Comparative Analysis of Arsenic Transport and Tolerance Mechanisms: Evolution from Prokaryote to Higher Plants
Abstract
1. Introduction
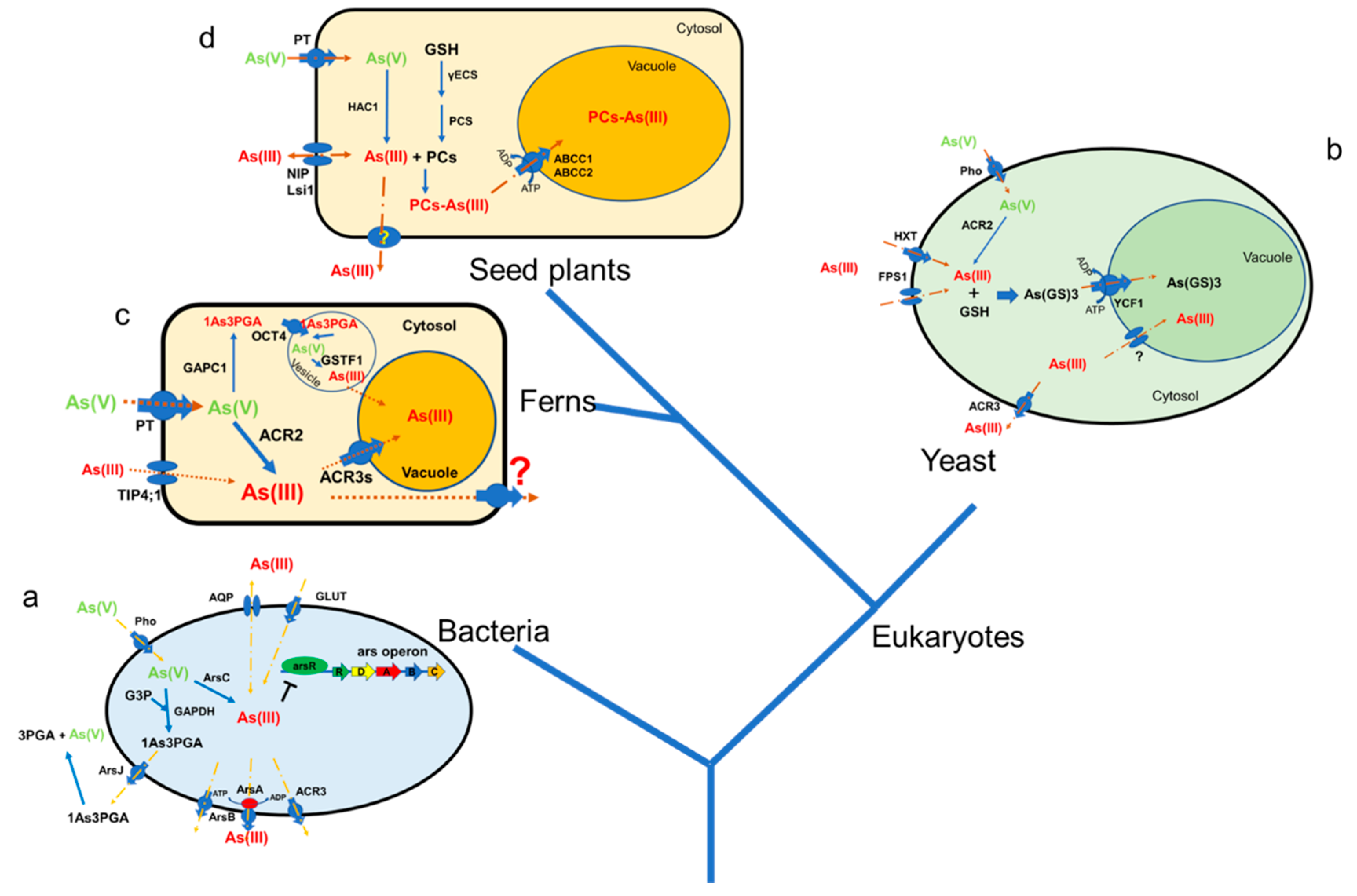
2. Arsenic Reduction Mechanisms, from Prokaryote to Higher Plants
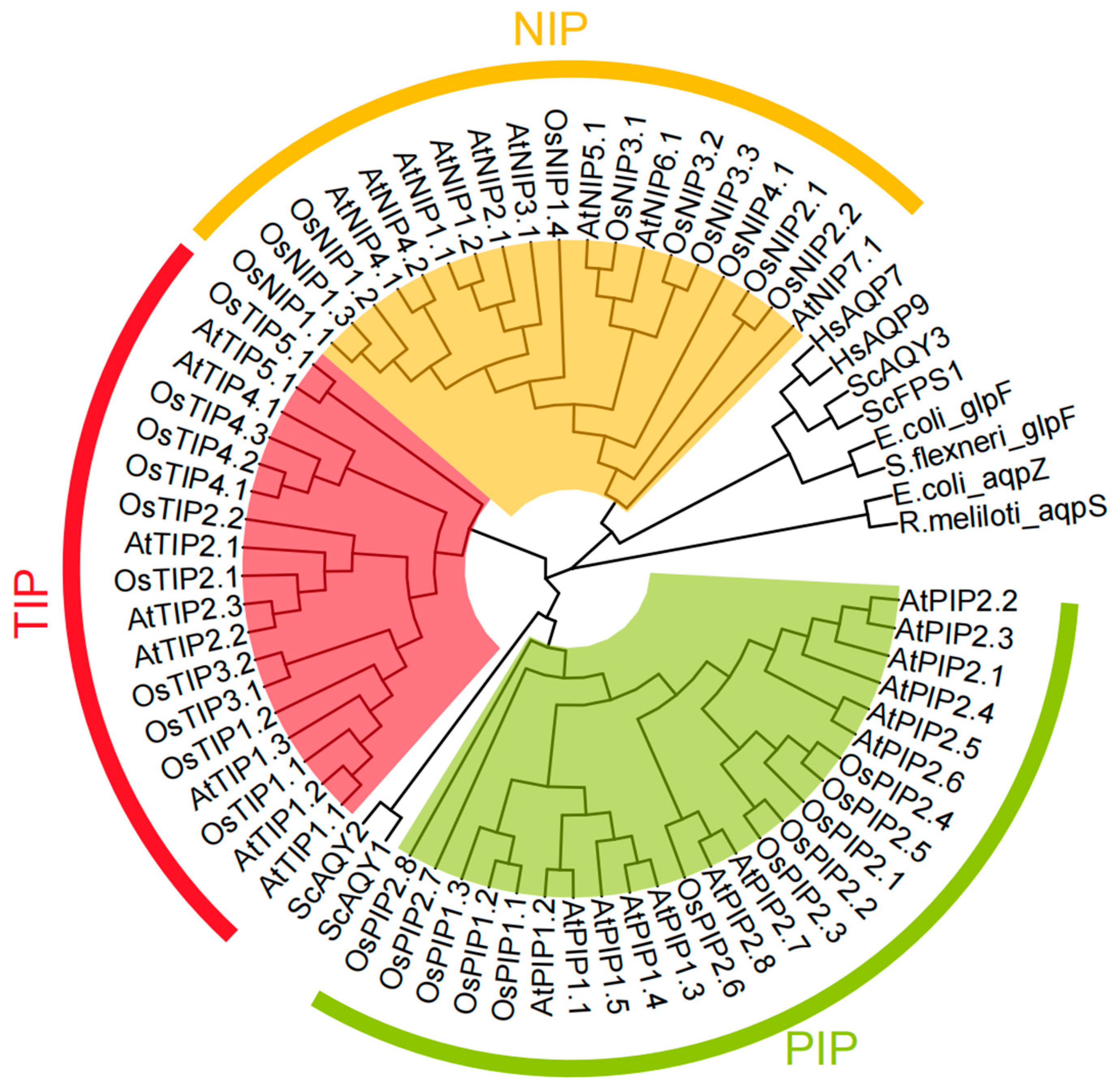
3. Arsenic Uptake and Extrusion Mechanisms, from Prokaryote to Eukaryote
4. Sequestration of As(III) Conjugated with Thiol Compounds in Yeast and Plants
5. Other as Resistance Mechanisms Present in Both Prokaryotes and Eukaryotes
6. Outstanding Questions to Be Solved in Future Research
- (1)
- How did the arsenic resistance trait evolve from prokaryotes to higher plants? For example, the arsenate reductases belong to different protein families in different organisms, such as ArsC protein in prokaryotes, ACR2 in yeast and fern, and HAC1 in higher plants. Although the ACR2 protein exists in higher plants, no clear evidence demonstrates its function as an arsenate reductase. It is interesting to know whether these proteins come from the same ancestor or have evolved separately.
- (2)
- Are there any new mechanisms for arsenic resistance in higher plants? In prokaryotes, a rare case of mechanism for the detoxification of arsenate by converting arsenate to As(V) phosphoglycerate and transporting it out of the cell was reported [16]. A similar strategy has been identified in the As hyperaccumulating fern P. vittata [15]. However, such a mechanism has not been identified in yeast or higher plants. Does this pathway only exist in prokaryotes and P. vittata? In addition, the present review focuses on the tolerance mechanisms for inorganic arsenic species, while organic arsenic species are also present in the soil and should be dealt with by living organisms.
- (3)
- We know that arsenate reductase, aquaglyceroporins, and ABC transporters participate directly in the transport of arsenic or arsenic-containing complexes. However, their regulation is still understood poorly, especially in higher plants. In bacteria arsRDABC operons, ArsR protein acts as a trans-acting transcriptional repressor that regulates the transcription of the operon [40]. Yeasts possess more complex regulation mechanisms for arsenic resistance. The AP-1-like protein Yap8p specifically contributes to arsenic resistance by mediating the arsenic-induced expression of ACR2 and ACR3 [93,94]. The stabilization of Yap8p was found to be regulated by the ubiquitin-proteasome pathway, which is mediated by the ubiquitin-conjugating enzyme Ubc4p [95]. The phosphorylation of As(III) uptake-transporter FPS1 by high-osmolarity glycerol kinase (HOG1) results in further ubiquitination and degradation in the vacuole, thus reducing arsenic uptake in yeast cells [96]. The most well-characterized member of the yeast AP-1 family, Yap1, is involved in the arsenic adaptation process through the regulation of the expression of the vacuolar pump encoded by YCF1 [93]. However, little is known about the regulatory mechanisms for the major components of arsenic resistance, such as ABCC1, aquaporins, PCS, and HAC1 in plants.
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singh, R.; Singh, S.; Parihar, P.; Singh, V.P.; Prasad, S.M. Arsenic contamination, consequences and remediation techniques: A review. Ecotoxicol. Environ. Saf. 2015, 112, 247–270. [Google Scholar] [CrossRef] [PubMed]
- Argos, M.; Kalra, T.; Rathouz, P.J.; Chen, Y.; Pierce, B.; Parvez, F.; Islam, T.; Ahmed, A.; Rakibuz-Zaman, M.; Hasan, R.; et al. Arsenic exposure from drinking water, and all-cause and chronic-disease mortalities in Bangladesh (HEALS): A prospective cohort study. Lancet 2010, 376, 252–258. [Google Scholar] [CrossRef]
- Basu, A.; Saha, D.; Saha, R.; Ghosh, T.; Saha, B. A review on sources, toxicity and remediation technologies for removing arsenic from drinking water. Res. Chem. Intermed. 2014, 40, 447–485. [Google Scholar] [CrossRef]
- Hettick, B.E.; Canas-Carrell, J.E.; French, A.D.; Klein, D.M. Arsenic: A Review of the Element’s Toxicity, Plant Interactions, and Potential Methods of Remediation. J. Agric. Food Chem. 2015, 63, 7097–7107. [Google Scholar] [CrossRef]
- Zhu, Y.G.; Yoshinaga, M.; Zhao, F.J.; Rosen, B.P. Earth Abides Arsenic Biotransformations. Annu. Rev. Earth Planet. Sci. 2014, 42, 443–467. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.J.; McGrath, S.P.; Meharg, A.A. Arsenic as a Food Chain Contaminant: Mechanisms of Plant Uptake and Metabolism and Mitigation Strategies. Annu. Rev. Plant Biol. 2010, 61, 535–559. [Google Scholar] [CrossRef]
- Ben Fekih, I.; Zhang, C.K.; Li, Y.P.; Zhao, Y.; Alwathnani, H.A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of Arsenic Resistance Genes in Prokaryotes. Front. Microbiol. 2018, 9, 2473. [Google Scholar] [CrossRef]
- Garbinski, L.D.; Rosen, B.P.; Chen, J. Pathways of arsenic uptake and efflux. Environ. Int. 2019, 126, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.C.; Sun, G.X.; Yan, Y.; Konstantinidis, K.T.; Zhang, S.Y.; Deng, Y.; Li, X.M.; Cui, H.L.; Musat, F.; Popp, D.; et al. The Great Oxidation Event expanded the genetic repertoire of arsenic metabolism and cycling. Proc. Natl. Acad. Sci. USA 2020, 117, 10414–10421. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Rosen, B.P. Arsenate reductases in prokaryotes and eukaryotes. Environ. Health Perspect. 2002, 110, 745–748. [Google Scholar] [CrossRef]
- Chao, D.Y.; Chen, Y.; Chen, J.G.; Shi, S.L.; Chen, Z.R.; Wang, C.C.; Danku, J.M.; Zhao, F.J.; Salt, D.E. Genome-wide Association Mapping Identifies a New Arsenate Reductase Enzyme Critical for Limiting Arsenic Accumulation in Plants. PLoS Biol. 2014, 12, e1002009. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, M.; Shen, J.; Rosen, B.P. Pathways of As(III) detoxification in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1999, 96, 5001–5006. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Bermejo, E.; Castrillo, G.; del Llano, B.; Navarro, C.; Zarco-Fernandez, S.; Martinez-Herrera, D.J.; del Puerto, Y.L.; Munoz, R.; Camara, C.; Paz-Ares, J.; et al. Natural variation in arsenate tolerance identifies an arsenate reductase in Arabidopsis thaliana. Nat. Commun. 2014, 5, 4617. [Google Scholar] [CrossRef]
- Song, W.Y.; Park, J.; Mendoza-Cozatl, D.G.; Suter-Grotemeyer, M.; Shim, D.; Hortensteiner, S.; Geisler, M.; Weder, B.; Rea, P.A.; Rentsch, D.; et al. Arsenic tolerance in Arabidopsis is mediated by two ABCC-type phytochelatin transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 21187–21192. [Google Scholar] [CrossRef]
- Cai, C.; Lanman, N.A.; Withers, K.A.; DeLeon, A.M.; Wu, Q.; Gribskov, M.; Salt, D.E.; Banks, J.A. Three Genes Define a Bacterial-Like Arsenic Tolerance Mechanism in the Arsenic Hyperaccumulating Fern Pteris vittata. Curr. Biol. 2019, 29, 1625–1633. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Yoshinaga, M.; Garbinski, L.D.; Rosen, B.P. Synergistic interaction of glyceraldehydes-3-phosphate dehydrogenase and ArsJ, a novel organoarsenical efflux permease, confers arsenate resistance. Mol. Microbiol. 2016, 100, 945–953. [Google Scholar] [CrossRef]
- Bhattacharjee, H.; Rosen, B.P. Arsenic Metabolism in Prokaryotic and Eukaryotic Microbes. In Molecular Microbiology of Heavy Metals; Nies, D.H., Silver, S., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 371–406. [Google Scholar]
- Yang, H.C.; Fu, H.L.; Lin, Y.F.; Rosen, B.P. Pathways of Arsenic Uptake and Efflux. Curr. Top. Membr. 2012, 69, 325–358. [Google Scholar]
- Messens, J.; Silver, S. Arsenate reduction: Thiol cascade chemistry with convergent evolution. J. Mol. Biol. 2006, 362, 1–17. [Google Scholar] [CrossRef]
- Saltikov, C.W.; Newman, D.K. Genetic identification of a respiratory arsenate reductase. Proc. Natl. Acad. Sci. USA 2003, 100, 10983–10988. [Google Scholar] [CrossRef]
- Ji, G.; Garber, E.A.; Armes, L.G.; Chen, C.M.; Fuchs, J.A.; Silver, S. Arsenate reductase of Staphylococcus aureus plasmid pI258. Biochemistry 1994, 33, 7294–7299. [Google Scholar] [CrossRef]
- Rosen, B.P. Families of arsenic transporters. Trends Microbiol. 1999, 7, 207–212. [Google Scholar] [CrossRef]
- Bennett, M.S.; Guan, Z.; Laurberg, M.; Su, X.D. Bacillus subtilis arsenate reductase is structurally and functionally similar to low molecular weight protein tyrosine phosphatases. Proc. Natl. Acad. Sci. USA 2001, 98, 13577–13582. [Google Scholar] [CrossRef]
- Bobrowicz, P.; Wysocki, R.; Owsianik, G.; Goffeau, A.; Ulaszewski, S. Isolation of three contiguous genes, ACR1, ACR2 and ACR3, involved in resistance to arsenic compounds in the yeast Saccharomyces cerevisiae. Yeast 1997, 13, 819–828. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Rosen, B.P. Saccharomyces cerevisiae ACR2 gene encodes an arsenate reductase. FEMS Microbiol. Lett. 1998, 168, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Fauman, E.B.; Cogswell, J.P.; Lovejoy, B.; Rocque, W.J.; Holmes, W.; Montana, V.G.; Piwnica-Worms, H.; Rink, M.J.; Saper, M.A. Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell 1998, 93, 617–625. [Google Scholar] [CrossRef]
- Dhankher, O.P.; Rosen, B.P.; McKinney, E.C.; Meagher, R.B. Hyperaccumulation of arsenic in the shoots of Arabidopsis silenced for arsenate reductase (ACR2). Proc. Natl. Acad. Sci. USA 2006, 103, 5413–5418. [Google Scholar] [CrossRef]
- Liu, W.; Schat, H.; Bliek, M.; Chen, Y.; McGrath, S.P.; George, G.; Salt, D.E.; Zhao, F.-J. Knocking Out ACR2 Does Not Affect Arsenic Redox Status in Arabidopsis thaliana: Implications for As Detoxification and Accumulation in Plants. PLoS ONE 2012, 7, e42408. [Google Scholar] [CrossRef]
- Ellis, D.R.; Gumaelius, L.; Indriolo, E.; Pickering, I.J.; Banks, J.A.; Salt, D.E. A novel arsenate reductase from the arsenic hyperaccumulating fern Pteris vittata. Plant Physiol. 2006, 141, 1544–1554. [Google Scholar] [CrossRef]
- Shi, S.L.; Wang, T.; Chen, Z.; Tang, Z.; Wu, Z.C.; Salt, D.E.; Chao, D.Y.; Zhao, F.J. OsHAC1;1 and OsHAC1;2 Function as Arsenate Reductases and Regulate Arsenic Accumulation. Plant Physiol. 2016, 172, 1708–1719. [Google Scholar] [CrossRef]
- Xu, J.M.; Shi, S.L.; Wang, L.; Tang, Z.; Lv, T.T.; Zhu, X.L.; Ding, X.M.; Wang, Y.F.; Zhao, F.J.; Wu, Z.C. OsHAC4 is critical for arsenate tolerance and regulates arsenic accumulation in rice. New Phytol. 2017, 215, 1090–1101. [Google Scholar] [CrossRef]
- Li, X.Y.; Sun, D.; Feng, H.Y.; Chen, J.X.; Chen, Y.S.; Li, H.B.; Cao, Y.; Ma, L.N.Q. Efficient arsenate reduction in As-hyperaccumulator Pteris vittata are mediated by novel arsenate reductases PvHAC1 and PvHAC2. J. Hazard. Mater. 2020, 399, 122895. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.R.; Dugas, S.L. Phylogenetic analysis of bacterial and archaeal arsC gene sequences suggests an ancient, common origin for arsenate reductase. BMC Evol. Biol. 2003, 3, 18. [Google Scholar] [CrossRef] [PubMed]
- Dunivin, T.K.; Yeh, S.Y.; Shade, A. A global survey of arsenic-related genes in soil microbiomes. BMC Biol. 2019, 17, 45. [Google Scholar] [CrossRef]
- Mukhopadhyay, R.; Rosen, B.P.; Pung, L.T.; Silver, S. Microbial arsenic: From geocycles to genes and enzymes. FEMS Microbiol. Rev. 2002, 26, 311–325. [Google Scholar] [CrossRef]
- Hofmann, K.; Bucher, P.; Kajava, A.V. A model of Cdc25 phosphatase catalytic domain and CDK-interaction surface based on the presence of a rhodanese homology domain. J. Mol. Biol. 1998, 282, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Meharg, A.A.; Macnair, M.R. An altered phosphate uptake system in arsenate-tolerant Holcus lanatus L. New Phytol. 1990, 116, 29–35. [Google Scholar] [CrossRef]
- Clemens, S.; Ma, J.F. Toxic Heavy Metal and Metalloid Accumulation in Crop Plants and Foods. Annu. Rev. Plant Biol. 2016, 67, 489–512. [Google Scholar] [CrossRef]
- Zhao, F.J.; Ago, Y.; Mitani, N.; Li, R.Y.; Su, Y.H.; Yamaji, N.; McGrath, S.P.; Ma, J.F. The role of the rice aquaporin Lsi1 in arsenite efflux from roots. New Phytol. 2010, 186, 392–399. [Google Scholar] [CrossRef]
- Busenlehner, L.S.; Pennella, M.A.; Giedroc, D.P. The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 2003, 27, 131–143. [Google Scholar] [CrossRef]
- Sato, T.; Kobayashi, Y. The ars operon in the skin element of Bacillus subtilis confers resistance to arsenate and arsenite. J. Bacteriol. 1998, 180, 1655–1661. [Google Scholar] [CrossRef]
- Achour, A.R.; Bauda, P.; Billard, P. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res. Microbiol. 2007, 158, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Dey, S.; Rosen, B.P. Dual-Mode of Energy Coupling by the Oxyanion-Translocating Arsb Protein. J. Bacteriol. 1995, 177, 385–389. [Google Scholar] [CrossRef]
- Castillo, R.; Saier, M.H. Functional Promiscuity of Homologues of the Bacterial ArsA ATPases. Int. J. Microbiol. 2010, 2010, 187373. [Google Scholar] [CrossRef] [PubMed]
- Luyten, K.; Albertyn, J.; Skibbe, W.F.; Prior, B.A.; Ramos, J.; Thevelein, J.M.; Hohmann, S. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 1995, 14, 1360–1371. [Google Scholar] [CrossRef]
- Sutherland, F.C.; Lages, F.; Lucas, C.; Luyten, K.; Albertyn, J.; Hohmann, S.; Prior, B.A.; Kilian, S.G. Characteristics of Fps1-dependent and -independent glycerol transport in Saccharomyces cerevisiae. J. Bacteriol. 1997, 179, 7790–7795. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wysocki, R.; Bobrowicz, P.; Ulaszewski, S. The Saccharomyces cerevisiae ACR3 gene encodes a putative membrane protein involved in arsenite transport. J. Biol. Chem. 1997, 272, 30061–30066. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Tisa, L.S.; Rosen, B.P. Membrane topology of the ArsB protein, the membrane subunit of an anion-translocating ATPase. J. Biol. Chem. 1992, 267, 12570–12576. [Google Scholar] [CrossRef]
- Indriolo, E.; Na, G.; Ellis, D.; Salt, D.E.; Banks, J.A. A Vacuolar Arsenite Transporter Necessary for Arsenic Tolerance in the Arsenic Hyperaccumulating Fern Pteris vittata Is Missing in Flowering Plants. Plant Cell 2010, 22, 2045–2057. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, W.; Shen, H.; Yan, H.; Xu, W.; He, Z.; Ma, M. Engineering arsenic tolerance and hyperaccumulation in plants for phytoremediation by a PvACR3 transgenic approach. Environ. Sci. Technol. 2013, 47, 9355–9362. [Google Scholar] [CrossRef]
- Wang, C.; Na, G.; Bermejo, E.S.; Chen, Y.; Banks, J.A.; Salt, D.E.; Zhao, F.J. Dissecting the components controlling root-to-shoot arsenic translocation in Arabidopsis thaliana. New Phytol. 2018, 217, 206–218. [Google Scholar] [CrossRef]
- Chen, Y.S.; Hua, C.Y.; Chen, J.X.; Rathinasabapathi, B.; Cao, Y.; Ma, L.Q. Expressing Arsenite Antiporter PvACR3;1 in Rice (Oryza sativa L.) Decreases Inorganic Arsenic Content in Rice Grains. Environ. Sci. Technol. 2019, 53, 10062–10069. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Hua, C.Y.; Jia, M.R.; Fu, J.W.; Liu, X.; Han, Y.H.; Liu, Y.G.; Rathinasabapathi, B.; Cao, Y.; Ma, L.Q. Heterologous Expression of Pteris vittata Arsenite Antiporter PvACR3;1 Reduces Arsenic Accumulation in Plant Shoots. Environ. Sci. Technol. 2017, 51, 10387–10395. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.X.; Cao, Y.; Yan, X.; Chen, Y.; Ma, L.Q. Novel PvACR3;2 and PvACR3;3 genes from arsenic-hyperaccumulator Pteris vittata and their roles in manipulating plant arsenic accumulation. J. Hazard. Mater. 2021, 415, 125647. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.L.; Liu, X.; Chen, Y.S.; Rathinasabapathi, B.; Rensing, C.; Chen, J.; Bi, J.; Xian, P.; Ma, L.N.Q. Aquaporins mediated arsenite transport in plants: Molecular mechanisms and applications in crop improvement. Crit. Rev. Environ. Sci. Technol. 2020, 50, 1613–1639. [Google Scholar] [CrossRef]
- Deshmukh, R.K.; Sonah, H.; Belanger, R.R. Plant Aquaporins: Genome-Wide Identification, Transcriptomics, Proteomics, and Advanced Analytical Tools. Front. Plant Sci. 2016, 7, 1896. [Google Scholar] [CrossRef]
- Madeira, A.; Moura, T.F.; Soveral, G. Detecting Aquaporin Function and Regulation. Front. Chem. 2016, 4, 3. [Google Scholar] [CrossRef]
- Wysocki, R.; Chery, C.C.; Wawrzycka, D.; Van Hulle, M.; Cornelis, R.; Thevelein, J.M.; Tamas, M.J. The glycerol channel Fps1p mediates the uptake of arsenite and antimonite in Saccharomyces cerevisiae. Mol. Microbiol. 2001, 40, 1391–1401. [Google Scholar] [CrossRef]
- Sabir, F.; Loureiro-Dias, M.C.; Soveral, G.; Prista, C. Functional relevance of water and glycerol channels in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2017, 364, fnx080. [Google Scholar] [CrossRef]
- Maciaszczyk-Dziubinska, E.; Migdal, I.; Migocka, M.; Bocer, T.; Wysocki, R. The yeast aquaglyceroporin Fps1p is a bidirectional arsenite channel. FEBS Lett. 2010, 584, 726–732. [Google Scholar] [CrossRef]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in Plants. Physiol. Rev. 2015, 95, 1321–1358. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. The Arabidopsis thaliana aquaglyceroporin AtNIP7;1 is a pathway for arsenite uptake. FEBS Lett. 2008, 582, 1625–1628. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, T.; Tanaka, M.; Mitani, N.; Ma, J.F.; Maeshima, M.; Fujiwara, T. NIP1;1, an Aquaporin Homolog, Determines the Arsenite Sensitivity of Arabidopsis thaliana. J. Biol. Chem. 2009, 284, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Mitani-Ueno, N.; Yamaji, N.; Zhao, F.J.; Ma, J.F. The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 2011, 62, 4391–4398. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.Z.; Dai, W.T.; Yan, H.L.; Li, S.; Shen, H.L.; Chen, Y.S.; Xu, H.; Sun, Y.Y.; He, Z.Y.; Ma, M. Arabidopsis NIP3;1 Plays an Important Role in Arsenic Uptake and Root-to-Shoot Translocation under Arsenite Stress Conditions. Mol. Plant 2015, 8, 722–733. [Google Scholar] [CrossRef] [PubMed]
- Bienert, G.P.; Thorsen, M.; Schussler, M.D.; Nilsson, H.R.; Wagner, A.; Tamas, M.J.; Jahn, T.P. A subgroup of plant aquaporins facilitate the bi-directional diffusion of As(OH)3 and Sb(OH)3 across membranes. BMC Biol. 2008, 6, 26. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Fujiwara, T. Three regions of the NIP5;1 promoter are required for expression in different cell types in Arabidopsis thaliana root. Plant Signal. Behav. 2021, 16, 1993654. [Google Scholar] [CrossRef]
- Takano, J.; Wada, M.; Ludewig, U.; Schaaf, G.; von Wiren, N.; Fujiwara, T. The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 2006, 18, 1498–1509. [Google Scholar] [CrossRef]
- Wang, Y.; Li, R.; Li, D.; Jia, X.; Zhou, D.; Li, J.; Lyi, S.M.; Hou, S.; Huang, Y.; Kochian, L.V.; et al. NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, 5047–5052. [Google Scholar] [CrossRef]
- Carey, A.M.; Scheckel, K.G.; Lombi, E.; Newville, M.; Choi, Y.; Norton, G.J.; Charnock, J.M.; Feldmann, J.; Price, A.H.; Meharg, A.A. Grain unloading of arsenic species in rice. Plant Physiol. 2010, 152, 309–319. [Google Scholar] [CrossRef]
- Li, N.; Wang, J.; Song, W.Y. Arsenic Uptake and Translocation in Plants. Plant Cell Physiol. 2016, 57, 4–13. [Google Scholar] [CrossRef]
- Tanaka, M.; Wallace, I.S.; Takano, J.; Roberts, D.M.; Fujiwara, T. NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 2008, 20, 2860–2875. [Google Scholar] [CrossRef]
- Lindsay, E.R.; Maathuis, F.J.M. New Molecular Mechanisms to Reduce Arsenic in Crops. Trends Plant Sci. 2017, 22, 1016–1026. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Tamai, K.; Yamaji, N.; Mitani, N.; Konishi, S.; Katsuhara, M.; Ishiguro, M.; Murata, Y.; Yano, M. A silicon transporter in rice. Nature 2006, 440, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Yamaji, N.; Mitani, N.; Xu, X.Y.; Su, Y.H.; McGrath, S.P.; Zhao, F.J. Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. USA 2008, 105, 9931–9935. [Google Scholar] [CrossRef] [PubMed]
- Li, R.Y.; Ago, Y.; Liu, W.J.; Mitani, N.; Feldmann, J.; McGrath, S.P.; Ma, J.F.; Zhao, F.J. The rice aquaporin Lsi1 mediates uptake of methylated arsenic species. Plant Physiol. 2009, 150, 2071–2080. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.K.; Chen, Y.; Che, J.; Konishi, N.; Tang, Z.; Miller, A.J.; Ma, J.F.; Zhao, F.J. Decreasing arsenic accumulation in rice by overexpressing OsNIP1;1 and OsNIP3;3 through disrupting arsenite radial transport in roots. New Phytol. 2018, 219, 641–653. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, S.K.; Tang, Z.; Liu, G.; Moore, K.L.; Maathuis, F.J.M.; Miller, A.J.; McGrath, S.P.; Zhao, F.J. The Nodulin 26-like intrinsic membrane protein OsNIP3;2 is involved in arsenite uptake by lateral roots in rice. J. Exp. Bot. 2017, 68, 3007–3016. [Google Scholar] [CrossRef]
- He, Z.Y.; Yan, H.L.; Chen, Y.S.; Shen, H.L.; Xu, W.X.; Zhang, H.Y.; Shi, L.; Zhu, Y.G.; Ma, M. An aquaporin PvTIP4;1 from Pteris vittata may mediate arsenite uptake. New Phytol. 2016, 209, 746–761. [Google Scholar] [CrossRef]
- Su, Y.H.; McGrath, S.P.; Zhao, F.J. Rice is more efficient in arsenite uptake and translocation than wheat and barley. Plant Soil 2010, 328, 27–34. [Google Scholar] [CrossRef]
- Li, Z.S.; Lu, Y.P.; Zhen, R.G.; Szczypka, M.; Thiele, D.J.; Rea, P.A. A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 1997, 94, 42–47. [Google Scholar] [CrossRef]
- Pickering, I.J.; Prince, R.C.; George, M.J.; Smith, R.D.; George, G.N.; Salt, D.E. Reduction and coordination of arsenic in Indian mustard. Plant Physiol. 2000, 122, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Raab, A.; Schat, H.; Meharg, A.A.; Feldmann, J. Uptake, translocation and transformation of arsenate and arsenite in sunflower (Helianthus annuus): Formation of arsenic-phytochelatin complexes during exposure to high arsenic concentrations. New Phytol. 2005, 168, 551–558. [Google Scholar] [CrossRef] [PubMed]
- Schmoger, M.E.V.; Oven, M.; Grill, E. Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 2000, 122, 793–801. [Google Scholar] [CrossRef]
- Sneller, F.E.C.; Van Heerwaarden, L.M.; Kraaijeveld-Smit, F.J.L.; Ten Bookum, W.M.; Koevoets, P.L.M.; Schat, H.; Verkleij, J.A.C. Toxicity of arsenate in Silene vulgaris, accumulation and degradation of arsenate-induced phytochelatins. New Phytol. 1999, 144, 223–232. [Google Scholar] [CrossRef]
- Ha, S.B.; Smith, A.P.; Howden, R.; Dietrich, W.M.; Bugg, S.; O’Connell, M.J.; Goldsbrough, P.B.; Cobbett, C.S. Phytochelatin synthase genes from arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell 1999, 11, 1153–1163. [Google Scholar] [CrossRef]
- Li, Y.J.; Dankher, O.P.; Carreira, L.; Smith, A.P.; Meagher, R.B. The shoot-specific expression of gamma-glutamylcysteine synthetase directs the long-distance transport of thiol-peptides to roots conferring tolerance to mercury and arsenic. Plant Physiol. 2006, 141, 288–298. [Google Scholar] [CrossRef]
- Dhankher, O.P.; Li, Y.J.; Rosen, B.P.; Shi, J.; Salt, D.; Senecoff, J.F.; Sashti, N.A.; Meagher, R.B. Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat. Biotechnol. 2002, 20, 1140–1145. [Google Scholar] [CrossRef]
- Gasic, K.; Korban, S.S. Transgenic Indian mustard (Brassica juncea) plants expressing an Arabidopsis phytochelatin synthase (AtPCS1) exhibit enhanced As and Cd tolerance. Plant Mol. Biol. 2007, 64, 361–369. [Google Scholar] [CrossRef]
- Li, Y.J.; Dhankher, O.P.; Carreira, L.; Lee, D.; Chen, A.; Schroeder, J.I.; Balish, R.S.; Meagher, R.B. Overexpression of phytochelatin synthase in Arabidopsis leads to enhanced arsenic tolerance and cadmium hypersensitivity. Plant Cell Physiol. 2004, 45, 1787–1797. [Google Scholar] [CrossRef]
- Song, W.Y.; Yamaki, T.; Yamaji, N.; Ko, D.; Jung, K.H.; Fujii-Kashino, M.; An, G.; Martinoia, E.; Lee, Y.; Ma, J.F. A rice ABC transporter, OsABCC1, reduces arsenic accumulation in the grain. Proc. Natl. Acad. Sci. USA 2014, 111, 15699–15704. [Google Scholar] [CrossRef]
- Tang, Z.; Chen, Y.; Miller, A.J.; Zhao, F.J. The C-type ATP-Binding Cassette Transporter OsABCC7 Is Involved in the Root-to-Shoot Translocation of Arsenic in Rice. Plant Cell Physiol. 2019, 60, 1525–1535. [Google Scholar] [CrossRef]
- Wysocki, R.; Fortier, P.K.; Maciaszczyk, E.; Thorsen, M.; Leduc, A.; Odhagen, A.; Owsianik, G.; Ulaszewski, S.; Ramotar, D.; Tamas, M.J. Transcriptional activation of metalloid tolerance genes in Saccharomyces cerevisiae requires the AP-1-like proteins Yap1p and Yap8p. Mol. Biol. Cell 2004, 15, 2049–2060. [Google Scholar] [CrossRef]
- Kumar, N.V.; Yang, J.B.; Pillai, J.K.; Rawat, S.; Solano, C.; Kumar, A.; Grotli, M.; Stemmler, T.L.; Rosen, B.P.; Tamas, M.J. Arsenic Directly Binds to and Activates the Yeast AP-1-Like Transcription Factor Yap8. Mol. Cell Biol. 2016, 36, 913–922. [Google Scholar] [CrossRef]
- Di, Y.J.; Tamas, M.J. Regulation of the arsenic-responsive transcription factor Yap8p involves the ubiquitin-proteasome pathway. J. Cell Sci. 2007, 120, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Mollapour, M.; Piper, P.W. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol. Cell Biol. 2007, 27, 6446–6456. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Liu, J.; Zheng, F.; Yu, M.; Shabala, S.; Song, W.-Y. Comparative Analysis of Arsenic Transport and Tolerance Mechanisms: Evolution from Prokaryote to Higher Plants. Cells 2022, 11, 2741. https://doi.org/10.3390/cells11172741
Zhang J, Liu J, Zheng F, Yu M, Shabala S, Song W-Y. Comparative Analysis of Arsenic Transport and Tolerance Mechanisms: Evolution from Prokaryote to Higher Plants. Cells. 2022; 11(17):2741. https://doi.org/10.3390/cells11172741
Chicago/Turabian StyleZhang, Jie, Jiayou Liu, Fubin Zheng, Min Yu, Sergey Shabala, and Won-Yong Song. 2022. "Comparative Analysis of Arsenic Transport and Tolerance Mechanisms: Evolution from Prokaryote to Higher Plants" Cells 11, no. 17: 2741. https://doi.org/10.3390/cells11172741
APA StyleZhang, J., Liu, J., Zheng, F., Yu, M., Shabala, S., & Song, W.-Y. (2022). Comparative Analysis of Arsenic Transport and Tolerance Mechanisms: Evolution from Prokaryote to Higher Plants. Cells, 11(17), 2741. https://doi.org/10.3390/cells11172741









