Influence of the Postmortem/Storage Time of Human Corneas on the Properties of Cultured Limbal Epithelial Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells Isolation and Culture
2.2. Growth Rate Measurements
2.3. Colony-Forming Efficiency Assays
2.4. Human Corneal Epithelial Stem Cells Count
(over 0.120)/total number of cells × 100
2.5. Scratch Wound Assay
2.6. Production of the Human-Tissue-Engineered Corneas and Wound Healing Assays
2.7. Histological Analyses
2.8. Immunofluorescence Analyses
2.9. Western Blot
2.10. Gene Expression Profiling
2.11. Bioinformatics and Statistical Analyses
3. Results
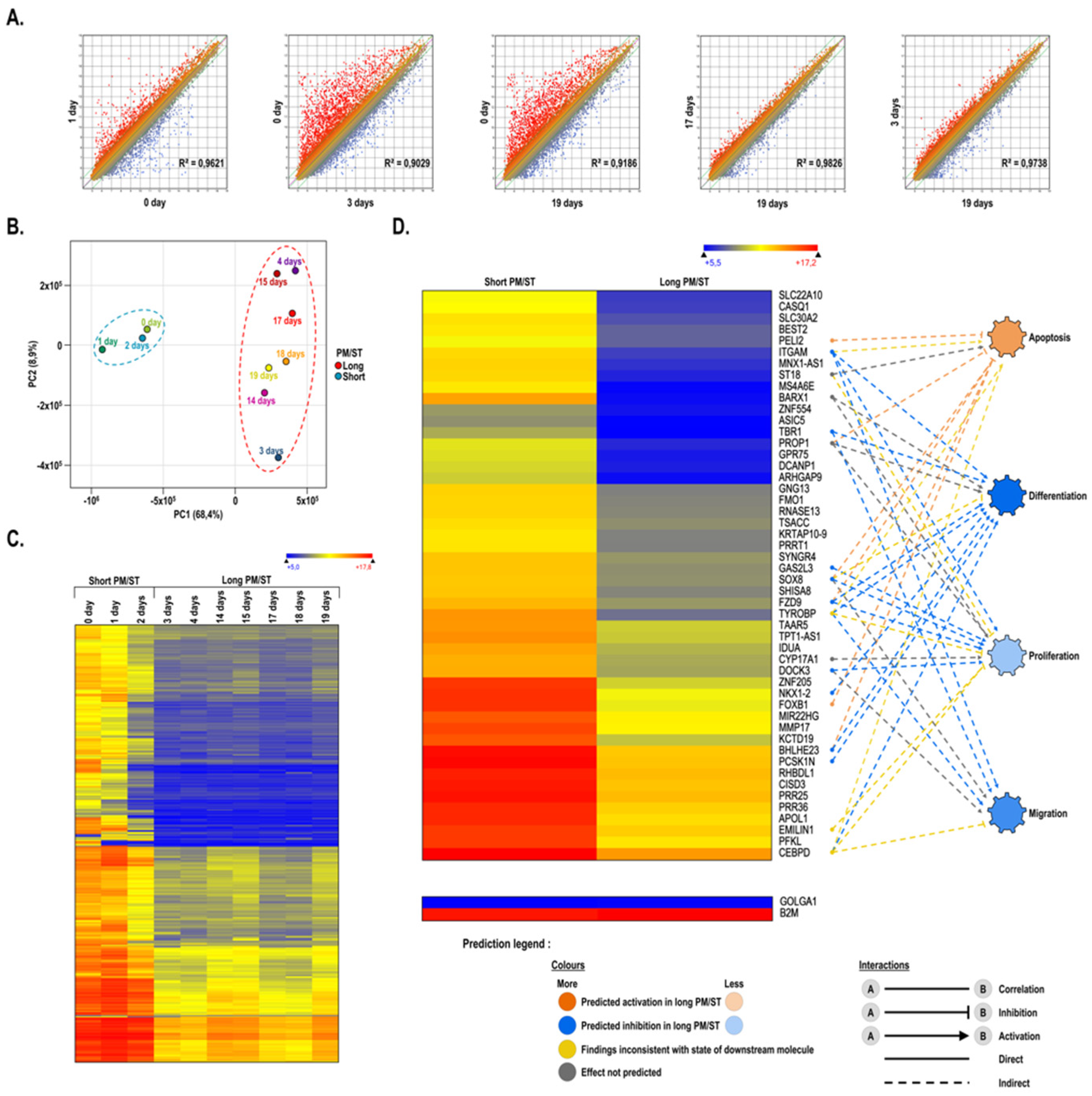
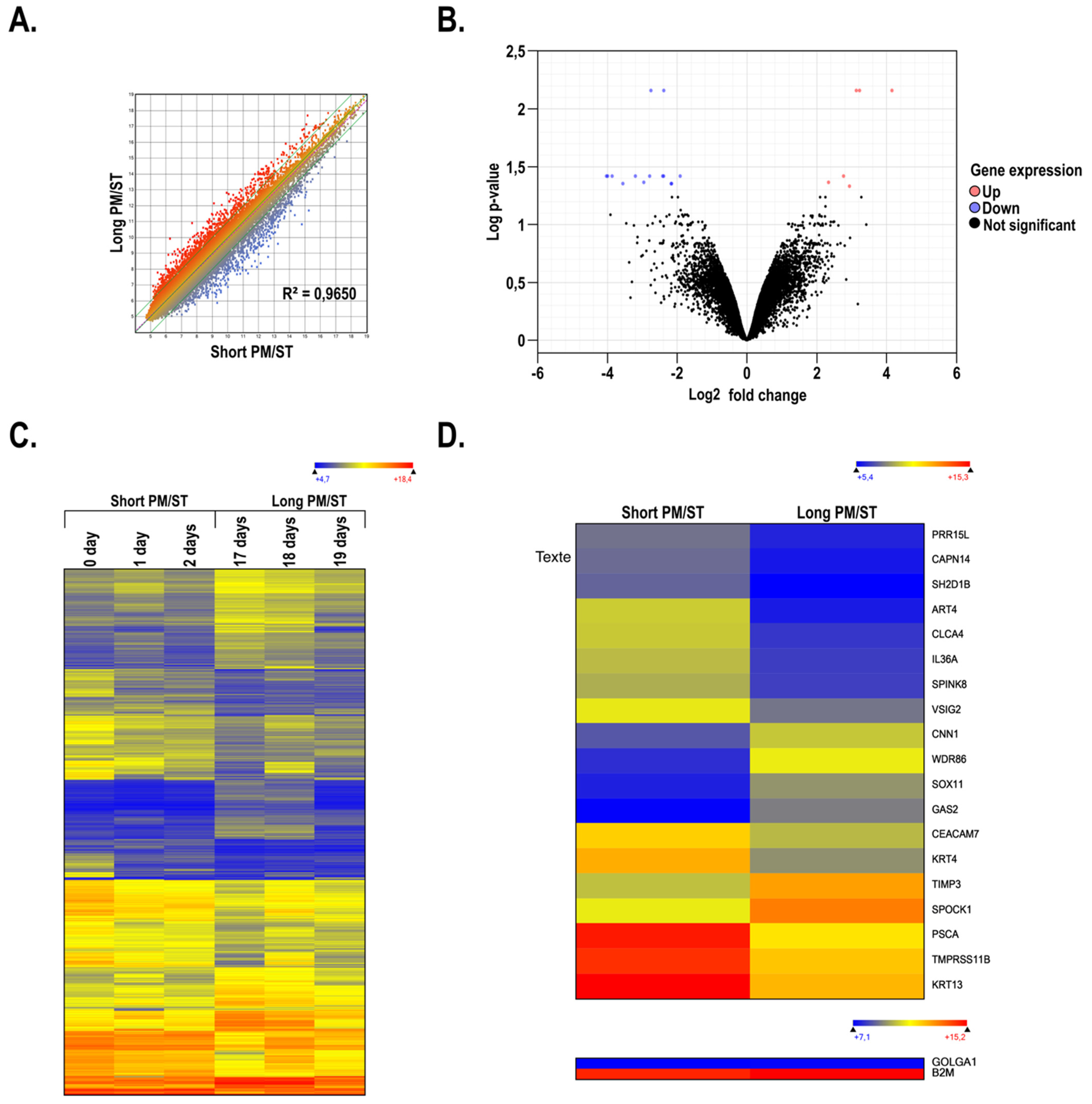
3.1. Gene Profiling of Short and Long PM/ST hLECs Grown in Monolayer
3.2. Gene Interaction Network Assisted in Silico Predictions of Biological Functions Regulation in Short and Long PM/ST hLECs Cultured in Monolayers
3.3. Growth Rate, Colony-Forming Efficiency, Cell Size, Morphology, and Stem Cell Analysis of Short and Long PM/ST hLECs Grown in Monolayer
3.4. Wound Closure of Short and Long PM/ST hLECs Grown in Monolayer
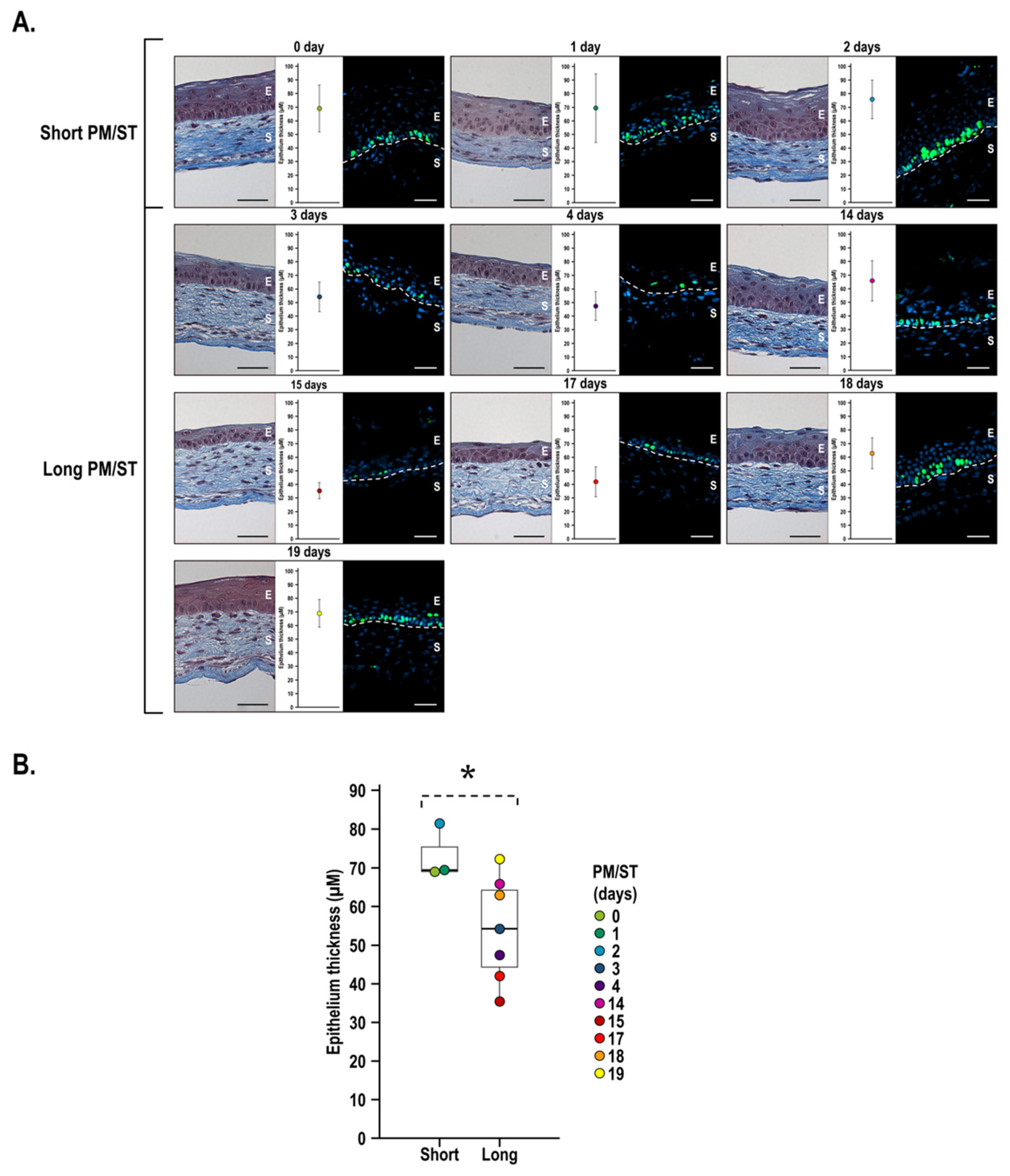
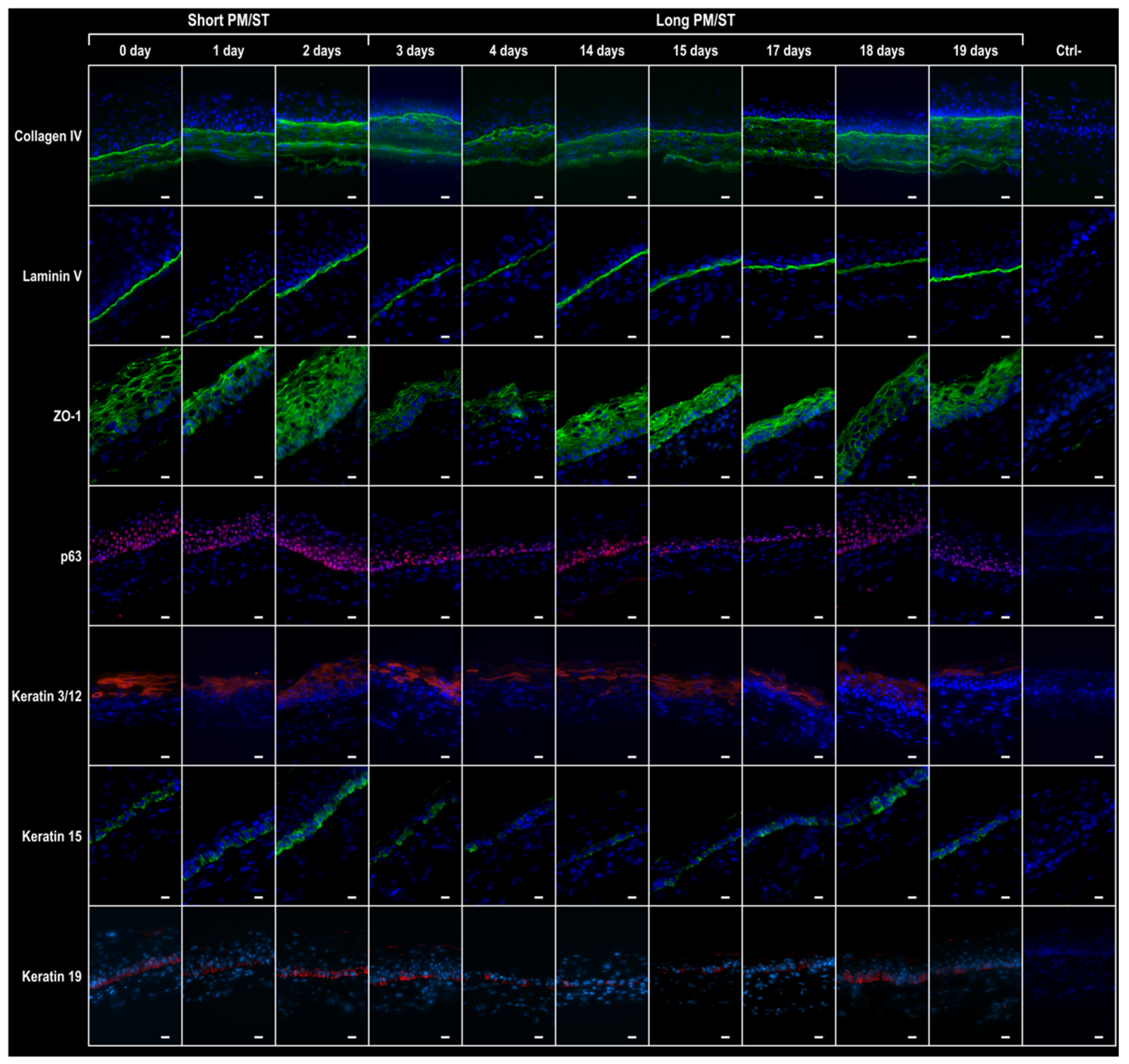
3.5. Morphology and Thickness of the Stratified Corneal Epithelium When Short and Long PM/ST hLECs Are Used for the Production of Human-Tissue-Engineered Corneas (hTECs)
3.6. Integrity of the hTECs Produced Using Short and Long PM/ST hLECs
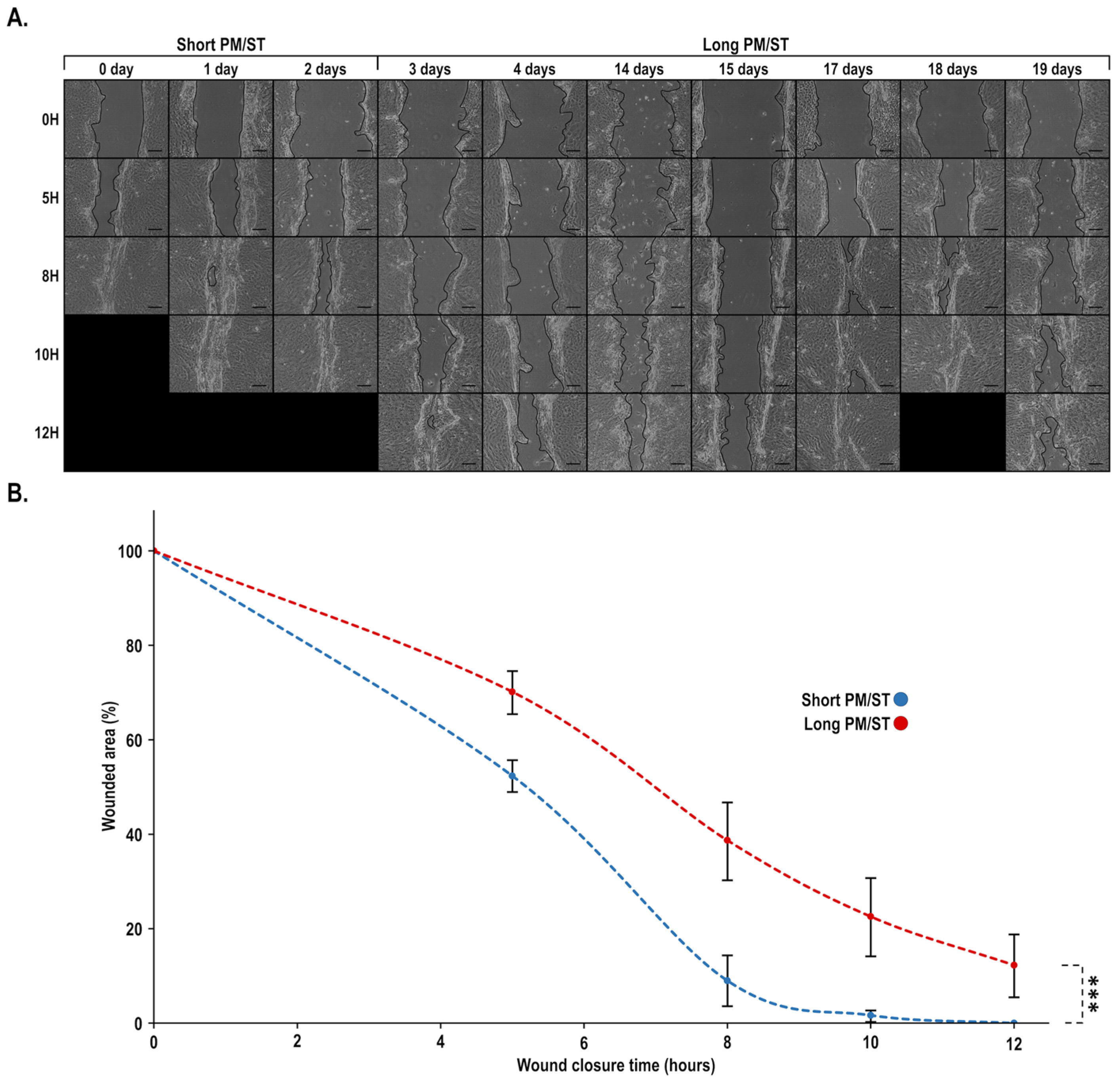
3.7. Wound Closure Dynamic of hTECs Produced Using Short and Long PM/ST hLECs
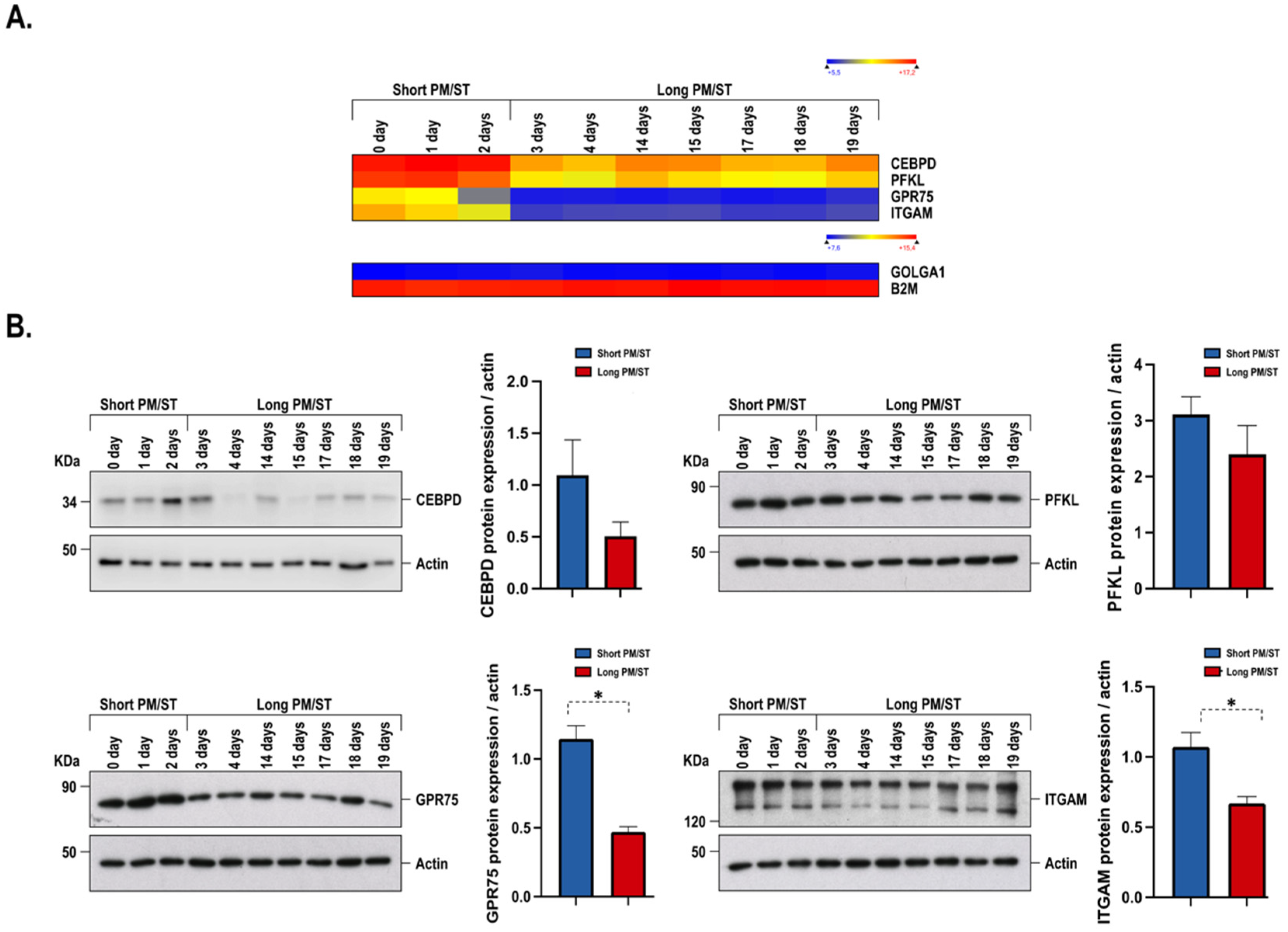
3.8. Gene Profiling on Microarrays of hTECs Produced Using Short and Long PM/ST hLECs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Atallah, M.R.; Palioura, S.; Perez, V.L.; Amescua, G. Limbal stem cell transplantation: Current perspectives. Clin. Ophthalmol. 2016, 10, 593–602. [Google Scholar] [PubMed]
- Daniels, J.T.; Dart, J.K.; Tuft, S.J.; Khaw, P.T. Corneal stem cells in review. Wound Rep. Regen. 2001, 9, 483–494. [Google Scholar] [CrossRef]
- Shapiro, M.S.; Friend, J.; Thoft, R.A. Corneal re-epithelialization from the conjunctiva. Investig. Ophthalmol. Vis. Sci. 1981, 21 Pt 1, 135–142. [Google Scholar]
- Dua, H.S.; Azuara-Blanco, A. Limbal stem cells of the corneal epithelium. Surv. Ophthalmol. 2000, 44, 415–425. [Google Scholar] [CrossRef]
- Rama, P.; Matuska, S.; Paganoni, G.; Spinelli, A.; De Luca, M.; Pellegrini, G. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010, 363, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Basu, S.; Fernandez, M.M.; Das, S.; Gaddipati, S.; Vemuganti, G.K.; Sangwan, V.S. Clinical outcomes of xeno-free allogeneic cultivated limbal epithelial transplantation for bilateral limbal stem cell deficiency. Br. J. Ophthalmol. 2012, 96, 1504–1509. [Google Scholar] [CrossRef] [PubMed]
- Shortt, A.J.; Bunce, C.; Levis, H.J.; Blows, P.; Dore, C.J.; Vernon, A.; Secker, G.A.; Tuft, S.J.; Daniels, J.T. Three-year outcomes of cultured limbal epithelial allografts in aniridia and Stevens-Johnson syndrome evaluated using the Clinical Outcome Assessment in Surgical Trials assessment tool. Stem Cells Transl. Med. 2014, 3, 265–275. [Google Scholar] [CrossRef]
- Titiyal, J.S.; Sharma, N.; Agarwal, A.K.; Prakash, G.; Tandon, R.; Vajpayee, R. Live Related versus Cadaveric Limbal Allograft in Limbal Stem Cell Deficiency. Ocul. Immunol. Inflamm. 2015, 23, 232–239. [Google Scholar] [CrossRef]
- Feizi, S.; Javadi, M.A.; Kanavi, M.R.; Javadi, F. Effect of donor graft quality on clinical outcomes after deep anterior lamellar keratoplasty. Cornea 2014, 33, 795–800. [Google Scholar] [CrossRef]
- Nioi, M.; Napoli, P.E.; Demontis, R.; Locci, E.; Fossarello, M.; d’Aloja, E. Morphological analysis of corneal findings modifications after death: A preliminary OCT study on an animal model. Exp. Eye Res. 2018, 169, 20–27. [Google Scholar] [CrossRef]
- Brett, S.J. Immunodepression in Giardia muris and Spironucleus muris infections in mice. Parasitology 1983, 87 Pt 3, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Van Meter, W.S.; Katz, D.G.; White, H.; Gayheart, R. Effect of death-to-preservation time on donor corneal epithelium. Trans. Am. Ophthalmol. Soc. 2005, 103, 209–222. [Google Scholar] [PubMed]
- Scott, A.; Olack, B.; Rouch, J.D.; Khalil, H.A.; Kokubun, B.A.; Lei, N.Y.; Wang, J.; Solorzano, S.; Lewis, M.; Dunn, J.C.Y.; et al. Comparison of Surgical and Cadaveric Intestine as a Source of Crypt Culture in Humans. Cell Transpl. 2020, 29, 963689720903709. [Google Scholar] [CrossRef]
- Yesus, Y.W.; Kenneally, C.; Taylor, H.M. Preservation of hematopoietic stem cells in cadaveric marrow. Am. J. Clin. Pathol. 1981, 76, 205–207. [Google Scholar] [CrossRef] [PubMed]
- Pels, E.; Beele, H.; Claerhout, I. Eye bank issues: II. Preservation techniques: Warm versus cold storage. Int. Ophthalmol. 2008, 28, 155–163. [Google Scholar] [PubMed]
- Pels, E.; Schuchard, Y. Organ-culture preservation of human corneas. Doc. Ophthalmol. 1983, 56, 147–153. [Google Scholar] [CrossRef]
- Ehlers, H.; Ehlers, N.; Hjortdal, J.O. Corneal transplantation with donor tissue kept in organ culture for 7 weeks. Acta Ophthalmol. Scand. 1999, 77, 277–278. [Google Scholar] [CrossRef]
- Lindstrom, R.L.; Kaufman, H.E.; Skelnik, D.L.; Laing, R.A.; Lass, J.H.; Musch, D.C.; Trousdale, M.D.; Reinhart, W.J.; Burris, T.E.; Sugar, A.; et al. Optisol corneal storage medium. Am. J. Ophthalmol. 1992, 114, 345–356. [Google Scholar] [CrossRef]
- Armitage, W.J. Preservation of Human Cornea. Transfus. Med. Hemother. 2011, 38, 143–147. [Google Scholar] [CrossRef]
- Andersen, J.; Ehlers, N. The influence of donor age and post mortem time on corneal graft survival and thickness when employing banked donor material (A five-year follow-up). Acta Ophthalmol. 1988, 66, 313–317. [Google Scholar] [CrossRef]
- Ehlers, N. Corneal banking and grafting: The background to the Danish Eye Bank System, where corneas await their patients. Acta Ophthalmol. Scand. 2002, 80, 572–578. [Google Scholar] [CrossRef] [PubMed]
- Slettedal, J.K.; Lyberg, T.; Ramstad, H.; Nicolaissen, B. Donor corneas for transplantation: A scanning electron microscopic study of the epithelium. Acta Ophthalmol. Scand. 2006, 84, 516–521. [Google Scholar] [CrossRef]
- Kim, T.; Palay, D.A.; Lynn, M. Donor factors associated with epithelial defects after penetrating keratoplasty. Cornea 1996, 15, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Schaub, F.; Enders, P.; Adler, W.; Bachmann, B.O.; Cursiefen, C.; Heindl, L.M. Impact of donor graft quality on deep anterior lamellar Keratoplasty (DALK). BMC Ophthalmol. 2017, 17, 204. [Google Scholar] [CrossRef]
- Borderie, V.M.; Sandali, O.; Basli, E.; Goldschmidt, P.; Laroche, L. Donor tissue selection for anterior lamellar keratoplasty. Cornea 2013, 32, 1105–1109. [Google Scholar] [CrossRef]
- Shanmuganathan, V.A.; Rotchford, A.P.; Tullo, A.B.; Joseph, A.; Zambrano, I.; Dua, H.S. Epithelial proliferative potential of organ cultured corneoscleral rims; implications for allo-limbal transplantation and eye banking. Br. J. Ophthalmol. 2006, 90, 55–58. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Jun Song, X.; de Paiva, C.S.; Chen, Z.; Pflugfelder, S.C.; Li, D.Q. Phenotypic characterization of human corneal epithelial cells expanded ex vivo from limbal explant and single cell cultures. Exp. Eye Res. 2004, 79, 41–49. [Google Scholar] [CrossRef]
- Baylis, O.; Rooney, P.; Figueiredo, F.; Lako, M.; Ahmad, S. An investigation of donor and culture parameters which influence epithelial outgrowths from cultured human cadaveric limbal explants. J. Cell. Physiol. 2013, 228, 1025–1030. [Google Scholar] [CrossRef]
- Liu, T.; Wang, Y.; Duan, H.Y.; Qu, M.L.; Yang, L.L.; Xu, Y.Y.; Zang, X.J.; Zhou, Q.J. Effects of preservation time on proliferative potential of human limbal stem/progenitor cells. Int. J. Ophthalmol. 2012, 5, 549–554. [Google Scholar]
- Romo-Valera, C.; Perez-Garrastachu, M.; Hernaez-Moya, R.; Rodriguez-Astigarraga, M.; Romano-Ruiz, P.; Etxebarria, J.; Arluzea, J.; Andollo, N. Characterisation of corneas following different time and storage methods for their use as a source of stem-like limbal epithelial cells. Exp. Eye Res. 2021, 211, 108720. [Google Scholar] [CrossRef]
- Carrier, P.; Deschambeault, A.; Talbot, M.; Giasson, C.J.; Auger, F.A.; Guerin, S.L.; Germain, L. Characterization of wound reepithelialization using a new human tissue-engineered corneal wound healing model. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1376–1385. [Google Scholar] [CrossRef] [PubMed]
- Bisson, F.; Rochefort, E.; Lavoie, A.; Larouche, D.; Zaniolo, K.; Simard-Bisson, C.; Damour, O.; Auger, F.A.; Guerin, S.L.; Germain, L. Irradiated human dermal fibroblasts are as efficient as mouse fibroblasts as a feeder layer to improve human epidermal cell culture lifespan. Int. J. Mol. Sci. 2013, 14, 4684–4704. [Google Scholar] [CrossRef]
- Le-Bel, G.; Cortez Ghio, S.; Guerin, L.P.; Bisson, F.; Germain, L.; Guerin, S.L. Irradiated Human Fibroblasts as a Substitute Feeder Layer to Irradiated Mouse 3T3 for the Culture of Human Corneal Epithelial Cells: Impact on the Stability of the Transcription Factors Sp1 and NFI. Int. J. Mol. Sci. 2019, 20, 6296 (p.1–21). [Google Scholar] [CrossRef] [PubMed]
- Germain, L.; Auger, F.A.; Grandbois, E.; Guignard, R.; Giasson, M.; Boisjoly, H.; Guerin, S.L. Reconstructed human cornea produced in vitro by tissue engineering. Pathobiology 1999, 67, 140–147. [Google Scholar] [CrossRef]
- Couture, C.; Zaniolo, K.; Carrier, P.; Lake, J.; Patenaude, J.; Germain, L.; Guerin, S.L. The tissue-engineered human cornea as a model to study expression of matrix metalloproteinases during corneal wound healing. Biomaterials 2016, 78, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, P.; Couture, C.; Germain, L.; Guerin, S.L. Contribution of the WNK1 kinase to corneal wound healing using the tissue-engineered human cornea as an in vitro model. J. Tissue Eng. Regen. Med. 2019, 13, 1595–1608. [Google Scholar] [CrossRef]
- Le-Bel, G.; Cortez Ghio, S.; Larouche, D.; Germain, L.; Guerin, S.L. Qualitatively Monitoring Binding and Expression of the Transcription Factors Sp1 and NFI as a Useful Tool to Evaluate the Quality of Primary Cultured Epithelial Stem Cells in Tissue Reconstruction. Methods Mol. Biol. 2019, 1879, 43–73. [Google Scholar]
- Kramer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Yang, J.; Siqueira, M.F.; Behl, Y.; Alikhani, M.; Graves, D.T. The transcription factor ST18 regulates proapoptotic and proinflammatory gene expression in fibroblasts. FASEB J. 2008, 22, 3956–3967. [Google Scholar] [CrossRef]
- Li, Y.; Wang, X.; Huang, Z.; Zhou, Y.; Xia, J.; Hu, W.; Wang, X.; Du, J.; Tong, X.; Wang, Y. CISD3 inhibition drives cystine-deprivation induced ferroptosis. Cell Death Dis. 2021, 12, 839. [Google Scholar] [CrossRef]
- Hevner, R.F.; Shi, L.; Justice, N.; Hsueh, Y.; Sheng, M.; Smiga, S.; Bulfone, A.; Goffinet, A.M.; Campagnoni, A.T.; Rubenstein, J.L. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 2001, 29, 353–366. [Google Scholar] [CrossRef]
- Elsafadi, M.; Manikandan, M.; Alajez, N.M.; Hamam, R.; Dawud, R.A.; Aldahmash, A.; Iqbal, Z.; Alfayez, M.; Kassem, M.; Mahmood, A. MicroRNA-4739 regulates osteogenic and adipocytic differentiation of immortalized human bone marrow stromal cells via targeting LRP3. Stem Cell Res. 2017, 20, 94–104. [Google Scholar] [CrossRef] [PubMed]
- Miletich, I.; Buchner, G.; Sharpe, P.T. Barx1 and evolutionary changes in feeding. J. Anat. 2005, 207, 619–622. [Google Scholar] [CrossRef] [PubMed]
- Barrow, A.D.; Raynal, N.; Andersen, T.L.; Slatter, D.A.; Bihan, D.; Pugh, N.; Cella, M.; Kim, T.; Rho, J.; Negishi-Koga, T.; et al. OSCAR is a collagen receptor that costimulates osteoclastogenesis in DAP12-deficient humans and mice. J. Clin. Investig. 2011, 121, 3505–3516. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Schill, R.L.; O’Donnell, M.; Xu, K.; Bagchi, D.P.; MacDougald, O.A.; Koenig, R.J.; Xu, B. The transcription factor NKX1–2 promotes adipogenesis and may contribute to a balance between adipocyte and osteoblast differentiation. J. Biol. Chem. 2019, 294, 18408–18420. [Google Scholar] [CrossRef]
- Siu, M.K.; Wong, E.S.; Kong, D.S.; Chan, H.Y.; Jiang, L.; Wong, O.G.; Lam, E.W.; Chan, K.K.; Ngan, H.Y.; Le, X.F.; et al. Stem cell transcription factor NANOG controls cell migration and invasion via dysregulation of E-cadherin and FoxJ1 and contributes to adverse clinical outcome in ovarian cancers. Oncogene 2013, 32, 3500–3509. [Google Scholar] [CrossRef]
- Musah, S.; Dimitrakakis, N.; Camacho, D.M.; Church, G.M.; Ingber, D.E. Directed differentiation of human induced pluripotent stem cells into mature kidney podocytes and establishment of a Glomerulus Chip. Nat. Protoc. 2018, 13, 1662–1685. [Google Scholar] [CrossRef]
- Perez Millan, M.I.; Brinkmeier, M.L.; Mortensen, A.H.; Camper, S.A. PROP1 triggers epithelial-mesenchymal transition-like process in pituitary stem cells. Elife 2016, 5, e14470. [Google Scholar] [CrossRef]
- He, H.; Yu, F.; Shen, W.; Chen, K.; Zhang, L.; Lou, S.; Zhang, Q.; Chen, S.; Yuan, X.; Jia, X.; et al. The Novel Key Genes of Non-obstructive Azoospermia Affect Spermatogenesis: Transcriptomic Analysis Based on RNA-Seq and scRNA-Seq Data. Front. Genet. 2021, 12, 608629. [Google Scholar] [CrossRef]
- Barbaro, V.; Testa, A.; Di Iorio, E.; Mavilio, F.; Pellegrini, G.; De Luca, M. C/EBPdelta regulates cell cycle and self-renewal of human limbal stem cells. J. Cell Biol. 2007, 177, 1037–1049. [Google Scholar] [CrossRef]
- Feng, Y.; Zhang, Y.; Cai, Y.; Liu, R.; Lu, M.; Li, T.; Fu, Y.; Guo, M.; Huang, H.; Ou, Y.; et al. A20 targets PFKL and glycolysis to inhibit the progression of hepatocellular carcinoma. Cell Death Dis. 2020, 11, 89. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, R.B.; Mostaghel, E.A.; Vessella, R.; Hess, D.L.; Kalhorn, T.F.; Higano, C.S.; True, L.D.; Nelson, P.S. Maintenance of intratumoral androgens in metastatic prostate cancer: A mechanism for castration-resistant tumor growth. Cancer Res. 2008, 68, 4447–4454. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zhou, H. Clinical effects and molecular mechanisms of lncRNA MNX1-AS1 in malignant tumors. Am. J. Transl. Res. 2020, 12, 7593–7602. [Google Scholar]
- Su, W.; Guo, C.; Wang, L.; Wang, Z.; Yang, X.; Niu, F.; Tzou, D.; Yang, X.; Huang, X.; Wu, J.; et al. LncRNA MIR22HG abrogation inhibits proliferation and induces apoptosis in esophageal adenocarcinoma cells via activation of the STAT3/c-Myc/FAK signaling. Aging 2019, 11, 4587–4596. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Huang, F.; Wang, H.; Cheng, F.; Pi, Y.; Zhao, J.; Li, Z. Knockdown of TPT1-AS1 inhibits cell proliferation, cell cycle G1/S transition, and epithelial-mesenchymal transition in gastric cancer. Bosn. J. Basic Med. Sci. 2021, 21, 39–46. [Google Scholar] [CrossRef]
- Wolter, P.; Schmitt, K.; Fackler, M.; Kremling, H.; Probst, L.; Hauser, S.; Gruss, O.J.; Gaubatz, S. GAS2L3, a target gene of the DREAM complex, is required for proper cytokinesis and genomic stability. J. Cell Sci. 2012, 125 Pt 10, 2393–2406. [Google Scholar] [CrossRef]
- Zacarias-Fluck, M.F.; Jauset, T.; Martinez-Martin, S.; Kaur, J.; Casacuberta-Serra, S.; Masso-Valles, D.; Serrano Del Pozo, E.; Martin-Fernandez, G.; Gonzalez-Larreategui, I.; Lopez-Estevez, S.; et al. The Wnt signaling receptor Fzd9 is essential for Myc-driven tumorigenesis in pancreatic islets. Life Sci. Alliance 2021, 4, e201900490. [Google Scholar] [CrossRef]
- Zhang, C.; Booz, G.W.; Yu, Q.; He, X.; Wang, S.; Fan, F. Conflicting roles of 20-HETE in hypertension and renal end organ damage. Eur. J. Pharm. 2018, 833, 190–200. [Google Scholar] [CrossRef]
- Chen, S.; Li, H.; Li, X.; Chen, W.; Zhang, X.; Yang, Z.; Chen, Z.; Chen, J.; Zhang, Y.; Shi, D.; et al. High SOX8 expression promotes tumor growth and predicts poor prognosis through GOLPH3 signaling in tongue squamous cell carcinoma. Cancer Med. 2020, 9, 4274–4289. [Google Scholar] [CrossRef]
- MacPherson, M.; Lek, H.S.; Prescott, A.; Fagerholm, S.C. A systemic lupus erythematosus-associated R77H substitution in the CD11b chain of the Mac-1 integrin compromises leukocyte adhesion and phagocytosis. J. Biol. Chem. 2011, 286, 17303–17310. [Google Scholar] [CrossRef]
- Spessotto, P.; Bulla, R.; Danussi, C.; Radillo, O.; Cervi, M.; Monami, G.; Bossi, F.; Tedesco, F.; Doliana, R.; Colombatti, A. EMILIN1 represents a major stromal element determining human trophoblast invasion of the uterine wall. J. Cell Sci. 2006, 119 Pt 21, 4574–4584. [Google Scholar] [CrossRef] [PubMed]
- Than, N.G.; Romero, R.; Tarca, A.L.; Kekesi, K.A.; Xu, Y.; Xu, Z.; Juhasz, K.; Bhatti, G.; Leavitt, R.J.; Gelencser, Z.; et al. Integrated Systems Biology Approach Identifies Novel Maternal and Placental Pathways of Preeclampsia. Front. Immunol. 2018, 9, 1661. [Google Scholar] [CrossRef]
- Wang, T.; Ha, M. Silencing ARHGAP9 correlates with the risk of breast cancer and inhibits the proliferation, migration, and invasion of breast cancer. J. Cell. Biochem. 2018, 119, 7747–7756. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.T.; Green, H. Differentiation of the epidermal keratinocyte in cell culture: Formation of the cornified envelope. Cell 1976, 9 Pt 1, 511–521. [Google Scholar] [CrossRef]
- Bocock, J.P.; Edgell, C.J.; Marr, H.S.; Erickson, A.H. Human proteoglycan testican-1 inhibits the lysosomal cysteine protease cathepsin L. Eur. J. Biochem. 2003, 270, 4008–4015. [Google Scholar] [CrossRef]
- Miller, G.S.; Zoratti, G.L.; Murray, A.S.; Bergum, C.; Tanabe, L.M.; List, K. HATL5: A cell surface serine protease differentially expressed in epithelial cancers. PLoS ONE 2014, 9, e87675. [Google Scholar] [CrossRef] [PubMed]
- Pawlowski, K.; Lepisto, M.; Meinander, N.; Sivars, U.; Varga, M.; Wieslander, E. Novel conserved hydrolase domain in the CLCA family of alleged calcium-activated chloride channels. Proteins 2006, 63, 424–439. [Google Scholar] [CrossRef]
- Jalkanen, J.; Kotimaki, M.; Huhtaniemi, I.; Poutanen, M. Novel epididymal protease inhibitors with Kazal or WAP family domain. Biochem. Biophys. Res. Commun. 2006, 349, 245–254. [Google Scholar] [CrossRef]
- Davis, B.P.; Stucke, E.M.; Khorki, M.E.; Litosh, V.A.; Rymer, J.K.; Rochman, M.; Travers, J.; Kottyan, L.C.; Rothenberg, M.E. Eosinophilic esophagitis-linked calpain 14 is an IL-13-induced protease that mediates esophageal epithelial barrier impairment. JCI Insight 2016, 1, e86355. [Google Scholar] [CrossRef]
- Foster, A.M.; Baliwag, J.; Chen, C.S.; Guzman, A.M.; Stoll, S.W.; Gudjonsson, J.E.; Ward, N.L.; Johnston, A. IL-36 promotes myeloid cell infiltration, activation, and inflammatory activity in skin. J. Immunol. 2014, 192, 6053–6061. [Google Scholar] [CrossRef]
- Liu, Y.; Peng, K.; Xie, R.; Zheng, J.; Guo, J.; Wei, R.; Yang, H.; Cai, C.; Wei, Q. Protocadherin gamma-A7 is down-regulated in colorectal cancer and associated with the prognosis in patients with wild-type KRAS. Hum. Pathol. 2019, 83, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, N.; Ni Dhubhghaill, S.; Taal, M.; Berneman, Z.; Koppen, C.; Tassignon, M.J. Optical Coherence Tomography in Cultivated Limbal Epithelial Stem Cell Transplantation Surgery. Asia-Pac. J. Ophthalmol. 2015, 4, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Baylis, O.; Figueiredo, F.; Henein, C.; Lako, M.; Ahmad, S. 13 years of cultured limbal epithelial cell therapy: A review of the outcomes. J. Cell. Biochem. 2011, 112, 993–1002. [Google Scholar] [CrossRef]
- Pellegrini, G.; Rama, P.; Matuska, S.; Lambiase, A.; Bonini, S.; Pocobelli, A.; Colabelli, R.G.; Spadea, L.; Fasciani, R.; Balestrazzi, E.; et al. Biological parameters determining the clinical outcome of autologous cultures of limbal stem cells. Regen. Med. 2013, 8, 553–567. [Google Scholar] [CrossRef] [PubMed]
- Di Iorio, E.; Barbaro, V.; Ferrari, S.; Ortolani, C.; De Luca, M.; Pellegrini, G. Q-FIHC: Quantification of fluorescence immunohistochemistry to analyse p63 isoforms and cell cycle phases in human limbal stem cells. Microsc. Res. Tech. 2006, 69, 983–991. [Google Scholar] [CrossRef]
- Davanger, M.; Evensen, A. Role of the pericorneal papillary structure in renewal of corneal epithelium. Nature 1971, 229, 560–561. [Google Scholar] [CrossRef]
- Scadden, D.T. The stem-cell niche as an entity of action. Nature 2006, 441, 1075–1079. [Google Scholar] [CrossRef]
- Slettedal, J.K.; Lyberg, T.; Ramstad, H.; Beraki, K.; Nicolaissen, B. Regeneration of the epithelium in organ-cultured donor corneas with extended post-mortem time. Acta Ophthalmol. Scand. 2007, 85, 371–376. [Google Scholar] [CrossRef]
- Mason, S.L.; Stewart, R.M.; Sheridan, C.M.; Keshtkar, F.; Rooney, P.; Austin, E.; Schlotzer-Schrehardt, U.; Kruse, F.E.; Kaye, S.B. Yield and Viability of Human Limbal Stem Cells From Fresh and Stored Tissue. Investig. Ophthalmol. Vis. Sci. 2016, 57, 3708–3713. [Google Scholar] [CrossRef]
- Means, T.L.; Geroski, D.H.; L’Hernault, N.; Grossniklaus, H.E.; Kim, T.; Edelhauser, H.F. The corneal epithelium after optisol-GS storage. Cornea 1996, 15, 599–605. [Google Scholar] [CrossRef]
- Tovell, V.E.; Massie, I.; Kureshi, A.K.; Daniels, J.T. Functional limbal epithelial cells can be successfully isolated from organ culture rims following long-term storage. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3531–3540. [Google Scholar] [CrossRef] [PubMed]
- Zito-Abbad, E.; Borderie, V.M.; Baudrimont, M.; Bourcier, T.; Laroche, L.; Chapel, C.; Uzel, J.L. Corneal epithelial cultures generated from organ-cultured limbal tissue: Factors influencing epithelial cell growth. Curr. Eye Res. 2006, 31, 391–399. [Google Scholar] [CrossRef]
- James, S.E.; Rowe, A.; Ilari, L.; Daya, S.; Martin, R. The potential for eye bank limbal rings to generate cultured corneal epithelial allografts. Cornea 2001, 20, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Vemuganti, G.K.; Kashyap, S.; Sangwan, V.S.; Singh, S. Ex-vivo potential of cadaveric and fresh limbal tissues to regenerate cultured epithelium. Indian J. Ophthalmol. 2004, 52, 113–120. [Google Scholar]
- Crewe, J.M.; Armitage, W.J. Integrity of epithelium and endothelium in organ-cultured human corneas. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1757–1761. [Google Scholar]
- Redbrake, C.; Salla, S.; Frantz, A.; Reim, M. Metabolic changes of the human donor cornea during organ-culture. Acta Ophthalmol. Scand. 1999, 77, 266–272. [Google Scholar] [CrossRef]
- Ramji, D.P.; Foka, P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem. J. 2002, 365 Pt 3, 561–575. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.F. Molecular stop signs: Regulation of cell-cycle arrest by C/EBP transcription factors. J. Cell Sci. 2005, 118 Pt 12, 2545–2555. [Google Scholar] [CrossRef]
- O’Rourke, J.P.; Newbound, G.C.; Hutt, J.A.; DeWille, J. CCAAT/enhancer-binding protein delta regulates mammary epithelial cell G0 growth arrest and apoptosis. J. Biol. Chem. 1999, 274, 16582–16589. [Google Scholar] [CrossRef]
- Hutt, J.A.; O’Rourke, J.P.; DeWille, J. Signal transducer and activator of transcription 3 activates CCAAT enhancer-binding protein delta gene transcription in G0 growth-arrested mouse mammary epithelial cells and in involuting mouse mammary gland. J. Biol. Chem. 2000, 275, 29123–29131. [Google Scholar] [CrossRef]
- Scandura, J.M.; Boccuni, P.; Massague, J.; Nimer, S.D. Transforming growth factor beta-induced cell cycle arrest of human hematopoietic cells requires p57KIP2 up-regulation. Proc. Natl. Acad. Sci. USA 2004, 101, 15231–15236. [Google Scholar] [CrossRef] [PubMed]
- Walkley, C.R.; Fero, M.L.; Chien, W.M.; Purton, L.E.; McArthur, G.A. Negative cell-cycle regulators cooperatively control self-renewal and differentiation of haematopoietic stem cells. Nat. Cell Biol. 2005, 7, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, S.; Iwama, A.; Takayanagi, S.; Morita, Y.; Eto, K.; Ema, H.; Nakauchi, H. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J. 2006, 25, 3515–3523. [Google Scholar] [CrossRef] [PubMed]
- Carroll, D.K.; Carroll, J.S.; Leong, C.O.; Cheng, F.; Brown, M.; Mills, A.A.; Brugge, J.S.; Ellisen, L.W. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat. Cell Biol. 2006, 8, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Vora, S.; Halper, J.P.; Knowles, D.M. Alterations in the activity and isozymic profile of human phosphofructokinase during malignant transformation in vivo and in vitro: Transformation- and progression-linked discriminants of malignancy. Cancer Res. 1985, 45, 2993–3001. [Google Scholar] [PubMed]
- Cheung, E.C.; Vousden, K.H. The role of p53 in glucose metabolism. Curr. Opin. Cell Biol. 2010, 22, 186–191. [Google Scholar] [CrossRef]
- Levine, A.J.; Puzio-Kuter, A.M. The control of the metabolic switch in cancers by oncogenes and tumor suppressor genes. Science 2010, 330, 1340–1344. [Google Scholar] [CrossRef]
- Yang, J.; Li, J.; Le, Y.; Zhou, C.; Zhang, S.; Gong, Z. PFKL/miR-128 axis regulates glycolysis by inhibiting AKT phosphorylation and predicts poor survival in lung cancer. Am. J. Cancer Res. 2016, 6, 473–485. [Google Scholar]
- Barthel, A.; Okino, S.T.; Liao, J.; Nakatani, K.; Li, J.; Whitlock, J.P., Jr.; Roth, R.A. Regulation of GLUT1 gene transcription by the serine/threonine kinase Akt1. J. Biol. Chem. 1999, 274, 20281–20286. [Google Scholar] [CrossRef]
- Frauwirth, K.A.; Riley, J.L.; Harris, M.H.; Parry, R.V.; Rathmell, J.C.; Plas, D.R.; Elstrom, R.L.; June, C.H.; Thompson, C.B. The CD28 signaling pathway regulates glucose metabolism. Immunity 2002, 16, 769–777. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; Plas, D.R.; Rathmell, J.C.; Fox, C.J.; Harris, M.H.; Thompson, C.B. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol. Cell. Biol. 2001, 21, 5899–5912. [Google Scholar] [CrossRef] [PubMed]
- Wieman, H.L.; Wofford, J.A.; Rathmell, J.C. Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell 2007, 18, 1437–1446. [Google Scholar] [CrossRef]
- Cantley, L.C. The phosphoinositide 3-kinase pathway. Science 2002, 296, 1655–1657. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.K.; Sellers, W.R. Akt-regulated pathways in prostate cancer. Oncogene 2005, 24, 7465–7474. [Google Scholar] [CrossRef]
- Kim, J.W.; Tchernyshyov, I.; Semenza, G.L.; Dang, C.V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. [Google Scholar] [CrossRef]
- Papandreou, I.; Cairns, R.A.; Fontana, L.; Lim, A.L.; Denko, N.C. HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab. 2006, 3, 187–197. [Google Scholar] [CrossRef] [Green Version]
- Dedoni, S.; Campbell, L.A.; Harvey, B.K.; Avdoshina, V.; Mocchetti, I. The orphan G-protein-coupled receptor 75 signaling is activated by the chemokine CCL5. J. Neurochem. 2018, 146, 526–539. [Google Scholar] [CrossRef]
- Garcia, V.; Gilani, A.; Shkolnik, B.; Pandey, V.; Zhang, F.F.; Dakarapu, R.; Gandham, S.K.; Reddy, N.R.; Graves, J.P.; Gruzdev, A.; et al. 20-HETE Signals Through G-Protein-Coupled Receptor GPR75 (Gq) to Affect Vascular Function and Trigger Hypertension. Circ. Res. 2017, 120, 1776–1788. [Google Scholar] [CrossRef]
- Gonzalez-Fernandez, E.; Staursky, D.; Lucas, K.; Nguyen, B.V.; Li, M.; Liu, Y.; Washington, C.; Coolen, L.M.; Fan, F.; Roman, R.J. 20-HETE Enzymes and Receptors in the Neurovascular Unit: Implications in Cerebrovascular Disease. Front. Neurol. 2020, 11, 983. [Google Scholar] [CrossRef]
- Liu, B.; Hassan, Z.; Amisten, S.; King, A.J.; Bowe, J.E.; Huang, G.C.; Jones, P.M.; Persaud, S.J. The novel chemokine receptor, G-protein-coupled receptor 75, is expressed by islets and is coupled to stimulation of insulin secretion and improved glucose homeostasis. Diabetologia 2013, 56, 2467–2476. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, A.; Robert, J.; Gregory-Evans, C.; Schaller, H.C. RANTES stimulates Ca2+ mobilization and inositol trisphosphate (IP3) formation in cells transfected with G protein-coupled receptor 75. Br. J. Pharm. 2006, 149, 490–497. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 2018, 14, 450–464. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V. MYC on the path to cancer. Cell 2012, 149, 22–35. [Google Scholar] [CrossRef] [PubMed]
- Cowling, V.H.; Cole, M.D. Turning the tables: Myc activates Wnt in breast cancer. Cell Cycle 2007, 6, 2625–2627. [Google Scholar] [CrossRef]
- Lawlor, E.R.; Soucek, L.; Brown-Swigart, L.; Shchors, K.; Bialucha, C.U.; Evan, G.I. Reversible kinetic analysis of Myc targets in vivo provides novel insights into Myc-mediated tumorigenesis. Cancer Res. 2006, 66, 4591–4601. [Google Scholar] [CrossRef]
- Kirikoshi, H.; Sekihara, H.; Katoh, M. Expression profiles of 10 members of Frizzled gene family in human gastric cancer. Int. J. Oncol. 2001, 19, 767–771. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, H.; Wang, Q.; Zhou, F.; Liu, Y.; Zhang, Y.; Ding, H.; Yuan, M.; Li, F.; Chen, Y. Involvement of c-Fos in cell proliferation, migration, and invasion in osteosarcoma cells accompanied by altered expression of Wnt2 and Fzd9. PLoS ONE 2017, 12, e0180558. [Google Scholar] [CrossRef]
- Zhang, Z.; Schittenhelm, J.; Guo, K.; Buhring, H.J.; Trautmann, K.; Meyermann, R.; Schluesener, H.J. Upregulation of frizzled 9 in astrocytomas. Neuropathol. Appl. Neurobiol. 2006, 32, 615–624. [Google Scholar] [CrossRef]
- Fujimoto, T.; Tomizawa, M.; Yokosuka, O. SiRNA of frizzled-9 suppresses proliferation and motility of hepatoma cells. Int. J. Oncol. 2009, 35, 861–866. [Google Scholar]
- Ahuja, P.; Zhao, P.; Angelis, E.; Ruan, H.; Korge, P.; Olson, A.; Wang, Y.; Jin, E.S.; Jeffrey, F.M.; Portman, M.; et al. Myc controls transcriptional regulation of cardiac metabolism and mitochondrial biogenesis in response to pathological stress in mice. J. Clin. Investig. 2010, 120, 1494–1505. [Google Scholar] [CrossRef] [PubMed]
- Osthus, R.C.; Shim, H.; Kim, S.; Li, Q.; Reddy, R.; Mukherjee, M.; Xu, Y.; Wonsey, D.; Lee, L.A.; Dang, C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000, 275, 21797–21800. [Google Scholar] [CrossRef] [PubMed]
- Arrhenius, S. Über die Dissociationswärme und den Einfluss der Temperatur auf den Dissociationsgrad der Elektrolyte. Z. Für Phys. Chem. 1889, 4, 96–116. [Google Scholar] [CrossRef]
- Fuller, B.J.; Grout, B.W. Clinical Applications of Cryobiology; CRC Press: Boca Raton, FL, USA, 1991. [Google Scholar]
- Southard, J.H. Clinical applications of cryobiology. In Principles of Medical Biology; Bittar, E.E., Ed.; Elsevier: Amsterdam, The Netherlands, 1996; Volume 4, pp. 385–397. [Google Scholar]
- Li, C.; Dong, N.; Wu, H.; Dong, F.; Xu, Y.; Du, H.; He, H.; Liu, Z.; Li, W. A novel method for preservation of human corneal limbal tissue. Investig. Ophthalmol. Vis. Sci. 2013, 54, 4041–4047. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le-Bel, G.; Desjardins, P.; Gross, C.; Cortez Ghio, S.; Couture, C.; Germain, L.; Guérin, S.L. Influence of the Postmortem/Storage Time of Human Corneas on the Properties of Cultured Limbal Epithelial Cells. Cells 2022, 11, 2716. https://doi.org/10.3390/cells11172716
Le-Bel G, Desjardins P, Gross C, Cortez Ghio S, Couture C, Germain L, Guérin SL. Influence of the Postmortem/Storage Time of Human Corneas on the Properties of Cultured Limbal Epithelial Cells. Cells. 2022; 11(17):2716. https://doi.org/10.3390/cells11172716
Chicago/Turabian StyleLe-Bel, Gaëtan, Pascale Desjardins, Christelle Gross, Sergio Cortez Ghio, Camille Couture, Lucie Germain, and Sylvain L. Guérin. 2022. "Influence of the Postmortem/Storage Time of Human Corneas on the Properties of Cultured Limbal Epithelial Cells" Cells 11, no. 17: 2716. https://doi.org/10.3390/cells11172716






