CtIP Regulates Mitotic Spindle Assembly by Modulating the TPX2-Aurora A Signaling Axis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Transfection
2.2. Plasmids
2.3. RNA Interference and Transfection
2.4. Antibodies
2.5. Immunofluorescence Microscopy and Live-Cell Analysis
2.6. Immunoprecipitation and Western Blot Analyses
2.7. Cell Cycle Synchronization
2.8. Statistical Analysis
3. Results
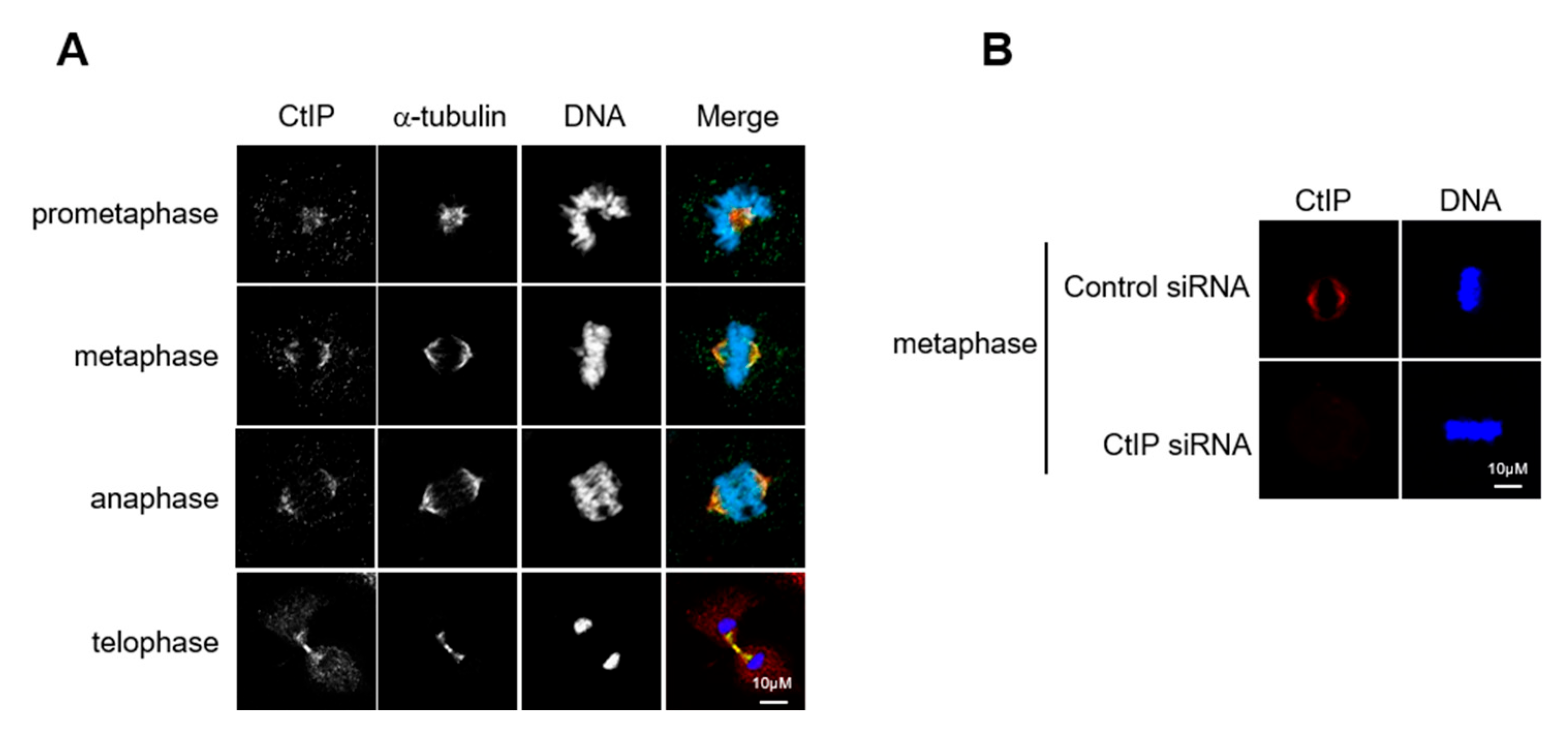
3.1. CtIP Is a Mitotic Spindle Associated Protein
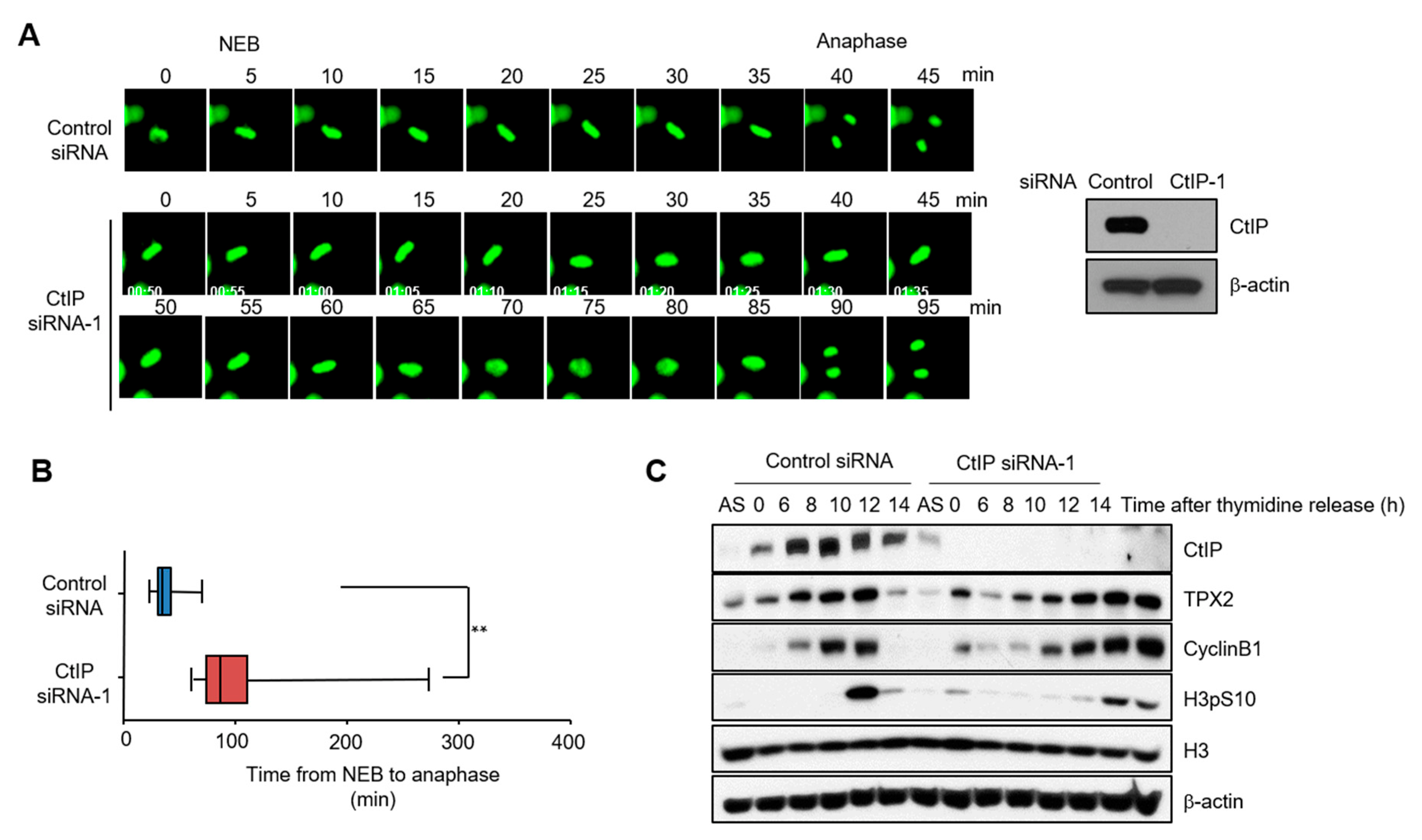
3.2. Knockdown of CtIP Results in Mitotic Delay
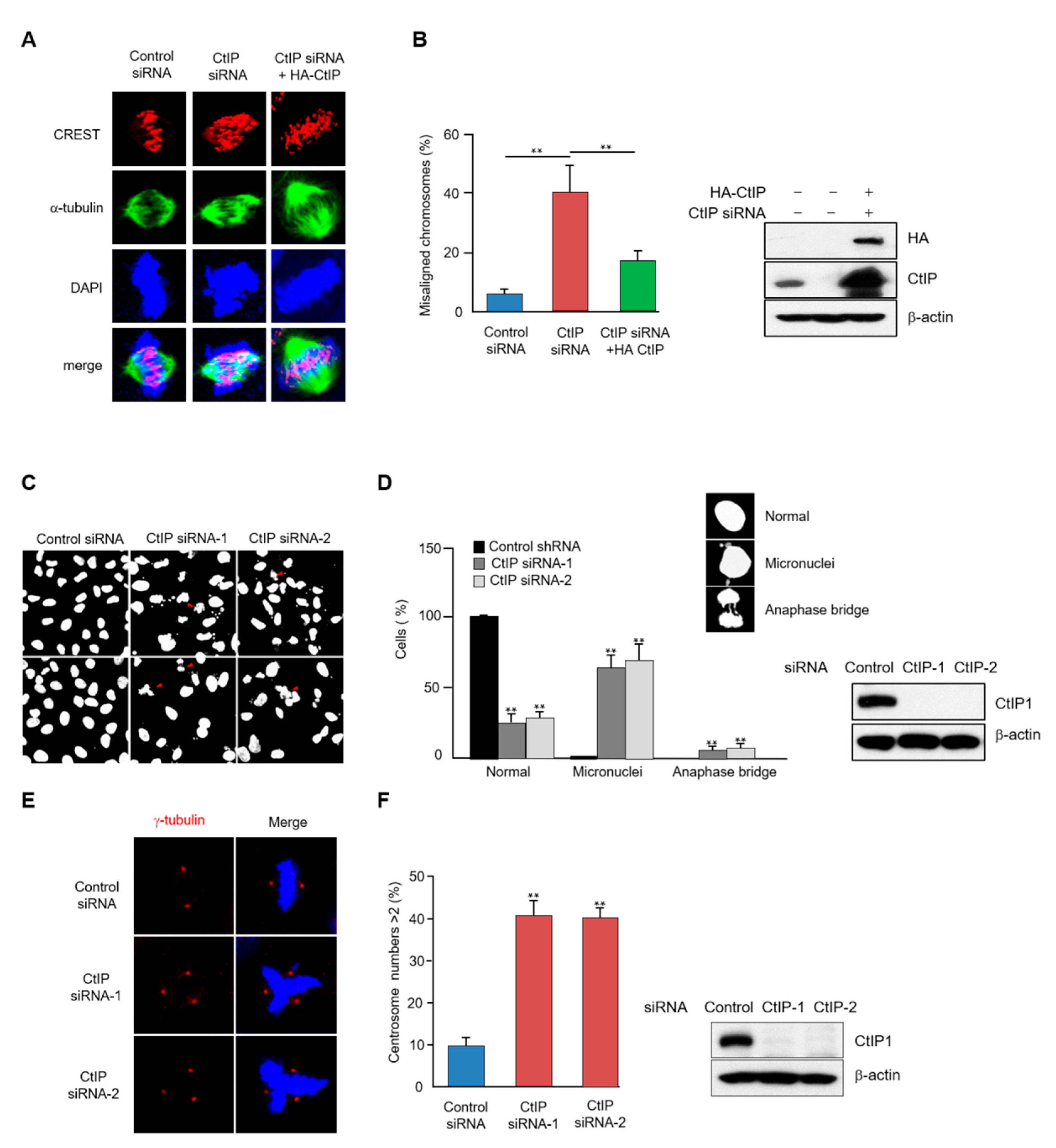
3.3. CtIP Depletion Leads to Chromosome Misalignment and Improper Spindle Formation during Mitosis
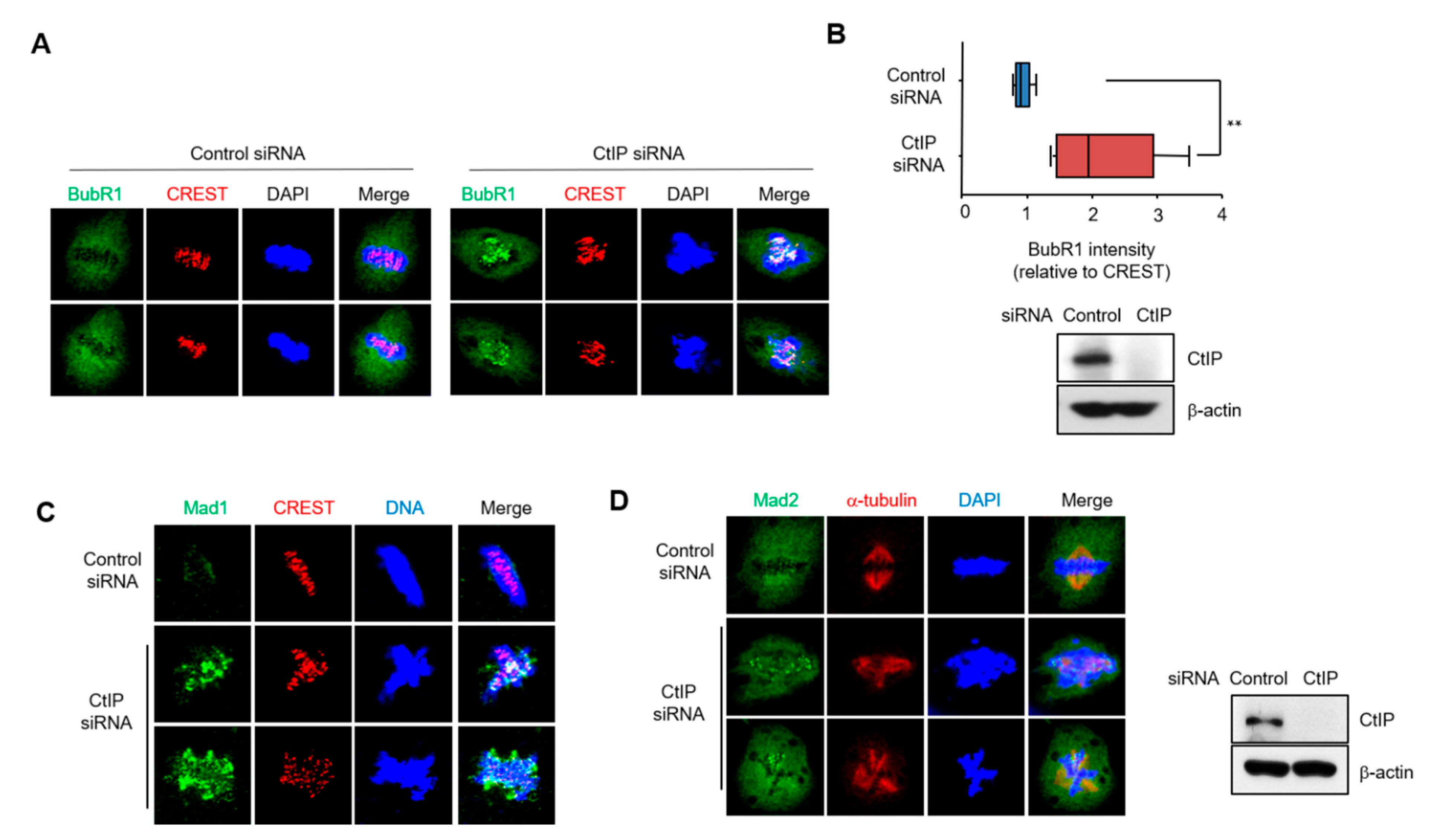
3.4. Depletion of CtIP Induces Activation of Spindle Assembly Checkpoint
3.5. CtIP Interacts and Colocalizes with TPX2
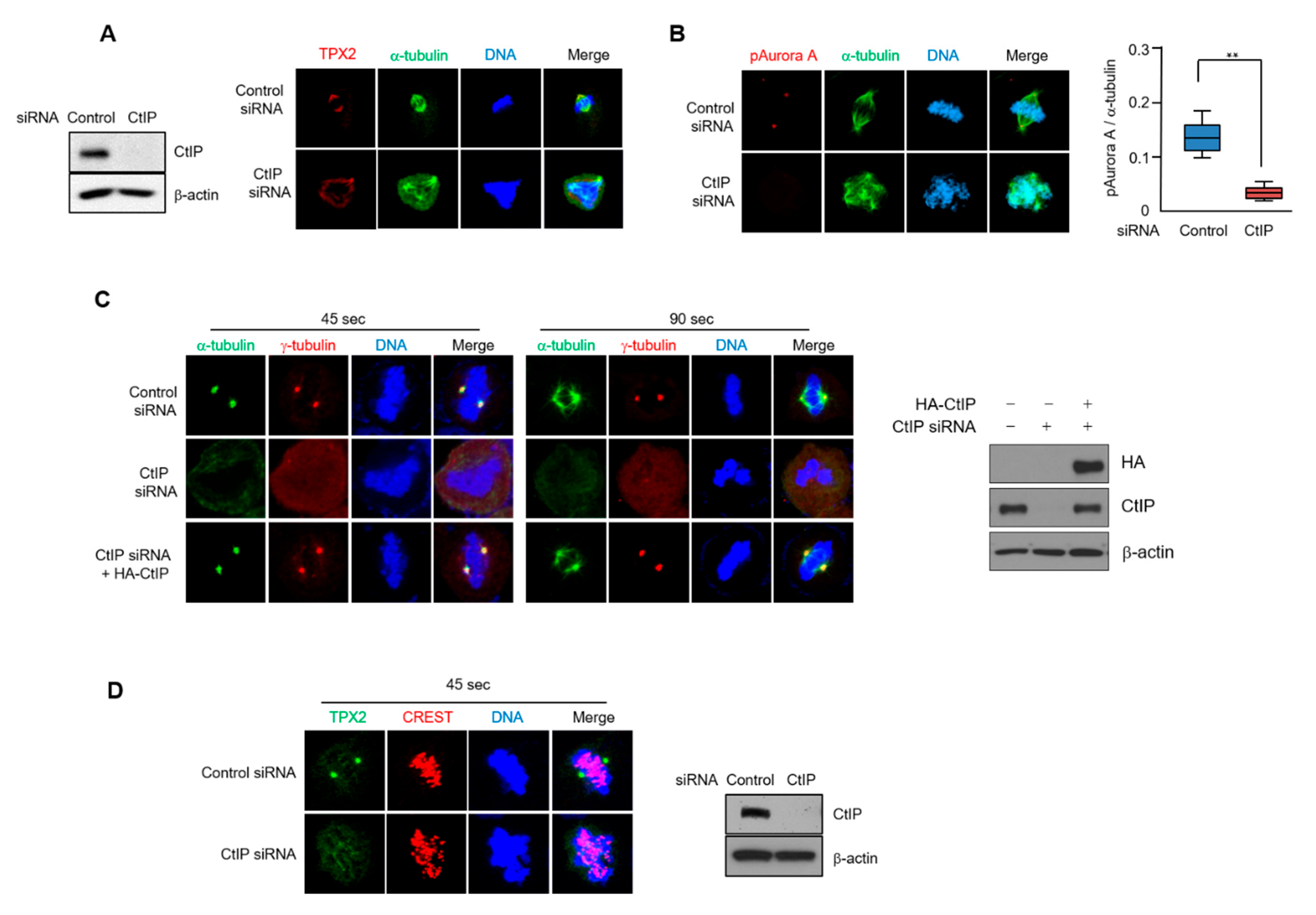
3.6. CtIP Depletion Leads to Impaired TPX2 Signaling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- You, Z.; Bailis, J.M. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends Cell Biol. 2010, 20, 402–409. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.L.; Liu, F.; Cai, S.; Lin, X.; Li, A.; Chen, Y.; Gu, B.; Lee, E.Y.; Lee, W.H. Inactivation of CtIP leads to early embry-onic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol. Cell Biol. 2005, 25, 3535–3542. [Google Scholar] [CrossRef] [PubMed]
- Makharashvili, N.; Paull, T.T. CtIP: A DNA damage response protein at the intersection of DNA metabolism. DNA Repair 2015, 32, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Schaeper, U.; Subramanian, T.; Lim, L.; Boyd, J.M.; Chinnadurai, G. Interaction between a Cellular Protein That Binds to the C-terminal Region of Adenovirus E1A (CtBP) and a Novel Cellular Protein Is Disrupted by E1A through a Conserved PLDLS Motif. J. Biol. Chem. 1998, 273, 8549–8552. [Google Scholar] [CrossRef]
- Liu, F.; Lee, W.H. CtIP activates its own and cyclin D1 promoters via the E2F/RB pathway during G1/S progression. Mol. Cell Biol. 2006, 26, 3124–3134. [Google Scholar] [CrossRef]
- Sum, E.Y.M.; Peng, B.; Yu, X.; Chen, J.; Byrne, J.; Lindeman, G.J.; Visvader, J.E. The LIM domain protein LMO4 interacts with the cofactor CtIP and the tumor suppressor BRCA1 and inhibits BRCA1 activity. J. Biol. Chem. 2002, 277, 7849–7856. [Google Scholar] [CrossRef]
- Koipally, J.; Georgopoulos, K. Ikaros-CtIP interactions do not require C-terminal binding protein and participate in a deacetylase-independent mode of repression. J. Biol. Chem. 2002, 277, 23143–23149. [Google Scholar] [CrossRef]
- Sartori, A.A.; Lukas, C.; Coates, J.; Mistrik, M.; Fu, S.; Bartek, J.; Baer, R.; Lukas, J.; Jackson, S.P. Human CtIP promotes DNA end resection. Nature 2007, 450, 509–514. [Google Scholar] [CrossRef]
- You, Z.; Shi, L.Z.; Zhu, Q.; Wu, P.; Zhang, Y.-W.; Basilio, A.; Tonnu, N.; Verma, I.M.; Berns, M.W.; Hunter, T. CtIP Links DNA Double-Strand Break Sensing to Resection. Mol. Cell 2009, 36, 954–969. [Google Scholar] [CrossRef]
- Chen, L.; Nievera, C.J.; Lee, A.Y.-L.; Wu, X. Cell Cycle-dependent Complex Formation of BRCA1·CtIP·MRN Is Important for DNA Double-strand Break Repair. J. Biol. Chem. 2008, 283, 7713–7720. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Fu, S.; Lai, M.; Baer, R.; Chen, J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006, 20, 1721–1726. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, T.; A Hyman, A.; Desai, A. The spindle: A dynamic assembly of microtubules and motors. Nat. Cell Biol. 2001, 3, E28–E34. [Google Scholar] [CrossRef] [PubMed]
- Gadde, S.; Heald, R. Mechanisms and molecules of the mitotic spindle. Curr. Biol. 2004, 14, R797–R805. [Google Scholar] [CrossRef] [PubMed]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Petry, S. Mechanisms of Mitotic Spindle Assembly. Annu. Rev. Biochem. 2016, 85, 659–683. [Google Scholar] [CrossRef]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef]
- Lara-Gonzalez, P.; Westhorpe, F.G.; Taylor, S.S. The spindle assembly checkpoint. Curr. Biol. 2012, 22, R966–R980. [Google Scholar] [CrossRef]
- Wittmann, T.; Wilm, M.; Karsenti, E.; Vernos, I. Tpx2, a novel xenopus map involved in spindle pole organization. J. Cell Biol. 2000, 149, 1405–1418. [Google Scholar] [CrossRef]
- Gruss, O.J.; Vernos, I. The mechanism of spindle assembly: Functions of Ran and its target TPX2. J. Cell Biol. 2004, 166, 949–955. [Google Scholar] [CrossRef]
- Kufer, T.A.; Silljé, H.H.; Körner, R.; Gruss, O.J.; Meraldi, P.; Nigg, E.A. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J. Cell Biol. 2002, 158, 617–623. [Google Scholar] [CrossRef] [Green Version]
- Eyers, P.A.; Erikson, E.; Chen, L.G.; Maller, J.L. A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 2003, 13, 691–697. [Google Scholar] [CrossRef]
- Tsai, M.Y.; Wiese, C.; Cao, K.; Martin, O.; Donovan, P.; Ruderman, J.; Prigent, C.; Zheng, Y. A Ran signalling pathway me-diated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 2003, 5, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Koffa, M.D.; Casanova, C.M.; Santarella, R.; Koecher, T.; Wilm, M.; Mattaj, I. HURP is part of a ran-dependent complex involved in spindle formation. Curr. Biol. 2006, 16, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Sillje, H.H.; Nagel, S.; Korner, R.; Nigg, E.A. HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006, 16, 731–742. [Google Scholar] [CrossRef]
- Wong, J.; Fang, G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006, 173, 879–891. [Google Scholar] [CrossRef]
- Ma, N.; Tulu, U.S.; Ferenz, N.P.; Fagerstrom, C.; Wilde, A.; Wadsworth, P. Poleward transport of TPX2 in the mammalian mitotic spindle requires dynein, Eg5, and microtubule flux. Mol. Biol. Cell 2010, 21, 979–988. [Google Scholar] [CrossRef]
- Hsu, W.-H.; Wang, W.; Lin, W.; Huang, Y.; Lai, C.; Liao, J.-C.; Chen, H. Adducin-1 is essential for spindle pole integrity through its interaction with TPX2. EMBO Rep. 2018, 19, 45607. [Google Scholar] [CrossRef]
- Eibes, S.; Gallisà-Suñé, N.; Rosas-Salvans, M.; Martínez-Delgado, P.; Vernos, I.; Roig, J. Nek9 Phosphorylation Defines a New Role for TPX2 in Eg5-Dependent Centrosome Separation before Nuclear Envelope Breakdown. Curr. Biol. 2017, 28, 121–129. [Google Scholar] [CrossRef]
- Eid, W.; Steger, M.; El-Shemerly, M.; Ferretti, L.P.; Pena-Diaz, J.; Konig, C.; Valtorta, E.; Sartori, A.A.; Ferrari, S. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010, 11, 962–968. [Google Scholar] [CrossRef]
- Yu, X.; Chen, J. DNA Damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 2004, 24, 9478–9486. [Google Scholar] [CrossRef] [Green Version]
- Rozier, L.; Guo, Y.; Peterson, S.; Sato, M.; Baer, R.; Gautier, J.; Mao, Y. The MRN-CtIP pathway is required for metaphase chromosome alignment. Mol. Cell 2013, 49, 1097–1107. [Google Scholar] [CrossRef] [PubMed]
- Vitre, B.D.; Cleveland, D.W. Centrosomes, chromosome instability (CIN) and aneuploidy. Curr. Opin. Cell Biol. 2012, 24, 809–815. [Google Scholar] [CrossRef] [PubMed]
- Foley, E.A.; Kapoor, T.M. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013, 14, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Hein, M.Y.; Hubner, N.C.; Poser, I.; Cox, J.; Nagaraj, N.; Toyoda, Y.; Gak, I.A.; Weisswange, I.; Mansfeld, J.; Buchholz, F.; et al. A human interactome in three quantitative dimensions organized by stoichiometries and abun-dances. Cell 2015, 163, 712–723. [Google Scholar] [CrossRef]
- Neumayer, G.; Belzil, C.; Gruss, O.J.; Nguyen, M.D. TPX2: Of spindle assembly, DNA damage response, and cancer. Experientia 2014, 71, 3027–3047. [Google Scholar] [CrossRef]
- A Compton, D. Mechanisms of aneuploidy. Curr. Opin. Cell Biol. 2011, 23, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Maiato, H.; DeLuca, J.; Salmon, E.D.; Earnshaw, W. The dynamic kinetochore-microtubule interface. J. Cell Sci. 2004, 117, 5461–5477. [Google Scholar] [CrossRef]
- Jallepalli, P.V.; Lengauer, C. Chromosome segregation and cancer: Cutting through the mystery. Nat. Cancer 2001, 1, 109–117. [Google Scholar] [CrossRef]
- Hubner, N.C.; Bird, A.W.; Cox, J.; Splettstoesser, B.; Bandilla, P.; Poser, I.; Hyman, A.; Mann, M. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J. Cell Biol. 2010, 189, 739–754. [Google Scholar] [CrossRef]
- Petry, S.; Groen, A.C.; Ishihara, K.; Mitchison, T.J.; Vale, R.D. Branching Microtubule Nucleation in Xenopus Egg Extracts Mediated by Augmin and TPX2. Cell 2013, 152, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Cavazza, T.; Vernos, I. The RanGTP Pathway: From Nucleo-Cytoplasmic Transport to Spindle Assembly and Beyond. Front. Cell Dev. Biol. 2016, 3, 82. [Google Scholar] [CrossRef] [PubMed]
- Ozlu, N.; Srayko, M.; Kinoshita, K.; Habermann, B.; O’Toole E, T.; Muller-Reichert, T.; Schmalz, N.; Desai, A.; Hyman, A.A. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev. Cell 2005, 9, 237–248. [Google Scholar] [CrossRef]
- Goshima, G. Identification of a TPX2-like microtubule-associated protein in drosophila. PLoS ONE 2011, 6, e28120. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, R.; Sardon, T.; Vernos, I.; Conti, E. Structural Basis of Aurora-A Activation by TPX2 at the Mitotic Spindle. Mol. Cell 2003, 12, 851–862. [Google Scholar] [CrossRef]
- Giubettini, M.; Asteriti, I.A.; Scrofani, J.; De Luca, M.; Lindon, C.; Lavia, P.; Guarguaglini, G. Control of Aurora-A stability through interaction with TPX2. J. Cell Sci. 2011, 124, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Pinyol, R.; Scrofani, J.; Vernos, I. The role of Nedd1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr. Biol. 2013, 23, 143–149. [Google Scholar] [CrossRef]
- Scrofani, J.; Sardon, T.; Meunier, S.; Vernos, I. Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr. Biol. 2015, 25, 131–140. [Google Scholar] [CrossRef]
- Tsai, M.Y.; Zheng, Y. Aurora A kinase-coated beads function as microtubule-organizing centers and enhance RanGTP-induced spindle assembly. Curr. Biol. 2005, 15, 2156–2163. [Google Scholar] [CrossRef]
- Alfaro-Aco, R.; Thawani, A.; Petry, S. Structural analysis of the role of TPX2 in branching microtubule nucleation. J. Cell Biol. 2017, 216, 983–997. [Google Scholar] [CrossRef]
- Sanchez-Pulido, L.; Perez, L.; Kuhn, S.; Vernos, I.; Andrade-Navarro, M.A. The C-terminal domain of TPX2 is made of al-pha-helical tandem repeats. BMC Struct. Biol. 2016, 16, 17. [Google Scholar] [CrossRef] [Green Version]
- Brunet, S.; Sardon, T.; Zimmerman, T.; Wittmann, T.; Pepperkok, R.; Karsenti, E.; Vernos, I. Characterization of the TPX2 domains involved in microtubule nucleation and spindle assembly in Xenopus egg extracts. Mol. Biol. Cell 2004, 15, 5318–5328. [Google Scholar] [CrossRef] [PubMed]
- Eckerdt, F.; Eyers, P.A.; Lewellyn, A.L.; Prigent, C.; Maller, J.L. Spindle pole regulation by a discrete Eg5-interacting do-main in TPX2. Curr. Biol. 2008, 18, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Tanenbaum, M.E.; Macůrek, L.; Janssen, A.; Geers, E.F.; Fernández, M.A.; Medema, R.H. Kif15 cooperates with Eg5 to promote bipolar spindle assembly. Curr. Biol. 2009, 19, 1703–1711. [Google Scholar] [CrossRef] [PubMed]
- Helmke, K.J.; Heald, R. TPX2 levels modulate meiotic spindle size and architecture in Xenopus egg extracts. J. Cell Biol. 2014, 206, 385–393. [Google Scholar] [CrossRef] [PubMed]
- Gruss, O.J.; E Carazo-Salas, R.; A Schatz, C.; Guarguaglini, G.; Kast, J.; Wilm, M.; Le Bot, N.; Vernos, I.; Karsenti, E.; Mattaj, I.W. Ran induces spindle assembly by reversing the inhibitory effect of importin α on TPX2 activity. Cell 2001, 104, 83–93. [Google Scholar] [CrossRef]
- Caudron, M.; Bunt, G.; Bastiaens, P.; Karsenti, E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science 2005, 309, 1373–1376. [Google Scholar] [CrossRef]
- Garrido, G.; Vernos, I. Non-centrosomal TPX2-Dependent Regulation of the Aurora A Kinase: Functional Implications for Healthy and Pathological Cell Division. Front. Oncol. 2016, 6, 88. [Google Scholar] [CrossRef]
- Neumayer, G.; Nguyen, M.D. TPX2 impacts acetylation of histone H4 at lysine 16: Implications for DNA damage response. PLoS ONE 2014, 9, e110994. [Google Scholar] [CrossRef]
- Byrum, A.K.; Carvajal-Maldonado, D.; Mudge, M.C.; Valle-Garcia, D.; Majid, M.C.; Patel, R.; Sowa, M.E.; Gygi, S.P.; Harper, J.W.; Shi, Y.; et al. Mitotic regulators TPX2 and Aurora A protect DNA forks during replication stress by counteracting 53BP1 function. J. Cell Biol. 2019, 218, 422–432. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oh, W.; Wu, T.T.; Jeong, S.-Y.; You, H.J.; Lee, J.-H. CtIP Regulates Mitotic Spindle Assembly by Modulating the TPX2-Aurora A Signaling Axis. Cells 2022, 11, 2814. https://doi.org/10.3390/cells11182814
Oh W, Wu TT, Jeong S-Y, You HJ, Lee J-H. CtIP Regulates Mitotic Spindle Assembly by Modulating the TPX2-Aurora A Signaling Axis. Cells. 2022; 11(18):2814. https://doi.org/10.3390/cells11182814
Chicago/Turabian StyleOh, Wonkyung, Ting Ting Wu, Seo-Yeon Jeong, Ho Jin You, and Jung-Hee Lee. 2022. "CtIP Regulates Mitotic Spindle Assembly by Modulating the TPX2-Aurora A Signaling Axis" Cells 11, no. 18: 2814. https://doi.org/10.3390/cells11182814





