Efficient Gene Expression in Human Stem Cell Derived-Cortical Organoids Using Adeno Associated Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Embryonic Stem Cells (hESCs) Maintenance and Generation of hESCs-Derived Cortical Organoids (hCOs)
2.2. Immunofluorescence Staining
2.3. Quantitative Real-Time PCR
2.4. Lipofectamine Transfection
2.5. Electroporation and BrainFectIN
2.6. AAV Production and Transduction
2.7. RIPA Extraction and Western Blotting
2.8. Calcium Imaging
2.9. Statistical Analysis
3. Results
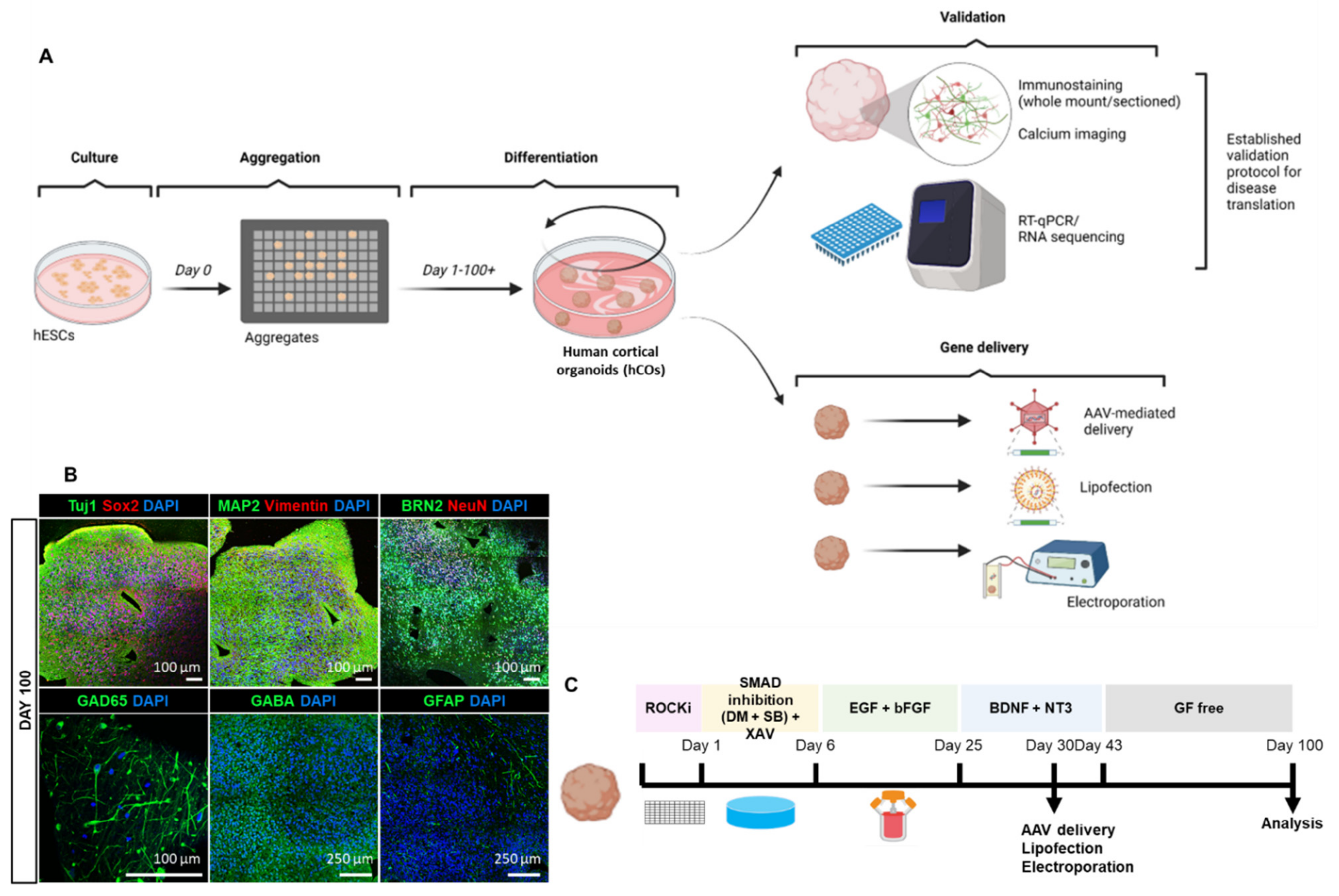
3.1. Reliable Production of hESCs-Derived Cortical Organoids (hCOs)
3.2. Homogenous Development of Diverse Cell Populations in hCOs
3.3. Exploring Various Transfection Delivery Methods in hCOs
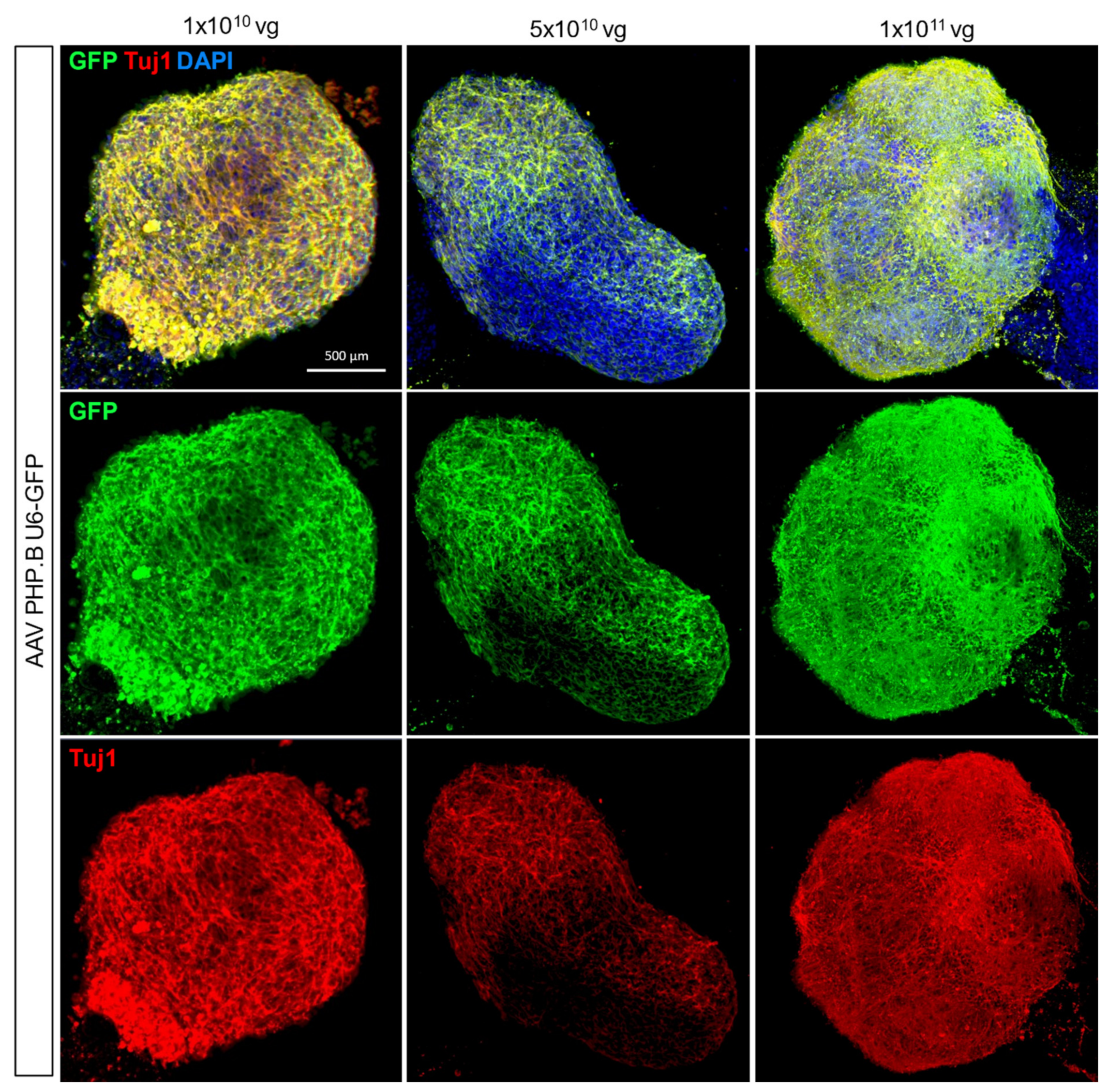
3.4. Adeno-Associated Virus-Mediated Gene Delivery in hCOs
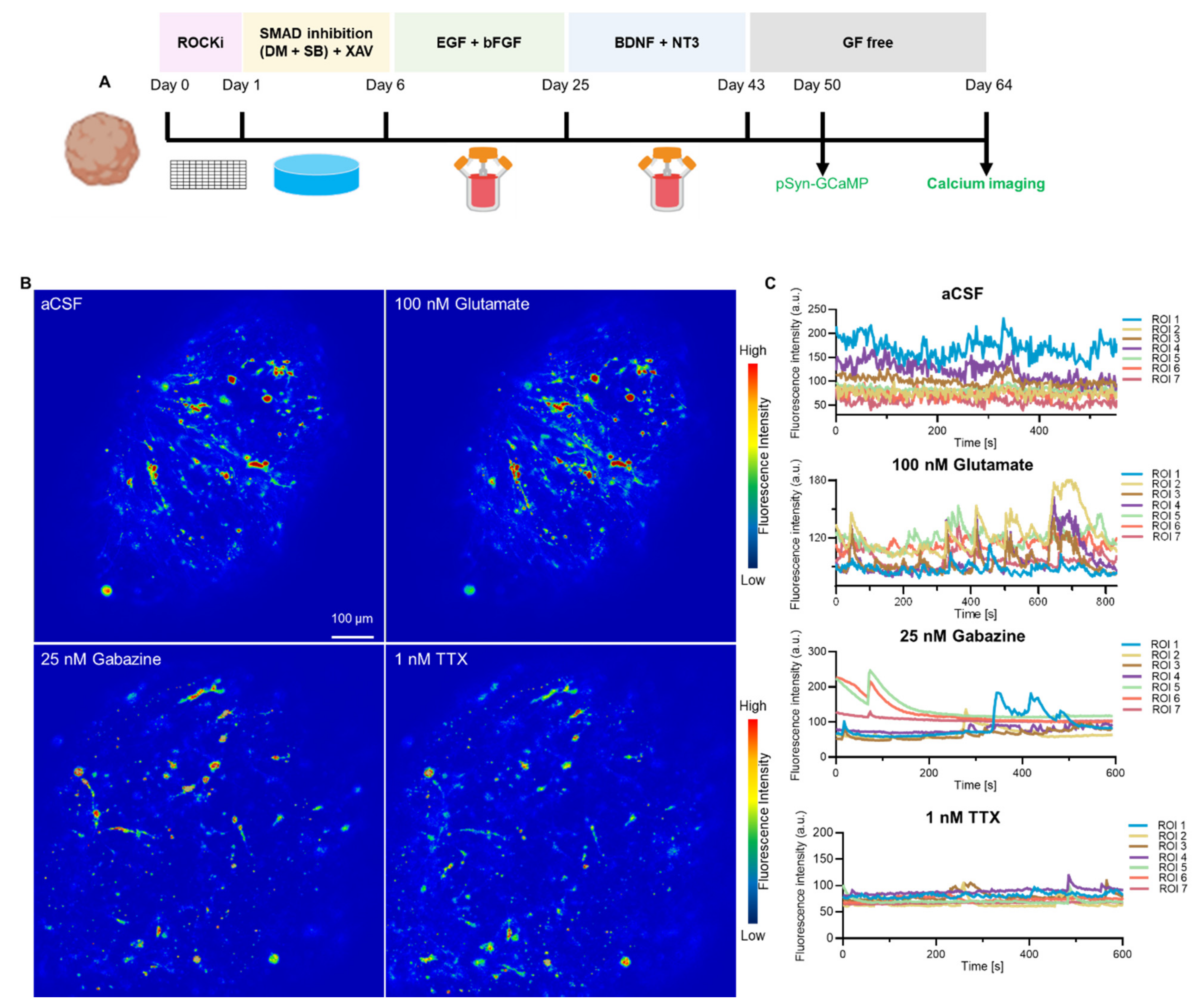
3.5. Neuron-Specific Calcium Functionality in hCOs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Yoon, S.-J.; Elahi, L.S.; Pașca, A.M.; Marton, R.M.; Gordon, A.; Revah, O.; Miura, Y.; Walczak, E.M.; Holdgate, G.M.; Fan, H.C.; et al. Reliability of human cortical organoid generation. Nat. Methods 2019, 16, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Giandomenico, S.L.; Sutcliffe, M.; Lancaster, M.A. Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nat. Protoc. 2020, 16, 579–602. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Corsini, N.S.; Wolfinger, S.; Gustafson, E.H.; Phillips, A.W.; Burkard, T.R.; Otani, T.; Livesey, F.J.; Knoblich, J.A. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017, 35, 659–666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szebényi, K.; Wenger, L.M.D.; Sun, Y.; Dunn, A.W.E.; Limegrover, C.A.; Gibbons, G.M.; Conci, E.; Paulsen, O.; Mierau, S.B.; Balmus, G.; et al. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci. 2021, 24, 1542–1554. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Shaker, M.R.; Aguado, J.; Chaggar, H.K.; Wolvetang, E.J. Klotho inhibits neuronal senescence in human brain organoids. npj Aging Mech. Dis. 2021, 7, 1–12. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F.; et al. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 2020, 26, 766–781.e9. [Google Scholar] [CrossRef]
- Ramani, A.; Müller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Müller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 targets neurons of 3D human brain organoids. EMBO J. 2020, 39, e106230. [Google Scholar] [CrossRef]
- Wulansari, N.; Darsono, W.H.W.; Woo, H.-J.; Chang, M.-Y.; Kim, J.; Bae, E.-J.; Sun, W.; Lee, J.-H.; Cho, I.-J.; Shin, H.; et al. Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci. Adv. 2021, 7, eabb1540. [Google Scholar] [CrossRef]
- Chen, X.; Sun, G.; Tian, E.; Zhang, M.; Davtyan, H.; Beach, T.G.; Reiman, E.M.; Blurton-Jones, M.; Holtzman, D.M.; Shi, Y. Modeling Sporadic Alzheimer’s Disease in Human Brain Organoids under Serum Exposure. Adv. Sci. 2021, 8, 2101462. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, Z.; Cao, J.; Yu, Y.; Pei, G. Chimeric cerebral organoids reveal the essentials of neuronal and astrocytic APOE4 for Alzheimer’s tau pathology. Signal Transduct. Target. Ther. 2022, 7, 1–10. [Google Scholar] [CrossRef]
- Bowles, K.R.; Silva, M.C.; Whitney, K.; Bertucci, T.; Berlind, J.E.; Lai, J.D.; Garza, J.C.; Boles, N.C.; Mahali, S.; Strang, K.H.; et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 2021, 184, 4547–4563.e17. [Google Scholar] [CrossRef]
- Pellegrini, L.; Lancaster, M.A. Modeling neurodegeneration with mutant-tau organoids. Cell 2021, 184, 4377–4379. [Google Scholar] [CrossRef]
- Camp, J.G.; Badsha, F.; Florio, M.; Kanton, S.; Gerber, T.; Wilsch-Bräuninger, M.; Lewitus, E.; Sykes, A.; Hevers, W.; Lancaster, M.A.; et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc. Natl. Acad. Sci. USA 2015, 112, 15672–15677. [Google Scholar] [CrossRef] [Green Version]
- Jabali, A.; Hoffrichter, A.; Uzquiano, A.; Marsoner, F.; Wilkens, R.; Siekmann, M.; Bohl, B.; Rossetti, A.C.; Horschitz, S.; Koch, P.; et al. Human cerebral organoids reveal progenitor pathology in EML1-linked cortical malformation. EMBO Rep. 2022, 23, e54027. [Google Scholar] [CrossRef]
- Samarasinghe, R.A.; Miranda, O.A.; Buth, J.E.; Mitchell, S.; Ferando, I.; Watanabe, M.; Allison, T.F.; Kurdian, A.; Fotion, N.N.; Gandal, M.J.; et al. Identification of neural oscillations and epileptiform changes in human brain organoids. Nat. Neurosci. 2021, 24, 1488–1500. [Google Scholar] [CrossRef]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef]
- Sharf, T.; van der Molen, T.; Glasauer, S.M.K.; Guzman, E.; Buccino, A.P.; Luna, G.; Cheng, Z.; Audouard, M.; Ranasinghe, K.G.; Kudo, K.; et al. Functional neuronal circuitry and oscillatory dynamics in human brain organoids. Nat. Commun. 2022, 13, 4403. [Google Scholar] [CrossRef]
- Andersen, J.; Revah, O.; Miura, Y.; Thom, N.; Amin, N.D.; Kelley, K.W.; Singh, M.; Chen, X.; Thete, M.V.; Walczak, E.M.; et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 2020, 183, 1913–1929.e26. [Google Scholar] [CrossRef]
- Bagley, J.A.; Reumann, D.; Bian, S.; Lévi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef]
- Ittner, L.M.; Klugmann, M.; Ke, Y.D. Adeno-associated virus-based Alzheimer’s disease mouse models and potential new therapeutic avenues. J. Cereb. Blood Flow Metab. 2019, 176, 3649–3665. [Google Scholar] [CrossRef]
- Kaspar, B.K.; Vissel, B.; Bengoechea, T.; Crone, S.; Randolph-Moore, L.; Muller, R.; Brandon, E.P.; Schaffer, D.; Verma, I.M.; Lee, K.-F.; et al. Adeno-associated virus effectively mediates conditional gene modification in the brain. Proc. Natl. Acad. Sci. USA 2002, 99, 2320–2325. [Google Scholar] [CrossRef] [Green Version]
- Marino, M.; Zhou, L.; Rincon, M.Y.; Callaerts-Vegh, Z.; Verhaert, J.; Wahis, J.; Creemers, E.; Yshii, L.; Wierda, K.; Saito, T.; et al. AAV-mediated delivery of an anti-BACE1 VHH alleviates pathology in an Alzheimer’s disease model. EMBO Mol. Med. 2022, 14, e09824. [Google Scholar] [CrossRef]
- Norante, R.P.; Massimino, M.L.; Lorenzon, P.; De Mario, A.; Peggion, C.; Vicario, M.; Albiero, M.; Sorgato, M.C.; Lopreiato, R.; Bertoli, A. Generation and validation of novel adeno-associated viral vectors for the analysis of Ca2+ homeostasis in motor neurons. Sci. Rep. 2017, 7, 6521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaj, T.; Ojala, D.S.; Ekman, F.K.; Byrne, L.C.; Limsirichai, P.; Schaffer, D.V. In vivo genome editing improves motor function and extends survival in a mouse model of ALS. Sci. Adv. 2017, 3, eaar3952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, T.; Chai, H.L.; Teramoto, S.; Tsuji, S.; Shimazaki, K.; Muramatsu, S.I.; Kwak, S. Rescue of amyotrophic lateral sclerosis phenotype in a mouse model by intravenous AAV9-ADAR2 delivery to motor neurons. EMBO Mol. Med. 2013, 5, 1710–1719. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ho, W.Y.; Lim, K.; Feng, J.; Tucker-Kellogg, G.; Nave, K.-A.; Ling, S.-C. Cell-autonomous requirement of TDP-43, an ALS/FTD signature protein, for oligodendrocyte survival and myelination. Proc. Natl. Acad. Sci. USA 2018, 115, E10941–E10950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, R.H.; Yang, Y.; Kim, W.S.; Dobson-Stone, C.; Kwok, J.B.; Kiernan, M.C.; Halliday, G.M. Distinct TDP-43 inclusion morphologies in frontotemporal lobar degeneration with and without amyotrophic lateral sclerosis. Acta Neuropathol. Commun. 2017, 5, 76. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, I.R.; Rademakers, R. The role of transactive response DNA-binding protein-43 in amyotrophic lateral sclerosis and frontotemporal dementia. Curr. Opin. Neurol. 2008, 21, 693–700. [Google Scholar] [CrossRef] [Green Version]
- Ke, Y.D.; Van Hummel, A.; Stevens, C.H.; Gladbach, A.; Ippati, S.; Bi, M.; Lee, W.S.; Kruger, S.; Van Der Hoven, J.; Volkerling, A.; et al. Short-term suppression of A315T mutant human TDP-43 expression improves functional deficits in a novel inducible transgenic mouse model of FTLD-TDP and ALS. Acta Neuropathol. 2015, 130, 661–678. [Google Scholar] [CrossRef]
- Kabashi, E.; Valdmanis, P.; Dion, P.; Spiegelman, D.; McConkey, B.J.; Velde, C.V.; Bouchard, J.-P.; Lacomblez, L.; Pochigaeva, K.; Salachas, F.; et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008, 40, 572–574. [Google Scholar] [CrossRef]
- Sreedharan, J.; Blair, I.P.; Tripathi, V.B.; Hu, X.; Vance, C.; Rogelj, B.; Ackerley, S.; Durnall, J.C.; Williams, K.L.; Buratti, E.; et al. TDP-43 Mutations in Familial and Sporadic Amyotrophic Lateral Sclerosis. Science 2008, 319, 1668–1672. [Google Scholar] [CrossRef]
- Ma, X.R.; Prudencio, M.; Koike, Y.; Vatsavayai, S.C.; Kim, G.; Harbinski, F.; Briner, A.; Rodriguez, C.M.; Guo, C.; Akiyama, T.; et al. TDP-43 represses cryptic exon inclusion in the FTD–ALS gene UNC13A. Nature 2022, 603, 124–130. [Google Scholar] [CrossRef]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Bi, M.; Delerue, F.; Forrest, S.L.; Chan, G.; van der Hoven, J.; van Hummel, A.; Feiten, A.F.; Lee, S.; Martinez-Valbuena, I.; et al. Loss of LAMP5 interneurons drives neuronal network dysfunction in Alzheimer’s disease. Acta Neuropathol. 2022, 144, 637–650. [Google Scholar] [CrossRef]
- Ke, Y.; Dramiga, J.; Schütz, U.; Kril, J.J.; Ittner, L.M.; Schröder, H.; Götz, J. Tau-mediated nuclear depletion and cytoplasmic accumulation of SFPQ in Alzheimer’s and Pick’s disease. PLoS ONE 2012, 7, e35678. [Google Scholar] [CrossRef]
- Zafeiriou, M.-P.; Bao, G.; Hudson, J.; Halder, R.; Blenkle, A.; Schreiber, M.-K.; Fischer, A.; Schild, D.; Zimmermann, W.-H. Developmental GABA polarity switch and neuronal plasticity in Bioengineered Neuronal Organoids. Nat. Commun. 2020, 11, 3791. [Google Scholar] [CrossRef]
- Miura, Y.; Li, M.-Y.; Birey, F.; Ikeda, K.; Revah, O.; Thete, M.V.; Park, J.-Y.; Puno, A.; Lee, S.H.; Porteus, M.H.; et al. Generation of human striatal organoids and cortico-striatal assembloids from human pluripotent stem cells. Nat. Biotechnol. 2020, 38, 1421–1430. [Google Scholar] [CrossRef]
- Kahle, K.T.; Staley, K.J.; Nahed, B.; Gamba, G.; Hebert, S.C.; Lifton, R.P.; Mount, D.B. Roles of the cation–chloride cotransporters in neurological disease. Nat. Clin. Pr. Cardiovasc. Med. 2008, 4, 490–503. [Google Scholar] [CrossRef]
- Moore, Y.E.; Kelley, M.R.; Brandon, N.J.; Deeb, T.Z.; Moss, S.J. Seizing Control of KCC2: A New Therapeutic Target for Epilepsy. Trends Neurosci. 2017, 40, 555–571. [Google Scholar] [CrossRef]
- Burrell, J.R.; Halliday, G.M.; Kril, J.J.; Ittner, L.M.; Götz, J.; Kiernan, M.C.; Hodges, J.R. The frontotemporal dementia-motor neuron disease continuum. Lancet 2016, 388, 919–931. [Google Scholar] [CrossRef]
- Martinez-Marmol, R.; Giordano-Santini, R.; Kaulich, E.; Cho, A.N.; Riyadh, M.A.; Robinson, E.; Balistreri, G.; Meunier, F.A.; Ke, Y.D.; Ittner, L.M.; et al. The SARS-CoV-2 spike (S) and the orthoreovirus p15 cause neuronal and glial fusion. bioRxiv 2021. [Google Scholar] [CrossRef]
- Renner, H.; Grabos, M.; Becker, K.J.; Kagermeier, T.E.; Wu, J.; Otto, M.; Peischard, S.; Zeuschner, D.; Tsytsyura, Y.; Disse, P.; et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. eLife 2020, 9, e52904. [Google Scholar] [CrossRef] [PubMed]
- Ippati, S.; Deng, Y.; van der Hoven, J.; Heu, C.; van Hummel, A.; Chua, S.W.; Paric, E.; Chan, G.; Feiten, A.; Fath, T.; et al. Rapid initiation of cell cycle reentry processes protects neurons from amyloid-β toxicity. Proc. Natl. Acad. Sci. USA 2021, 118, e2011876118. [Google Scholar] [CrossRef] [PubMed]
- Kanton, S.; Boyle, M.J.; He, Z.; Santel, M.; Weigert, A.; Sanchís-Calleja, F.; Guijarro, P.; Sidow, L.; Fleck, J.S.; Han, D.; et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 2019, 574, 418–422. [Google Scholar] [CrossRef]
- Velasco, S.; Kedaigle, A.J.; Simmons, S.K.; Nash, A.; Rocha, M.; Quadrato, G.; Paulsen, B.; Nguyen, L.; Adiconis, X.; Regev, A.; et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 2019, 570, 523–527. [Google Scholar] [CrossRef]
- Sawada, T.; Chater, T.E.; Sasagawa, Y.; Yoshimura, M.; Fujimori-Tonou, N.; Tanaka, K.; Benjamin, K.J.M.; Paquola, A.C.M.; Erwin, J.A.; Goda, Y.; et al. Developmental excitation-inhibition imbalance underlying psychoses revealed by single-cell analyses of discordant twins-derived cerebral organoids. Mol. Psychiatry 2020, 25, 2695–2711. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, A.-N.; Bright, F.; Morey, N.; Au, C.; Ittner, L.M.; Ke, Y.D. Efficient Gene Expression in Human Stem Cell Derived-Cortical Organoids Using Adeno Associated Virus. Cells 2022, 11, 3194. https://doi.org/10.3390/cells11203194
Cho A-N, Bright F, Morey N, Au C, Ittner LM, Ke YD. Efficient Gene Expression in Human Stem Cell Derived-Cortical Organoids Using Adeno Associated Virus. Cells. 2022; 11(20):3194. https://doi.org/10.3390/cells11203194
Chicago/Turabian StyleCho, Ann-Na, Fiona Bright, Nicolle Morey, Carol Au, Lars M. Ittner, and Yazi D. Ke. 2022. "Efficient Gene Expression in Human Stem Cell Derived-Cortical Organoids Using Adeno Associated Virus" Cells 11, no. 20: 3194. https://doi.org/10.3390/cells11203194
APA StyleCho, A. -N., Bright, F., Morey, N., Au, C., Ittner, L. M., & Ke, Y. D. (2022). Efficient Gene Expression in Human Stem Cell Derived-Cortical Organoids Using Adeno Associated Virus. Cells, 11(20), 3194. https://doi.org/10.3390/cells11203194





