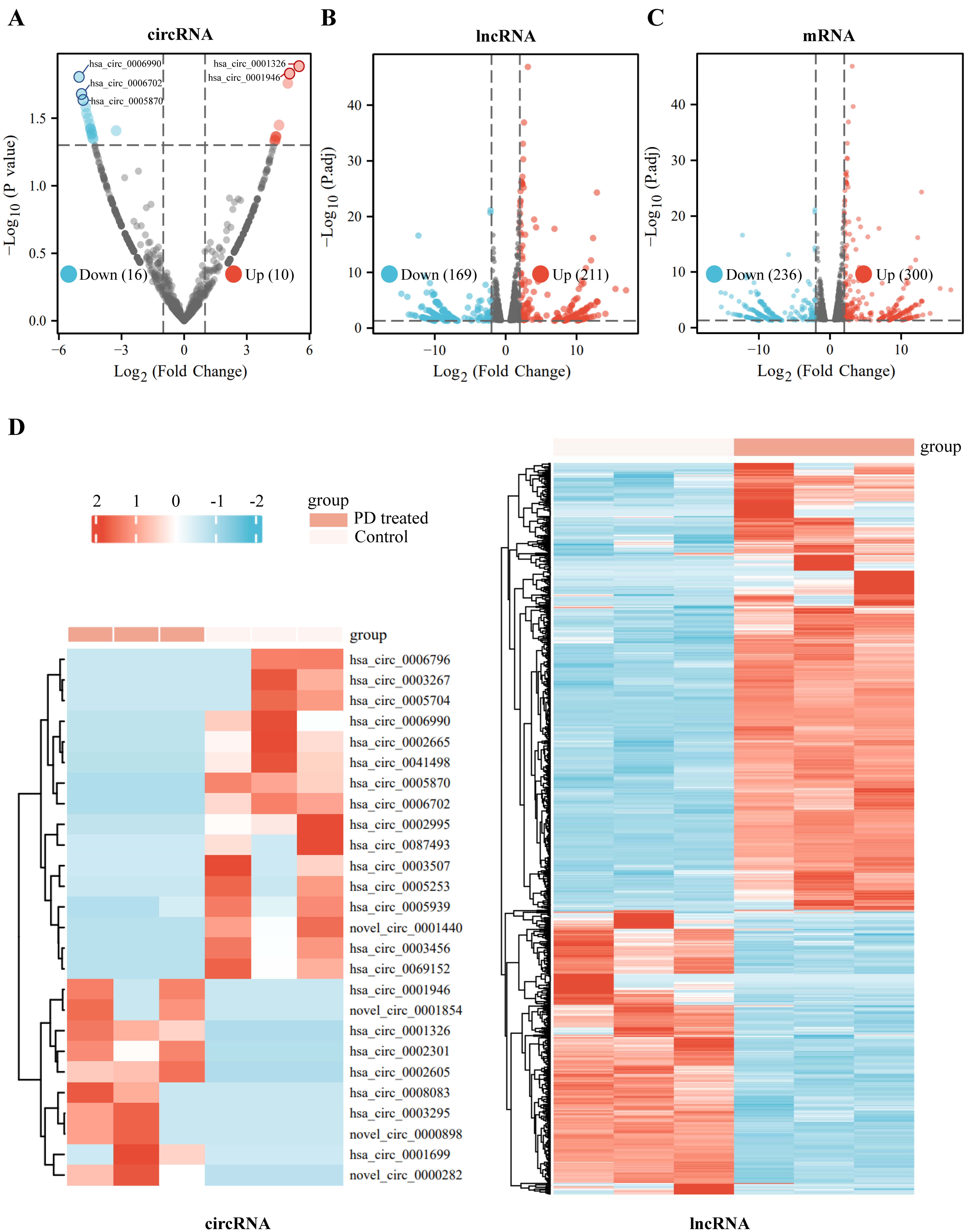
3.2. Identification of Differentially Expressed Genes
3.3. Enrichment and Functional Analysis of DE-ncRNAs-Related Target Genes
3.4. qPCR Validation of DE-ncRNAs
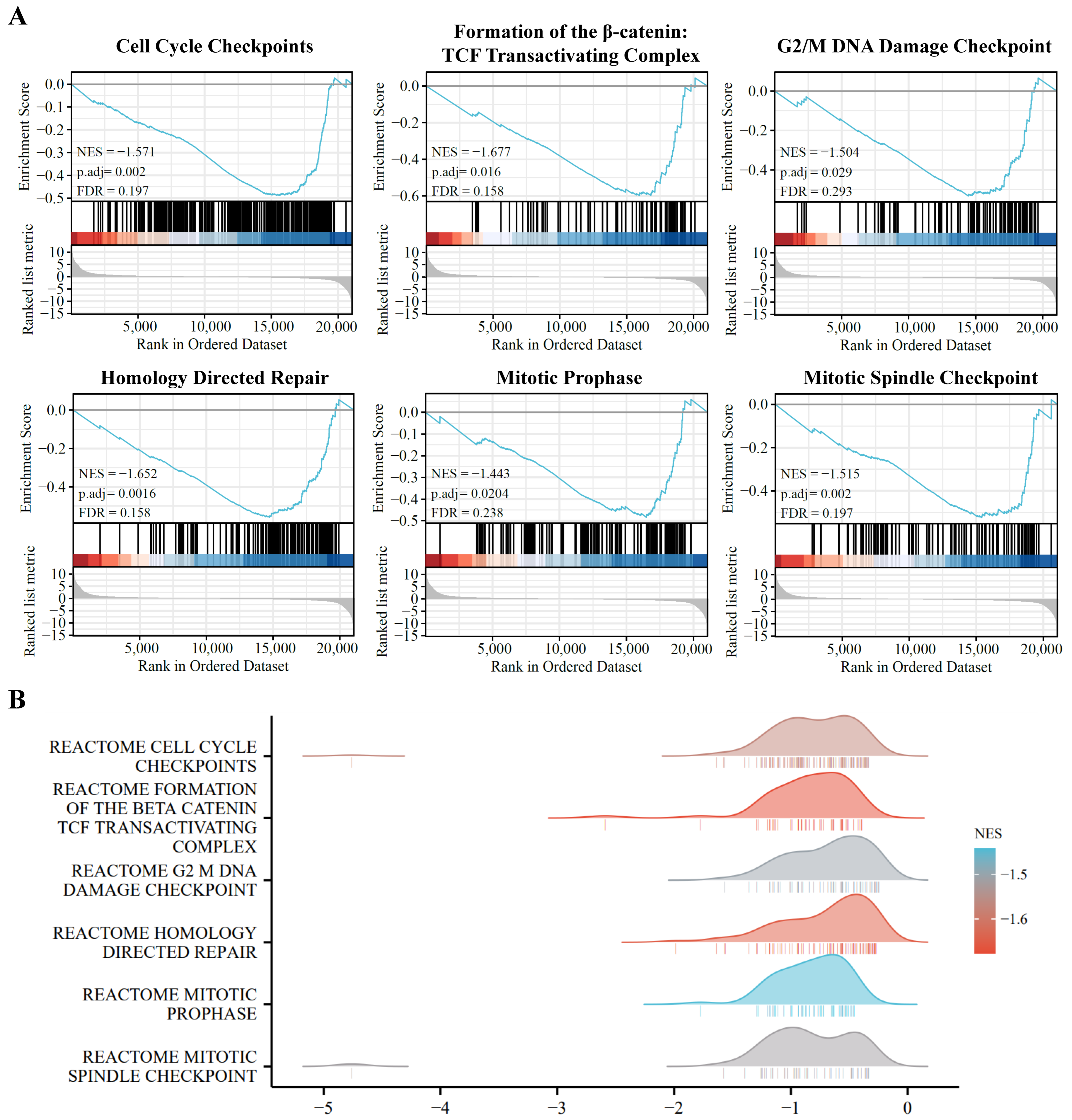
3.5. GSEA Enrichment Analysis
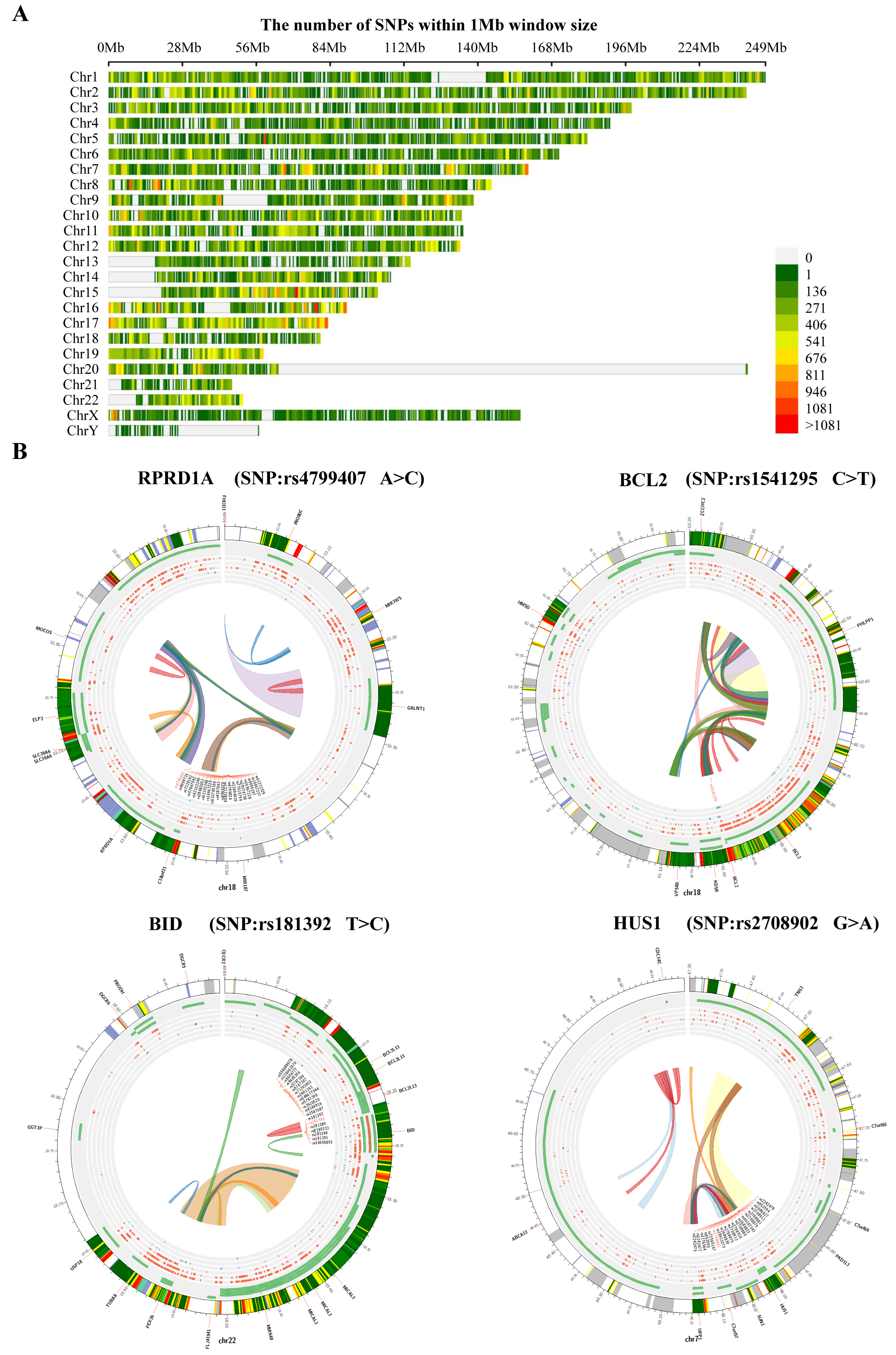
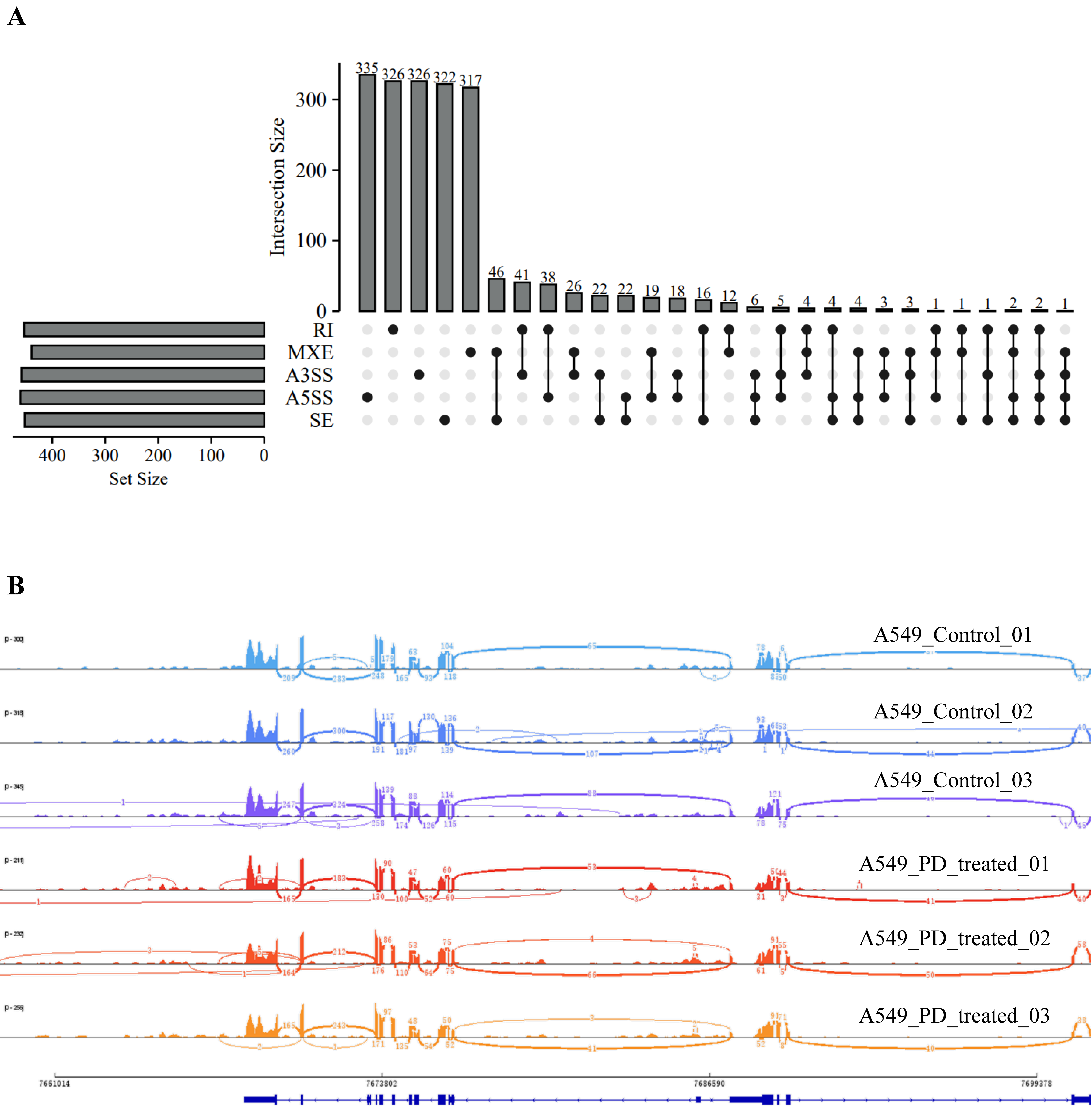
3.6. Structural Variant Analysis
3.7. Assessment in TCGA and GEO Cohorts
3.8. Molecular Docking
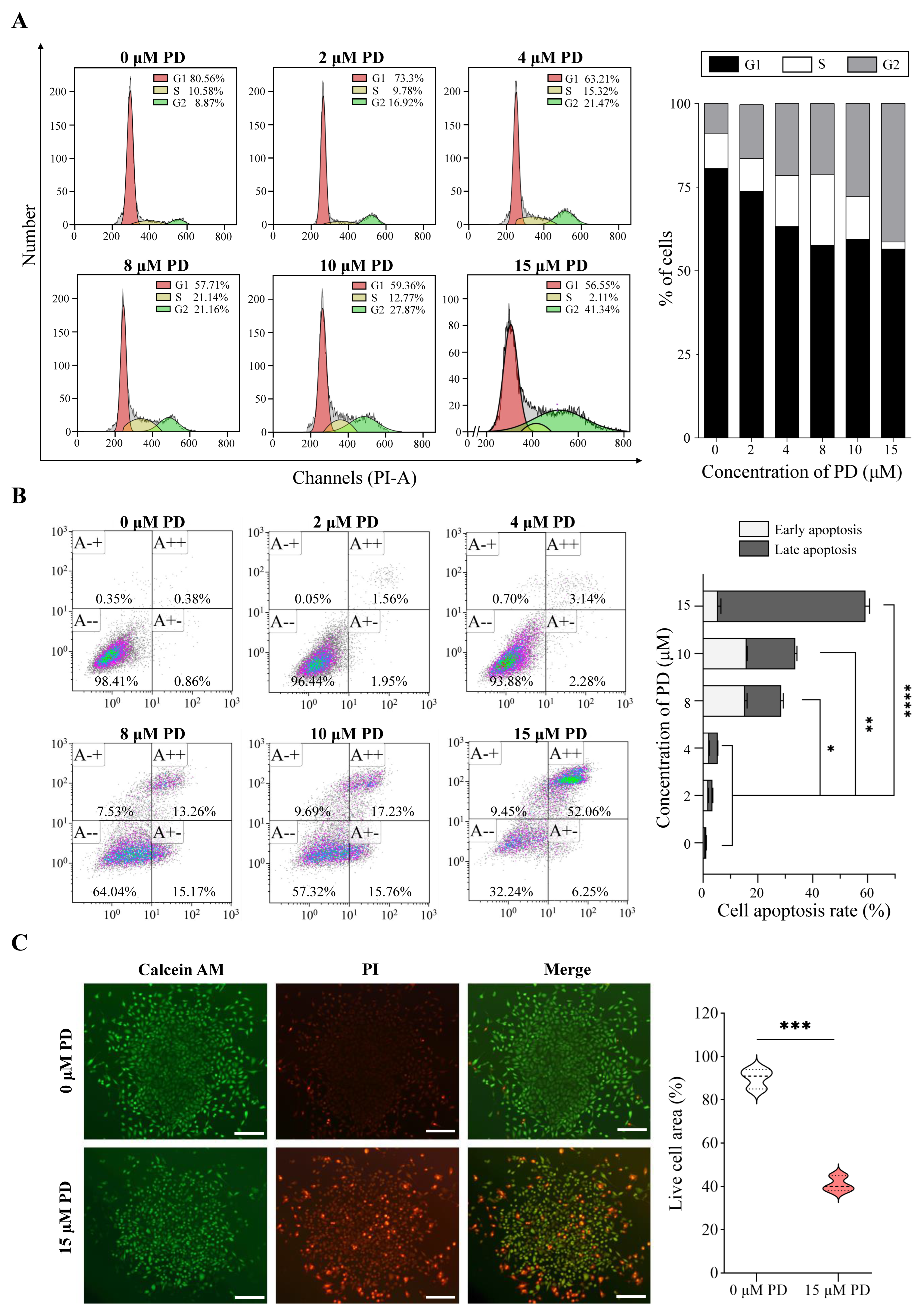
3.9. Evaluation the Effect of PD on Cell Proliferation, Cell Cycle, and Apoptosis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Soneji, S.; Tanner, N.T.; Silvestri, G.A.; Lathan, C.S.; Black, W. Racial and Ethnic Disparities in Early-Stage Lung Cancer Survival. Chest 2017, 152, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Schneider, B.J.; Ismaila, N.; Altorki, N. Lung Cancer Surveillance after Definitive Curative-Intent Therapy: ASCO Guideline Summary. JCO Oncol. Pract. 2020, 16, 83–86. [Google Scholar] [CrossRef]
- Daly, M.E.; Singh, N.; Ismaila, N.; Antonoff, M.B.; Arenberg, D.A.; Bradley, J.; David, E.; Detterbeck, F.; Früh, M.; Gubens, M.A.; et al. Management of Stage III Non-Small-Cell Lung Cancer: ASCO Guideline. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2022, 40, 1356–1384. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA A Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ye, Y.P.; Sun, H.X.; Li, D. Contribution of the glycidic moieties to the haemolytic and adjuvant activity of platycodigenin-type saponins from the root of Platycodon grandiflorum. Vaccine 2008, 26, 3452–3460. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Chen, T.; Guo, Y.; Wang, C.; Dong, L.; Lu, C. Platycodin D (PD) regulates LncRNA-XIST/miR-335 axis to slow down bladder cancer progression in vitro and in vivo. Exp. Cell Res. 2020, 396, 112281. [Google Scholar] [CrossRef]
- Kim, T.W.; Song, I.B.; Lee, H.K.; Lim, J.H.; Cho, E.S.; Son, H.Y.; Park, S.J.; Kim, J.W.; Yun, H.I. Platycodin D, a triterpenoid sapoinin from Platycodon grandiflorum, ameliorates cisplatin-induced nephrotoxicity in mice. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2012, 50, 4254–4259. [Google Scholar] [CrossRef]
- Tang, Z.H.; Li, T.; Gao, H.W.; Sun, W.; Chen, X.P.; Wang, Y.T.; Lu, J.J. Platycodin D from Platycodonis Radix enhances the anti-proliferative effects of doxorubicin on breast cancer MCF-7 and MDA-MB-231 cells. Chin. Med. 2014, 9, 16. [Google Scholar] [CrossRef] [Green Version]
- Seo, Y.S.; Kang, O.H.; Kong, R.; Zhou, T.; Kim, S.A.; Ryu, S.; Kim, H.R.; Kwon, D.Y. Polygalacin D induces apoptosis and cell cycle arrest via the PI3K/Akt pathway in non-small cell lung cancer. Oncol. Rep. 2018, 39, 1702–1710. [Google Scholar] [CrossRef]
- Yim, N.H.; Hwang, Y.H.; Liang, C.; Ma, J.Y. A platycoside-rich fraction from the root of Platycodon grandiflorum enhances cell death in A549 human lung carcinoma cells via mainly AMPK/mTOR/AKT signal-mediated autophagy induction. J. Ethnopharmacol. 2016, 194, 1060–1068. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-small-cell lung cancers: A heterogeneous set of diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; He, J.; Zhao, X. A new systematic computational approach to predicting target genes of transcription factors. Nucleic Acids Res. 2007, 35, 4433–4440. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.J.; Gao, G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat. Biotechnol. 2022. advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Nogales, C.; Mamdouh, Z.M.; List, M.; Kiel, C.; Casas, A.I.; Schmidt, H. Network pharmacology: Curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 2022, 43, 136–150. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef] [Green Version]
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C.; et al. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Imyanitov, E.N.; Iyevleva, A.G.; Levchenko, E.V. Molecular testing and targeted therapy for non-small cell lung cancer: Current status and perspectives. Crit. Rev. Oncol./Hematol. 2021, 157, 103194. [Google Scholar] [CrossRef]
- Li, Z.; Feiyue, Z.; Gaofeng, L. Traditional Chinese medicine and lung cancer--From theory to practice. Biomed. Pharmacother. 2021, 137, 111381. [Google Scholar] [CrossRef]
- Wang, S.; Long, S.; Deng, Z.; Wu, W. Positive Role of Chinese Herbal Medicine in Cancer Immune Regulation. Am. J. Chin. Med. 2020, 48, 1577–1592. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [Green Version]
- Teng, P.C.; Liang, Y.; Yarmishyn, A.A.; Hsiao, Y.J.; Lin, T.Y.; Lin, T.W.; Teng, Y.C.; Yang, Y.P.; Wang, M.L.; Chien, C.S.; et al. RNA Modifications and Epigenetics in Modulation of Lung Cancer and Pulmonary Diseases. Int. J. Mol. Sci. 2021, 22, 10592. [Google Scholar] [CrossRef]
- De Martino, M.; Esposito, F.; Pallante, P. Long non-coding RNAs regulating multiple proliferative pathways in cancer cell. Transl. Cancer Res. 2021, 10, 3140–3157. [Google Scholar] [CrossRef]
- Hou, S.; Li, Z.; Zheng, X.; Gao, Y.; Dong, J.; Ni, Y.; Wang, X.; Li, Y.; Ding, X.; Chang, Z.; et al. Embryonic endothelial evolution towards first hematopoietic stem cells revealed by single-cell transcriptomic and functional analyses. Cell Res. 2020, 30, 376–392. [Google Scholar] [CrossRef] [Green Version]
- Cen, X.; Huang, Y.; Lu, Z.; Shao, W.; Zhuo, C.; Bao, C.; Feng, S.; Wei, C.; Tang, X.; Cen, L.; et al. LncRNA IGFL2-AS1 Promotes the Proliferation, Migration, and Invasion of Colon Cancer Cells and is Associated with Patient Prognosis. Cancer Manag. Res. 2021, 13, 5957–5968. [Google Scholar] [CrossRef]
- Zhao, R.; Wang, S.; Tan, L.; Li, H.; Liu, J.; Zhang, S. IGFL2-AS1 facilitates tongue squamous cell carcinoma progression via Wnt/β-catenin signaling pathway. Oral Dis. 2021. advance online publication. [Google Scholar] [CrossRef]
- Wang, H.; Shi, Y.; Chen, C.H.; Wen, Y.; Zhou, Z.; Yang, C.; Sun, J.; Du, G.; Wu, J.; Mao, X.; et al. KLF5-induced lncRNA IGFL2-AS1 promotes basal-like breast cancer cell growth and survival by upregulating the expression of IGFL1. Cancer Lett. 2021, 515, 49–62. [Google Scholar] [CrossRef]
- Chen, L.; Shan, G. CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Lett. 2021, 505, 49–57. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- Zhang, M.; Wen, F.; Zhao, K. Circular RNA_0001946 is insufficiently expressed in tumor tissues, while its higher expression correlates with less lymph node metastasis, lower TNM stage, and improved prognosis in NSCLC patients. J. Clin. Lab. Anal. 2021, 35, e23625. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Cao, Q.; Liu, J.; Zhang, J.; Li, B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol. Cancer 2019, 18, 16. [Google Scholar] [CrossRef]
- Yao, Y.; Hua, Q.; Zhou, Y.; Shen, H. CircRNA has_circ_0001946 promotes cell growth in lung adenocarcinoma by regulating miR-135a-5p/SIRT1 axis and activating Wnt/β-catenin signaling pathway. Biomed. Pharmacother. 2019, 111, 1367–1375. [Google Scholar] [CrossRef]
- Martin, B.; Paesmans, M.; Berghmans, T.; Branle, F.; Ghisdal, L.; Mascaux, C.; Meert, A.P.; Steels, E.; Vallot, F.; Verdebout, J.M.; et al. Role of Bcl-2 as a prognostic factor for survival in lung cancer: A systematic review of the literature with meta-analysis. Br. J. Cancer 2003, 89, 55–64. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Pei, F.; Yang, F.; Li, L.; Amin, A.D.; Liu, S.; Buchan, J.R.; Cho, W.C. Role of Autophagy and Apoptosis in Non-Small-Cell Lung Cancer. Int. J. Mol. Sci. 2017, 18, 367. [Google Scholar] [CrossRef] [PubMed]
- Nor Hisam, N.S.; Ugusman, A.; Rajab, N.F.; Ahmad, M.F.; Fenech, M.; Liew, S.L.; Mohamad Anuar, N.N. Combination Therapy of Navitoclax with Chemotherapeutic Agents in Solid Tumors and Blood Cancer: A Review of Current Evidence. Pharmaceutics 2021, 13, 1353. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.; Faazil, S.; Malik, M.S. Apoptosis-inducing agents: A patent review (2010–2013). Expert Opin. Ther. Pat. 2014, 24, 339–354. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Xie, Z.; Xin, Y.; Lu, W.; Yang, H.; Lu, T.; Li, J.; Wang, S.; Cheng, K.; Yang, X.; et al. Whole Transcriptome Analysis Identifies Platycodin D-Mediated RNA Regulatory Network in Non–Small-Cell Lung Cancer. Cells 2022, 11, 2360. https://doi.org/10.3390/cells11152360
Zheng S, Xie Z, Xin Y, Lu W, Yang H, Lu T, Li J, Wang S, Cheng K, Yang X, et al. Whole Transcriptome Analysis Identifies Platycodin D-Mediated RNA Regulatory Network in Non–Small-Cell Lung Cancer. Cells. 2022; 11(15):2360. https://doi.org/10.3390/cells11152360
Chicago/Turabian StyleZheng, Shuyu, Zejuan Xie, Yanlin Xin, Wenli Lu, Hao Yang, Tianming Lu, Jun Li, Shanshan Wang, Keyu Cheng, Xi Yang, and et al. 2022. "Whole Transcriptome Analysis Identifies Platycodin D-Mediated RNA Regulatory Network in Non–Small-Cell Lung Cancer" Cells 11, no. 15: 2360. https://doi.org/10.3390/cells11152360
APA StyleZheng, S., Xie, Z., Xin, Y., Lu, W., Yang, H., Lu, T., Li, J., Wang, S., Cheng, K., Yang, X., Qi, R., Qiu, Y., & Guo, Y. (2022). Whole Transcriptome Analysis Identifies Platycodin D-Mediated RNA Regulatory Network in Non–Small-Cell Lung Cancer. Cells, 11(15), 2360. https://doi.org/10.3390/cells11152360






