Retinal Ganglion Cells: Global Number, Density and Vulnerability to Glaucomatous Injury in Common Laboratory Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Glaucoma Model
2.3. Retrograde Tracing, Electroretinography and Optical Coherence Tomography
2.4. Tissue Collection and RBPMS Immunohistochemistry
2.5. Imaging, RGC Counting and Statistics
3. Results
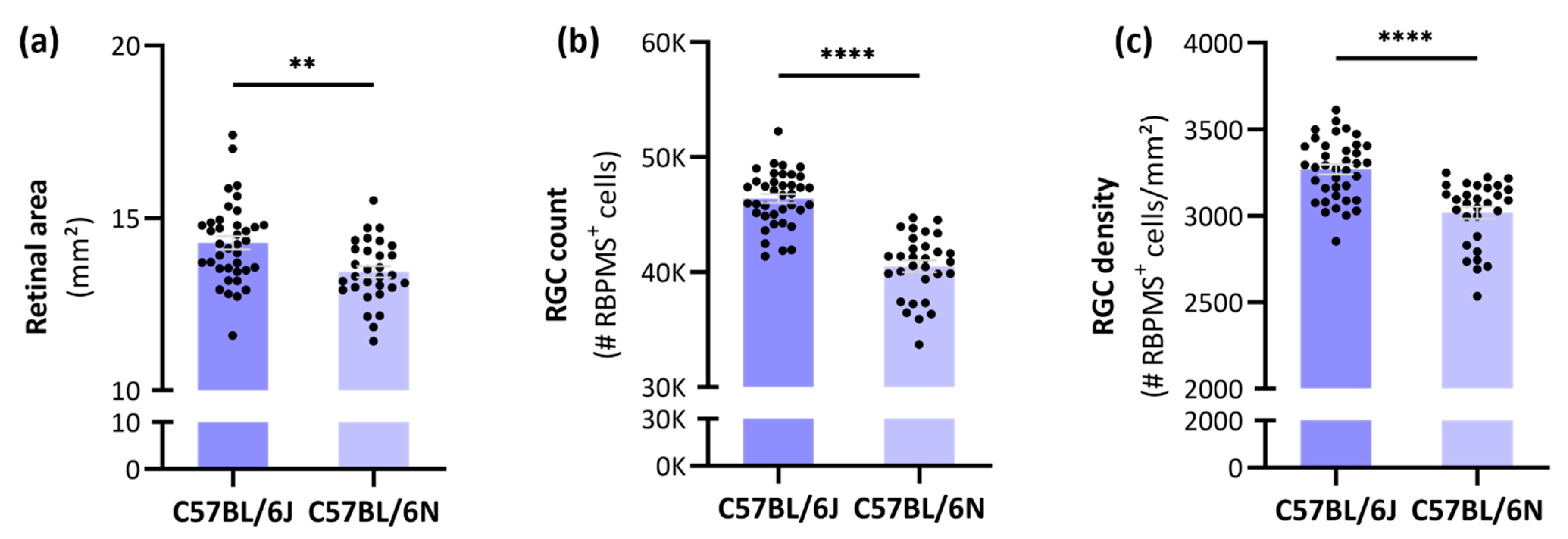
3.1. Differences in Retinal Area, RGC Count and -Density between C57BL/6J and -N mice
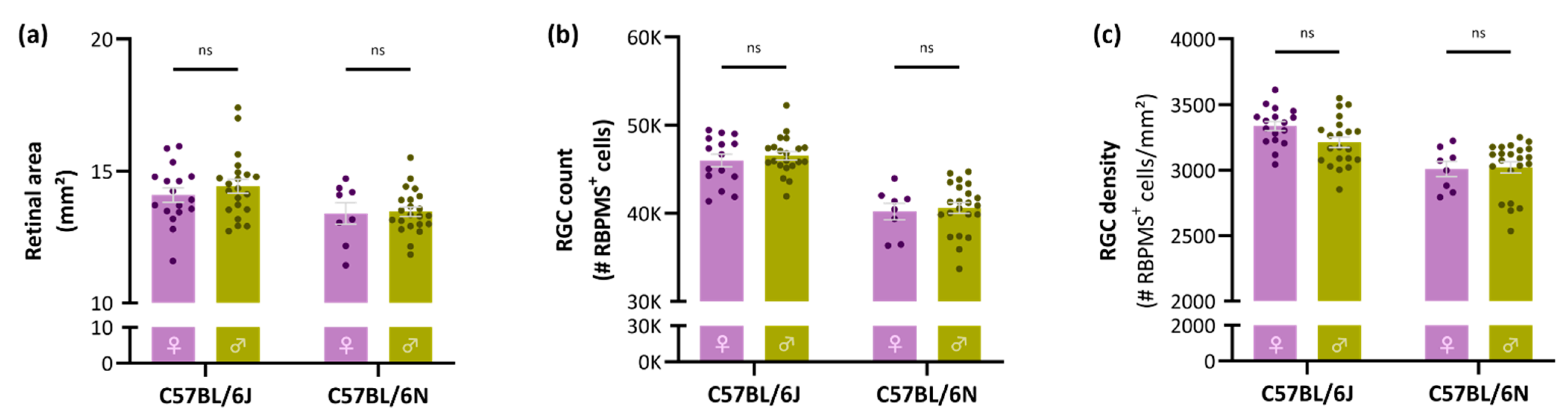
3.2. No Sex-Related Differences in Retinal Area, Global RGC Number or Density
3.3. Mild Photoreceptor Layer Thinning in C57BL/6N Mice but No Difference in RGC Functioning between C57BL/6 Substrains
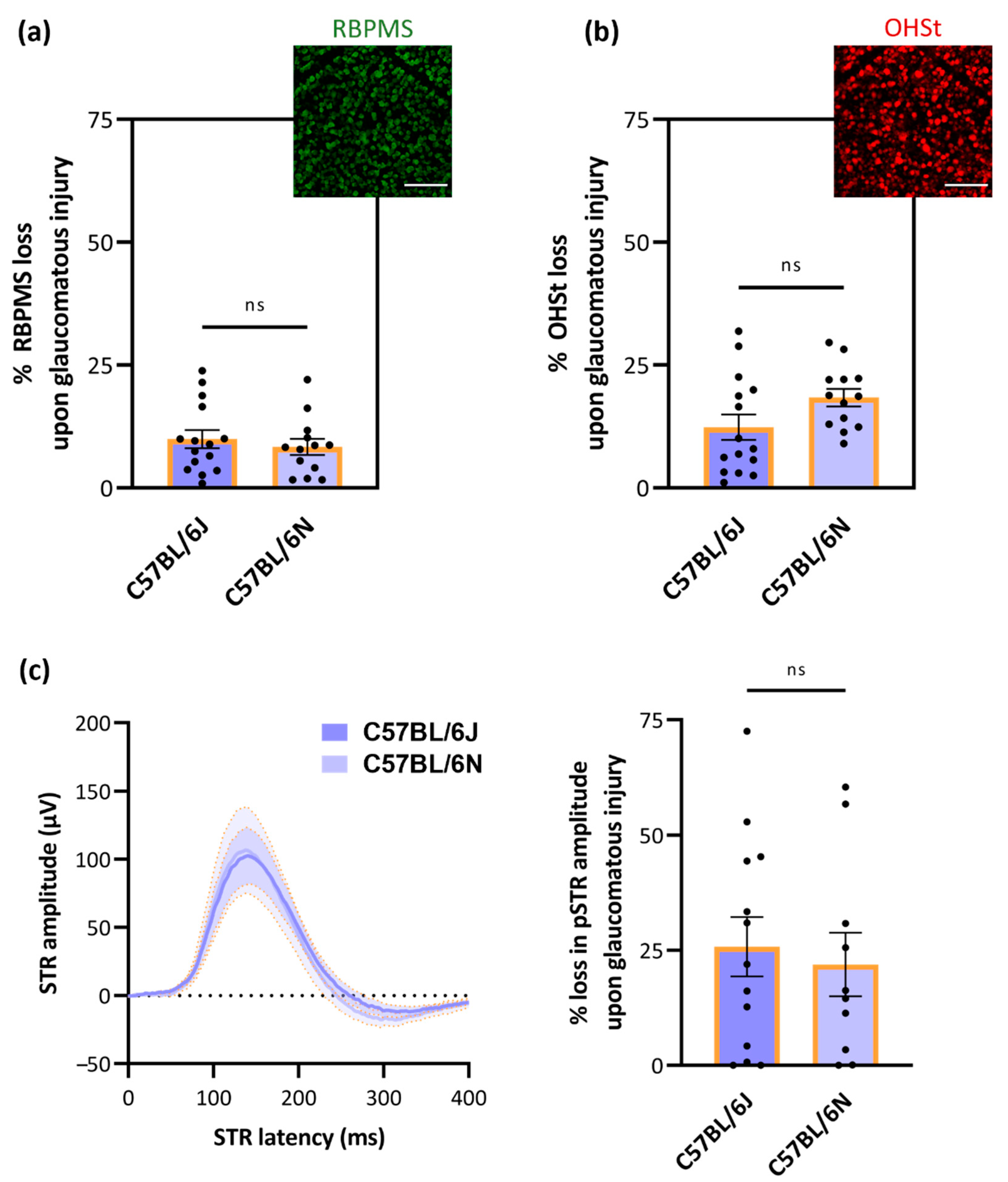
3.4. No Substrain-Dependent Differences in RGC Vulnerability to Glaucomatous Damage
4. Discussion
4.1. The Total Number of RGCs in C57BL/6J and -N Mice
| RGC Labeling Method | Mean Number of RGCs ± SD | C57BL/6 Substrain | Sex | Sample Size | References |
|---|---|---|---|---|---|
| Axon counts | 54,630 ± 3910 | J | Mixed | 21 | [24] |
| 44,857 ± 3125 | J | - | 4 | [5] | |
| 46,000 ± 1000 | - | Male | - | [25] | |
| 51,064 ± 5045 | - | Female | 97 | [27] | |
| 44,846 ± 3980 | J | Mixed | 7 | [28] | |
| 41,659 ± 2700 | J | Male | 10 | [30] | |
| Retrograde tracing from optic nerve | 50,920 ± 1161 | - | Mixed | 5 | [26] |
| 49,823 ± 1792 | J | Mixed | 9 | [31] | |
| 42,658 ± 1540 | N | Male | 10 | [23] | |
| Retrograde tracing from target area | 41,192 ± 3395 | N | Male | 42 | [23] |
| 40,437 ± 3196 | N | Female | 9 | [35] | |
| BRN3A counts on entire wholemounts | 34,627 ± 1821 | N | Female | 9 | [35] |
| 45,637 ± 2632 | J | Mixed | 8 | [12] | |
| RBPMS counts on entire wholemounts | 46,395 ± 2373 | J | Mixed | 38 | Current study |
| 40,501 ± 2788 | N | Mixed | 30 | Current study |
4.2. Substrain-Dependent Differences in Retinal Area, RGC Count and -Density
4.3. No Sex-Related Differences in Retinal Area, Global RGC Number or Density
4.4. Mild Photoreceptor Layer Thinning in C57BL/6N Mice but No Difference in RGC Functioning between C57BL/6 Substrains
4.5. No Substrain-Dependent Differences in RGC Vulnerability to Glaucomatous Damage
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moons, L.; De Groef, L. Multimodal retinal imaging to detect and understand Alzheimer’s and Parkinson’s disease. Curr. Opin. Neurobiol. 2021, 72, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Christinaki, E.; Kulenovic, H.; Hadoux, X.; Baldassini, N.; Van Eijgen, J.; De Groef, L.; Stalmans, I.; van Wijngaarden, P. Retinal imaging biomarkers of neurodegenerative diseases. Clin. Exp. Optom. 2021, 105, 194–204. [Google Scholar] [CrossRef] [PubMed]
- La Morgia, C.; Di Vito, L.; Carelli, V.; Carbonelli, M. Patterns of Retinal Ganglion Cell Damage in Neurodegenerative Disorders: Parvocellular vs. Magnocellular Degeneration in Optical Coherence Tomography Studies. Front. Neurol. 2017, 8, 710. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.L.; Gordon, L.K. Retinal ganglion cell disorders: Types and treatments. Prog. Retin. Eye Res. 2002, 21, 465–484. [Google Scholar] [CrossRef]
- Jeon, C.-J.; Strettoi, E.; Masland, R.H. The Major Cell Populations of the Mouse Retina. J. Neurosci. 1998, 18, 8936–8946. [Google Scholar] [CrossRef]
- Tran, N.M.; Shekhar, K.; Whitney, I.E.; Jacobi, A.; Benhar, I.; Hong, G.; Yan, W.; Adiconis, X.; Arnold, M.E.; Lee, J.M.; et al. Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron 2019, 104, 1039–1055.e12. [Google Scholar] [CrossRef]
- Claes, M.; Geeraerts, E.; Plaisance, S.; Mentens, S.; Haute, C.V.D.; De Groef, L.; Arckens, L.; Moons, L. Chronic Chemogenetic Activation of the Superior Colliculus in Glaucomatous Mice: Local and Retrograde Molecular Signature. Cells 2022, 11, 1784. [Google Scholar] [CrossRef]
- Jacobi, A.; Tran, N.M.; Yan, W.; Benhar, I.; Tian, F.; Schaffer, R.; He, Z.; Sanes, J.R. Overlapping transcriptional programs promote survival and axonal regeneration of injured retinal ganglion cells. Neuron 2022, 110, 2625–2645.e7. [Google Scholar] [CrossRef]
- Jiang, S.-M.; Zeng, L.-P.; Zeng, J.-H.; Tang, L.; Chen, X.-M.; Wei, X. β-III-Tubulin: A reliable marker for retinal ganglion cell labeling in experimental models of glaucoma. Int. J. Ophthalmol. 2015, 8, 643–652. [Google Scholar] [CrossRef]
- Nadal-Nicolás, F.M.; López, M.J.; Sobrado-Calvo, P.; Nieto-Lo´pez, L.; Ca´novas-Marti´nez, I.; Salinas-Navarro, M.; Vidal-Sanz, M.; Agudo-Barriuso, M. Brn3a as a Marker of Retinal Ganglion Cells: Qualitative and Quantitative Time Course Studies in Naïve and Optic Nerve–Injured Retinas. Investig. Opthalmol. Vis. Sci. 2009, 50, 3860–3868. [Google Scholar] [CrossRef] [Green Version]
- Kwong, J.M.K.; Caprioli, J.; Piri, N. RNA Binding Protein with Multiple Splicing: A New Marker for Retinal Ganglion Cells. Investig. Opthalmol. Vis. Sci. 2010, 51, 1052–1058. [Google Scholar] [CrossRef] [PubMed]
- Geeraerts, E.; Dekeyster, E.; Gaublomme, D.; Salinas-Navarro, M.; De Groef, L.; Moons, L. A freely available semi-automated method for quantifying retinal ganglion cells in entire retinal flatmounts. Exp. Eye Res. 2016, 147, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Guymer, C.; Damp, L.; Chidlow, G.; Wood, J.; Tang, Y.F.; Casson, R. Software for Quantifying and Batch Processing Images of Brn3a and RBPMS Immunolabelled Retinal Ganglion Cells in Retinal Wholemounts. Transl. Vis. Sci. Technol. 2020, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Masin, L.; Claes, M.; Bergmans, S.; Cools, L.; Andries, L.; Davis, B.M.; Moons, L.; De Groef, L. A novel retinal ganglion cell quantification tool based on deep learning. Sci. Rep. 2021, 11, 702. [Google Scholar] [CrossRef]
- Moore, B.A.; Roux, M.J.; Sebbag, L.; Cooper, A.; Edwards, S.G.; Leonard, B.C.; Imai, D.M.; Griffey, S.; Bower, L.; Clary, D.; et al. A Population Study of Common Ocular Abnormalities in C57BL/6Nrd8 Mice. Investig. Opthalmol. Vis. Sci. 2018, 59, 2252–2261. [Google Scholar] [CrossRef]
- Mattapallil, M.J.; Wawrousek, E.F.; Chan, C.-C.; Zhao, H.; Roychoudhury, J.; Ferguson, T.A.; Caspi, R.R. The Rd8 Mutation of the Crb1 Gene Is Present in Vendor Lines of C57BL/6N Mice and Embryonic Stem Cells, and Confounds Ocular Induced Mutant Phenotypes. Investig. Opthalmol. Vis. Sci. 2012, 53, 2921–2927. [Google Scholar] [CrossRef]
- Chang, B.; Hawes, N.; Hurd, R.; Davisson, M.; Nusinowitz, S.; Heckenlively, J. Retinal degeneration mutants in the mouse. Vis. Res. 2002, 42, 517–525. [Google Scholar] [CrossRef]
- Mehalow, A.K.; Kameya, S.; Smith, R.S.; Hawes, N.L.; Denegre, J.M.; Young, J.A.; Bechtold, L.; Haider, N.B.; Tepass, U.; Heckenlively, J.R.; et al. CRB1 is essential for external limiting membrane integrity and photoreceptor morphogenesis in the mammalian retina. Hum. Mol. Genet. 2003, 12, 2179–2189. [Google Scholar] [CrossRef]
- Stojic, A.; Fairless, R.; Beck, S.C.; Sothilingam, V.; Weissgerber, P.; Wissenbach, U.; Gimmy, V.; Seeliger, M.W.; Flockerzi, V.; Diem, R.; et al. Murine Autoimmune Optic Neuritis Is Not Phenotypically Altered by the Retinal Degeneration 8 Mutation. Investig. Opthalmol. Vis. Sci. 2017, 58, 318. [Google Scholar] [CrossRef]
- Ito, Y.A.; Belforte, N.; Vargas, J.L.C.; Di Polo, A. A Magnetic Microbead Occlusion Model to Induce Ocular Hypertension-Dependent Glaucoma in Mice. J. Vis. Exp. 2016, 109, e53731. [Google Scholar] [CrossRef] [Green Version]
- Claes, M.; Santos, J.; Masin, L.; Cools, L.; Davis, B.; Arckens, L.; Farrow, K.; De Groef, L.; Moons, L. A Fair Assessment of Evaluation Tools for the Murine Microbead Occlusion Model of Glaucoma. Int. J. Mol. Sci. 2021, 22, 5633. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Nicolás, F.M.; Salinas-Navarro, M.; Vidal-Sanz, M.; Agudo-Barriuso, M. Two methods to trace retinal ganglion cells with fluorogold: From the intact optic nerve or by stereotactic injection into the optic tract. Exp. Eye Res. 2015, 131, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Navarro, M.; Jiménez-López, M.; Valiente-Soriano, F.; Alarcón-Martínez, L.; Avilés-Trigueros, M.; Mayor, S.; Holmes, T.; Lund, R.; Villegas-Pérez, M.; Vidal-Sanz, M. Retinal ganglion cell population in adult albino and pigmented mice: A computerized analysis of the entire population and its spatial distribution. Vis. Res. 2009, 49, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.W.; Strom, R.C.; Rice, D.S.; Goldowitz, D. Genetic and Environmental Control of Variation in Retinal Ganglion Cell Number in Mice. J. Neurosci. 1996, 16, 7193–7205. [Google Scholar] [CrossRef]
- Sappington, R.; Carlson, B.J.; Crish, S.D.; Calkins, D.J. The Microbead Occlusion Model: A Paradigm for Induced Ocular Hypertension in Rats and Mice. Investig. Opthalmol. Vis. Sci. 2010, 51, 207–216. [Google Scholar] [CrossRef]
- Robinson, G.; Madison, R. Axotomized mouse retinal ganglion cells containing melanopsin show enhanced survival, but not enhanced axon regrowth into a peripheral nerve graft. Vis. Res. 2004, 44, 2667–2674. [Google Scholar] [CrossRef]
- Cone, F.E.; Gelman, S.E.; Son, J.L.; Pease, M.; Quigley, H.A. Differential susceptibility to experimental glaucoma among 3 mouse strains using bead and viscoelastic injection. Exp. Eye Res. 2010, 91, 415–424. [Google Scholar] [CrossRef]
- Templeton, J.P.; Struebing, F.L.; Lemmon, A.; Geisert, E.E. ImagePAD, a novel counting application for the Apple iPad®, used to quantify axons in the Mouse Optic Nerve. Exp. Eye Res. 2014, 128, 102–108. [Google Scholar] [CrossRef]
- Steinhart, M.R.; Cone, F.E.; Nguyen, C.; Nguyen, T.D.; Pease, M.E.; Puk, O.; Graw, J.; Oglesby, E.N.; Quigley, H.A. Mice with an induced mutation in collagen 8A2 develop larger eyes and are resistant to retinal ganglion cell damage in an experimental glaucoma model. Mol. Vis. 2012, 18, 1093–1106. [Google Scholar]
- Ward, N.J.; Ho, K.W.; Lambert, W.; Weitlauf, C.; Calkins, D.J. Absence of Transient Receptor Potential Vanilloid-1 Accelerates Stress-Induced Axonopathy in the Optic Projection. J. Neurosci. 2014, 34, 3161–3170. [Google Scholar] [CrossRef]
- Pang, J.-J.; Wu, S.M. Morphology and Immunoreactivity of Retrogradely Double-Labeled Ganglion Cells in the Mouse Retina. Investig. Opthalmol. Vis. Sci. 2011, 52, 4886–4896. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.K.; Tse, D.Y.; van der Heijden, M.E.; Shah, P.; Nusbaum, D.M.; Yang, Z.; Wu, S.M.; Frankfort, B.J. Prolonged elevation of intraocular pressure results in retinal ganglion cell loss and abnormal retinal function in mice. Exp. Eye Res. 2014, 130, 29–37. [Google Scholar] [CrossRef]
- Dibas, A.; Millar, C.; Al-Farra, A.; Yorio, T. Neuroprotective Effects of Psalmotoxin-1, an Acid-Sensing Ion Channel (ASIC) Inhibitor, in Ischemia Reperfusion in Mouse Eyes. Curr. Eye Res. 2018, 43, 921–933. [Google Scholar] [CrossRef] [PubMed]
- Stankowska, D.L.; Dibas, A.; Li, L.; Zhang, W.; Krishnamoorthy, V.R.; Chavala, S.H.; Nguyen, T.P.; Yorio, T.; Ellis, D.Z.; Acharya, S. Hybrid Compound SA-2 is Neuroprotective in Animal Models of Retinal Ganglion Cell Death. Investig. Opthalmol. Vis. Sci. 2019, 60, 3064–3073. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Romero, C.; Avilés-Trigueros, M.; Jiménez-López, M.; Valiente-Soriano, F.; Salinas-Navarro, M.; Nadal-Nicolás, F.; Villegas-Pérez, M.; Vidal-Sanz, M.; Agudo-Barriuso, M. Axotomy-induced retinal ganglion cell death in adult mice: Quantitative and topographic time course analyses. Exp. Eye Res. 2011, 92, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Zurita, E.; Chagoyen, M.; Cantero, M.; Alonso, R.; Gonzalez-Neira, A.; López-Jiménez, A.; Moreno, J.A.L.; Landel, C.P.; Benítez, J.; Pazos, F.; et al. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 2010, 20, 481–489. [Google Scholar] [CrossRef]
- Mekada, K.; Abe, K.; Murakami, A.; Nakamura, S.; Nakata, H.; Moriwaki, K.; Obata, Y.; Yoshiki, A. Genetic Differences among C57BL/6 Substrains. Exp. Anim. 2009, 58, 141–149. [Google Scholar] [CrossRef]
- Simon, M.M.; Greenaway, S.; White, J.K.; Fuchs, H.; Gailus-Durner, V.; Wells, S.; Sorg, T.; Wong, K.; Bedu, E.; Cartwright, E.J.; et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013, 14, R82. [Google Scholar] [CrossRef]
- Keane, T.M.; Goodstadt, L.; Danecek, P.; White, M.A.; Wong, K.; Yalcin, B.; Heger, A.; Agam, A.; Slater, G.; Goodson, M.; et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature 2011, 477, 289–294. [Google Scholar] [CrossRef]
- Nemoto, S.; Kubota, T. Metabolic differences and differentially expressed genes between C57BL/6J and C57BL/6N mice substrains. bioRxiv 2022. [Google Scholar] [CrossRef]
- Concas, D.; Cater, H.; Wells, S. A scoring system for the evaluation of the mutated Crb1/rd8-derived retinal lesions in C57BL/6N mice. F1000Research 2017, 6, 404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capri, K.M.; Maroni, M.; Deane, H.V.; Concepcion, H.A.; Decourcey, H.; Logan, R.W.; Seggio, J.A. Male C57BL6/N and C57BL6/J Mice Respond Differently to Constant Light and Running-Wheel Access. Front. Behav. Neurosci. 2019, 13, 268. [Google Scholar] [CrossRef] [PubMed]
- Aredo, B.; Zhang, K.; Chen, X.; Wang, C.X.-Z.; Li, T.; Ufret-Vincenty, R.L. Differences in the distribution, phenotype and gene expression of subretinal microglia/macrophages in C57BL/6N (Crb1rd8/rd8) versus C57BL6/J (Crb1wt/wt) mice. J. Neuroinflamm. 2015, 12, 6. [Google Scholar] [CrossRef] [PubMed]
- Schnabolk, G.; Stauffer, K.; O’Quinn, E.; Coughlin, B.; Kunchithapautham, K.; Rohrer, B. A comparative analysis of C57BL/6J and 6N substrains; chemokine/cytokine expression and susceptibility to laser-induced choroidal neovascularization. Exp. Eye Res. 2014, 129, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Barlek, M.H.; Rouan, J.R.; Wyatt, T.G.; Helenowski, I.; Kibbe, M.R. The Persistence of Sex Bias in High-Impact Clinical Research. J. Surg. Res. 2022, 278, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Higginbotham, E.J. Does Sex Matter in Glaucoma? Arch. Ophthalmol. 2004, 122, 374–375. [Google Scholar] [CrossRef]
- Vajaranant, T.S.; Nayak, S.; Wilensky, J.T.; Joslin, C.E. Gender and glaucoma: What we know and what we need to know. Curr. Opin. Ophthalmol. 2010, 21, 91–99. [Google Scholar] [CrossRef]
- Ooto, S.; Hangai, M.; Tomidokoro, A.; Saito, H.; Araie, M.; Otani, T.; Kishi, S.; Matsushita, K.; Maeda, N.; Shirakashi, M.; et al. Effects of Age, Sex, and Axial Length on the Three-Dimensional Profile of Normal Macular Layer Structures. Investig. Opthalmol. Vis. Sci. 2011, 52, 8769–8779. [Google Scholar] [CrossRef]
- Schuman, M.W.; Dubis, A.M.; Nordgren, R.N.; Lei, Y.; Odell, D.; Chiao, H.; Weh, E.; Fischer, W.; Sulai, Y.; Dubra, A.; et al. Race- and Sex-Related Differences in Retinal Thickness and Foveal Pit Morphology. Investig. Opthalmol. Vis. Sci. 2011, 52, 625–634. [Google Scholar] [CrossRef]
- Birch, D.G. Standardized Full-Field Electroretinography. Arch. Ophthalmol. 1992, 110, 1571–1576. [Google Scholar] [CrossRef]
- Brûlé, J.; Lavoie, M.-P.; Casanova, C.; Lachapelle, P.; Hébert, M. Evidence of a possible impact of the menstrual cycle on the reproducibility of scotopic ERGs in women. Doc. Ophthalmol. 2007, 114, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Van Alphen, B.; Winkelman, B.H.J.; Frens, M.A.; Owen, C.G.; Rudnicka, A.R.; Mullen, R.; Barman, S.A.; Monekosso, D.; Whincup, P.H.; Ng, J.; et al. Age- and Sex-Related Differences in Contrast Sensitivity in C57Bl/6 Mice. Investig. Opthalmol. Vis. Sci. 2009, 50, 2451–2458. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Mangold, C.A.; Bixler, G.V.; Brucklacher, R.M.; Masser, D.R.; Stout, M.B.; Elliott, M.H.; Freeman, W.M. Retinal gene expression responses to aging are sexually divergent. Mol. Vis. 2017, 23, 707–717. [Google Scholar] [PubMed]
- Li, B.; Gografe, S.; Munchow, A.; Lopez-Toledano, M.; Pan, Z.-H.; Shen, W. Sex-related differences in the progressive retinal degeneration of the rd10 mouse. Exp. Eye Res. 2019, 187, 107773. [Google Scholar] [CrossRef]
- Guarneri, R.; Russo, D.; Cascio, C.; D’Agostino, S.; Galizzi, G.; Bigini, P.; Mennini, T.; Guarneri, P. Retinal oxidation, apoptosis and age- and sex-differences in the mnd mutant mouse, a model of neuronal ceroid lipofuscinosis. Brain Res. 2004, 1014, 209–220. [Google Scholar] [CrossRef]
- Kumari, R.; Astafurov, K.; Genis, A.; Danias, J. Differential Effects of C1qa Ablation on Glaucomatous Damage in Two Sexes in DBA/2NNia Mice. PLoS ONE 2015, 10, e0142199. [Google Scholar] [CrossRef]
- Aleman, T.S.; Cideciyan, A.V.; Aguirre, G.K.; Huang, W.C.; Mullins, C.L.; Roman, A.J.; Sumaroka, A.; Olivares, M.B.; Tsai, F.F.; Schwartz, S.B.; et al. Human CRB1-Associated Retinal Degeneration: Comparison with the rd8 Crb1-Mutant Mouse Model. Investig. Opthalmol. Vis. Sci. 2011, 52, 6898–6910. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Ii, J.M.D.; Balaiya, S.; Grover, S.; Chalam, K.V. Retinal Thickness Normative Data in Wild-Type Mice Using Customized Miniature SD-OCT. PLoS ONE 2013, 8, e67265. [Google Scholar] [CrossRef]
- De Groef, L.; Dekeyster, E.; Geeraerts, E.; Lefevere, E.; Stalmans, I.; Salinas-Navarro, M.; Moons, L. Differential visual system organization and susceptibility to experimental models of optic neuropathies in three commonly used mouse strains. Exp. Eye Res. 2016, 145, 235–247. [Google Scholar] [CrossRef]
- Pak, J.S.; Lee, E.-J.; Craft, C.M. The retinal phenotype of Grk1-/- is compromised by a Crb1 rd8 mutation. Mol. Vis. 2015, 21, 1281–1294. [Google Scholar]
- Lucas-Ruiz, F.; Galindo-Romero, C.; Rodríguez-Ramírez, K.T.; Vidal-Sanz, M.; Agudo-Barriuso, M. Neuronal Death in the Contralateral Un-Injured Retina after Unilateral Axotomy: Role of Microglial Cells. Int. J. Mol. Sci. 2019, 20, 5733. [Google Scholar] [CrossRef] [PubMed]
- Lucas-Ruiz, F.; Galindo-Romero, C.; Albaladejo-García, V.; Vidal-Sanz, M.; Agudo-Barriuso, M. Mechanisms implicated in the contralateral effect in the central nervous system after unilateral injury: Focus on the visual system. Neural Regen. Res. 2021, 16, 2125–2131. [Google Scholar] [CrossRef] [PubMed]
- Macharadze, T.; Goldschmidt, J.; Marunde, M.; Wanger, T.; Scheich, H.; Zuschratter, W.; Gundelfinger, E.D.; Kreutz, M.R. Interretinal transduction of injury signals after unilateral optic nerve crush. NeuroReport 2009, 20, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Bodeutsch, N.; Siebert, H.; Dermon, C.; Thanos, S. Unilateral injury to the adult rat optic nerve causes multiple cellular responses in the contralateral site. J. Neurobiol. 1999, 38, 116–128. [Google Scholar] [CrossRef]
- Panagis, L.; Thanos, S.; Fischer, D.; Dermon, C.R. Unilateral optic nerve crush induces bilateral retinal glial cell proliferation. Eur. J. Neurosci. 2005, 21, 2305–2309. [Google Scholar] [CrossRef]
- Cen, L.; Han, M.; Zhou, L.; Tan, L.; Liang, J.; Pang, C.; Zhang, M. Bilateral retinal microglial response to unilateral optic nerve transection in rats. Neuroscience 2015, 311, 56–66. [Google Scholar] [CrossRef]
- Sobrado-Calvo, P.; Vidal-Sanz, M.; Villegas-Pérez, M.P. Rat retinal microglial cells under normal conditions, after optic nerve section, and after optic nerve section and intravitreal injection of trophic factors or macrophage inhibitory factor. J. Comp. Neurol. 2007, 501, 866–878. [Google Scholar] [CrossRef]
- Ananthakrishnan, L.; Gervasi, C.; Szaro, B. Dynamic regulation of middle neurofilament RNA pools during optic nerve regeneration. Neuroscience 2008, 153, 144–153. [Google Scholar] [CrossRef]
- Kanamori, A.; Nakamura, M.; Nakanishi, Y.; Yamada, Y.; Negi, A. Long-term glial reactivity in rat retinas ipsilateral and contralateral to experimental glaucoma. Exp. Eye Res. 2005, 81, 48–56. [Google Scholar] [CrossRef]
- Gallego, B.I.; Salazar, J.J.; De Hoz, R.; Rojas, B.; Ramírez, I.A.; Navarro, M.S.; Ortín-Martínez, A.; Soriano, F.J.V.; Avilés-Trigueros, M.; Pérez, M.P.V.; et al. IOP induces upregulation of GFAP and MHC-II and microglia reactivity in mice retina contralateral to experimental glaucoma. J. Neuroinflamm. 2012, 9, 92. [Google Scholar] [CrossRef]
- Cooper, M.L.; Pasini, S.; Lambert, W.S.; D’Alessandro, K.B.; Yao, V.; Risner, M.L.; Calkins, D.J. Redistribution of metabolic resources through astrocyte networks mitigates neurodegenerative stress. Proc. Natl. Acad. Sci. USA 2020, 117, 18810–18821. [Google Scholar] [CrossRef] [PubMed]

| C57BL/6J | C57BL/6N | |||
|---|---|---|---|---|
| Female | Male | Female | Male | |
| Sample size | 17 | 21 | 8 | 22 |
| Area (mm2) | 14.09 ± 1.11 | 14.43 ± 1.22 | 13.4 ± 1.16 | 13.46 ± 0.84 |
| 14.28 ± 1.17 | 13.44 ± 0.91 | |||
| Count (number of RBPMS+ cells) | 46,225 ± 2655 | 46,533 ± 2178 | 40,203 ± 2685 | 40,609 ± 2880 |
| 46,395 ± 2,373 | 40,501 ± 2788 | |||
| Density (number of RBPMS+ cells/mm2) | 3336 ± 144 | 3212 ± 184 | 3009 ± 161 | 3021 ± 201 |
| 3268 ± 177 | 3018 ± 189 | |||
| Retinal Layer Thickness (µm) | |||||||
|---|---|---|---|---|---|---|---|
| NFL + GCL | IPL | INL | OPL | ONL | PL | TOTAL | |
| C75BL/6J (n = 13) | 12.27 ± 0.70 | 49.25 ± 2.36 | 29.36 ± 1.40 | 9.11 ± 0.70 | 62.68 ± 1.44 | 45.30 ± 1.60 | 208.00 ± 3.63 |
| C75BL/6N (n = 10) | 12.80 ± 1.18 | 50.99 ± 1.20 | 29.59 ± 1.32 | 9.85 ± 0.94 | 61.58 ± 1.44 | 41.86 ± 1.91 | 206.60 ± 1.90 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Claes, M.; Moons, L. Retinal Ganglion Cells: Global Number, Density and Vulnerability to Glaucomatous Injury in Common Laboratory Mice. Cells 2022, 11, 2689. https://doi.org/10.3390/cells11172689
Claes M, Moons L. Retinal Ganglion Cells: Global Number, Density and Vulnerability to Glaucomatous Injury in Common Laboratory Mice. Cells. 2022; 11(17):2689. https://doi.org/10.3390/cells11172689
Chicago/Turabian StyleClaes, Marie, and Lieve Moons. 2022. "Retinal Ganglion Cells: Global Number, Density and Vulnerability to Glaucomatous Injury in Common Laboratory Mice" Cells 11, no. 17: 2689. https://doi.org/10.3390/cells11172689
APA StyleClaes, M., & Moons, L. (2022). Retinal Ganglion Cells: Global Number, Density and Vulnerability to Glaucomatous Injury in Common Laboratory Mice. Cells, 11(17), 2689. https://doi.org/10.3390/cells11172689






