Interactions of PARP1 Inhibitors with PARP1-Nucleosome Complexes
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. PARP1 Inhibitors Do Not Affect the Nucleosome Structure
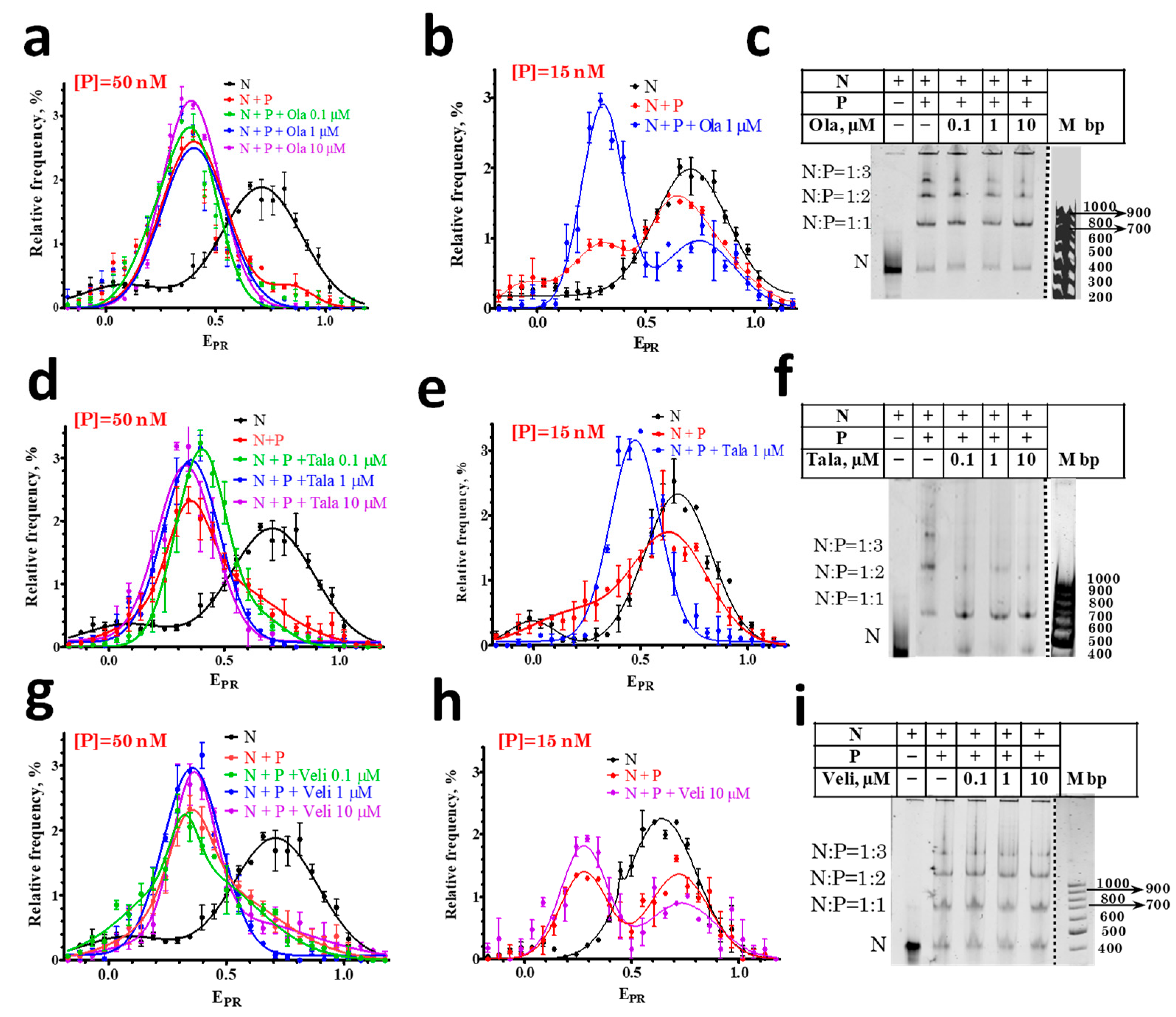
3.2. Interaction of PARPi with Nucleosome-PARP1 Complexes
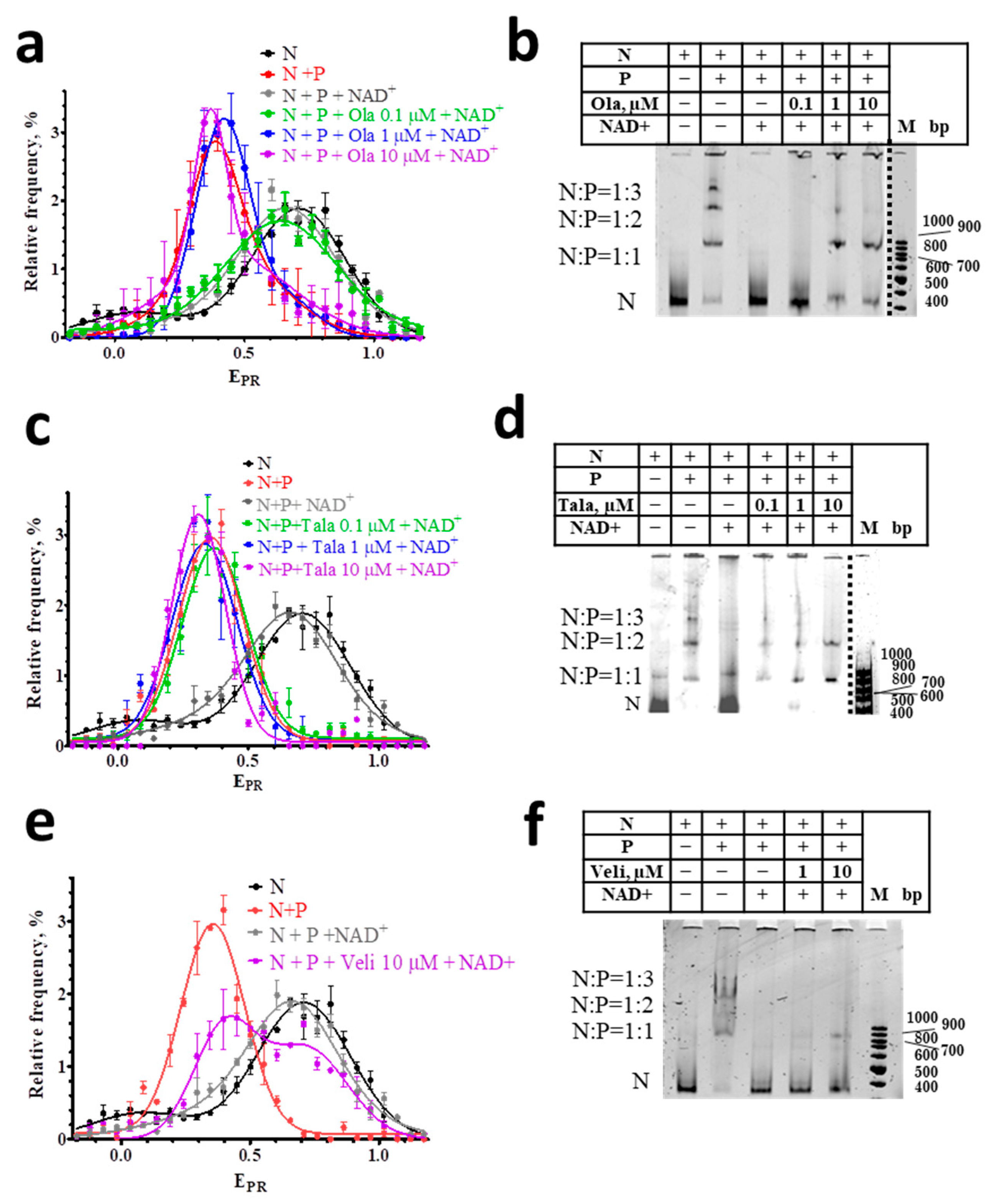
3.3. Effect of PARPi on the Interaction of NAD+ with Nucleosome-PARP1 Complexes
4. Discussion
PARP1 + N + PARPi ⇄ PARP1:N + PARPi ⇄ PARPi:PARP1:N,
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kraus, W.L. PARPs and ADP-ribosylation: 60 years on. Genes Dev. 2020, 34, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Eisemann, T.; Pascal, J.M. Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell. Mol. Life Sci. 2020, 77, 19–33. [Google Scholar] [CrossRef] [PubMed]
- Maluchenko, N.V.; Koshkina, D.O.; Feofanov, A.V.; Studitsky, V.M.; Kirpichnikov, M.P. Poly(ADP-Ribosyl) Code Functions. Acta Nat. 2021, 13, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Maluchenko, N.V.; Feofanov, A.V.; Studitsky, V.M. PARP-1-Associated Pathological Processes: Inhibition by Natural Polyphenols. Int. J. Mol. Sci. 2021, 22, 11441. [Google Scholar] [CrossRef] [PubMed]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear func-tions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, A.B.B.; Holtlund, J.; Hilz, H. Immunoquantitation and size determination of intrinsic poly(ADP-ribose) polymerase from acid precipitates. An analysis of the in vivo status in mammalian species and in lower eukaryotes. J. Biol. Chem. 1988, 263, 6993–6999. [Google Scholar] [CrossRef]
- Yamanaka, H.P.C.; Willis, E.H.; Wasson, D.B.; Carson, D.A. Characterization of human poly(ADP-ribose) polymerase with au-toantibodies. J. Biol. Chem. 1988, 263, 3879–3883. [Google Scholar] [CrossRef]
- Brady, P.N.; Goel, A.; Johnson, M.A. Poly(ADP-Ribose) Polymerases in Host-Pathogen Interactions, Inflammation, and Immunity. Microbiol. Mol. Biol. Rev. MMBR 2019, 83, e00038-18. [Google Scholar] [CrossRef] [Green Version]
- Mateo, J.; Lord, C.J.; Serra, V.; Tutt, A.; Balmana, J.; Castroviejo-Bermejo, M.; Cruz, C.; Oaknin, A.; Kaye, S.B.; de Bono, J.S. A decade of clinical development of PARP inhibitors in perspective. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1437–1447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtin, N.J.; Szabo, C. Poly(ADP-ribose) polymerase inhibition: Past, present and future. Nat. Rev. Drug Discov. 2020, 19, 711–736. [Google Scholar] [CrossRef]
- Murai, J. Targeting DNA repair and replication stress in the treatment of ovarian cancer. Int. J. Clin. Oncol. 2017, 22, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, T.A.; Shi, Y.; Rodriguez, L.E.; Solomon, L.R.; Donawho, C.K.; DiGiammarino, E.L.; Panchal, S.C.; Wilsbacher, J.L.; Gao, W.; Olson, A.M.; et al. Mechanistic Dissection of PARP1 Trapping and the Impact on In Vivo Tolerability and Efficacy of PARP Inhibitors. Mol. Cancer Res. 2015, 13, 1465–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashworth, A.; Lord, C.J. Synthetic lethal therapies for cancer: What’s next after PARP inhibitors? Nat. Rev. Clin. Oncol. 2018, 15, 564–576. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Eisemann, T.; Riccio, A.A.; Pascal, J.M. PARP family enzymes: Regulation and catalysis of the poly(ADP-ribose) posttranslational modification. Curr. Opin. Struct. Biol. 2018, 53, 187–198. [Google Scholar] [CrossRef]
- Rudolph, J.; Jung, K.; Luger, K. Inhibitors of PARP: Number crunching and structure gazing. Proc. Natl. Acad. Sci. USA 2022, 119, e2121979119. [Google Scholar] [CrossRef]
- Rudolph, J.; Mahadevan, J.; Luger, K. Probing the Conformational Changes Associated with DNA Binding to PARP1. Biochemistry 2020, 59, 2003–2011. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.H.S.; Das, B.B.; Renaud, A.; Zhang, Y.; Dorowhow, J.H. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012, 72, 5588–5599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murai, J.; Marchand, C.; Shahane, S.A.; Sun, H.; Huang, R.; Zhang, Y.; Chergui, A.; Ji, J.; Doroshow, J.H.; Jadhav, A.; et al. Identification of novel PARP inhibitors using a cell-based TDP1 inhibitory assay in a quantitative high-throughput screening platform. DNA Repair 2014, 21, 177–182. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, T.A.; Ainsworth, W.B.; Ellis, P.A.; Donawho, C.K.; DiGiammarino, E.L.; Panchal, S.C.; Abraham, V.C.; Algire, M.A.; Shi, Y.; Olson, A.M.; et al. PARP1 Trapping by PARP Inhibitors Drives Cyto-toxicity in Both Cancer Cells and Healthy Bone Marrow. Mol. Cancer Res. MCR 2019, 17, 409–419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zandarashvili, L.; Langelier, M.-F.; Velagapudi, U.K.; Hancock, M.A.; Steffen, J.D.; Billur, R.; Hannan, Z.M.; Wicks, A.J.; Krastev, D.B.; Pettitt, S.J.; et al. Structural basis for allosteric PARP-1 retention on DNA breaks. Science 2020, 368, eaax6367. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Pommier, Y. Classification of PARP Inhibitors Based on PARP Trapping and Catalytic Inhibition, and Rationale for Combinations with Topoisomerase I Inhibitors and Alkylating Agents. In PARP Inhibitors for Cancer Therapy; Curtin, N., Sharma, R., Eds.; Cancer Drug Discovery and Development, Humana Press: Cham, Switzerland, 2015; Volume 83, pp. 261–274. [Google Scholar]
- Langelier, M.-F.; Zandarashvili, L.; Aguiar, P.M.; Black, B.E.; Pascal, J.M. NAD+ analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xue, H.; Bhardwaj, A.; Yin, Y.; Fijen, C.; Ephstein, A.; Zhang, L.; Ding, X.; Pascal, J.M.; VanArsdale, T.L.; Rothenberg, E. A two-step mechanism governing PARP1-DNA retention by PARP inhibitors. Sci. Adv. 2022, 8, eabq0414. [Google Scholar] [CrossRef]
- Rudolph, J.; Roberts, G.; Luger, K. Histone Parylation factor 1 contributes to the inhibition of PARP1 by cancer drugs. Nat. Commun. 2021, 12, 736. [Google Scholar] [CrossRef] [PubMed]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and Analysis of Uniquely Positioned Mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [CrossRef] [Green Version]
- Langelier, M.-F.; Steffen, J.D.; Riccio, A.A.; McCauley, M.; Pascal, J.M. Purification of DNA Damage-Dependent PARPs from E. coli for Structural and Biochemical Analysis. Methods Mol. Biol. 2017, 1608, 431–444. [Google Scholar] [CrossRef]
- Kudryashova, K.S.; Nikitin, D.V.; Chertkov, O.V.; Gerasimova, N.S.; Valieva, M.E.; Studitsky, V.M.; Feofanov, A.V. Devel-opment of fluorescently labeled mononucleosomes to study transcription mechanisms by method of microscopy of single complexes. Mosc. Univ. Biol. Sci. Bull. 2015, 70, 189–193. [Google Scholar] [CrossRef]
- Valieva, M.E.; Armeev, G.A.; Kudryashova, K.S.; Gerasimova, N.S.; Shaytan, A.K.; Kulaeva, O.I.; McCullough, L.L.; Formosa, T.; Georgiev, P.G.; Kirpichnikov, M.P.; et al. Large-scale ATP-independent nucleo-some unfolding by a histone chaperone. Nat. Struct. Mol. Biol. 2016, 23, 1111–1116. [Google Scholar] [CrossRef] [Green Version]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.F.; Pascal, J.M.; Akhtar, M.S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. [Google Scholar] [CrossRef]
- Sultanov, D.C.; Gerasimova, N.S.; Kudryashova, K.S.; Maluchenko, N.V.; Kotova, E.Y.; Langelier, M.F.; Pascal, J.M.; Kirpichnikov, M.P.; Feofanov, A.V.; Studitsky, V.M. Unfolding of core nucleosomes by PARP-1 revealed by spFRET mi-croscopy. AIMS Genet. 2017, 4, 21–31. [Google Scholar]
- Messner, S.; Altmeyer, M.; Zhao, H.; Pozivil, A.; Roschitzki, B.; Gehrig, P.; Rutishauser, D.; Huang, D.; Caflisch, A.; O Hottiger, M. PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res. 2010, 38, 6350–6362. [Google Scholar] [CrossRef] [Green Version]
- Ciccarone, F.; Zampieri, M.; Caiafa, P. PARP1 orchestrates epigenetic events setting up chromatin domains. Semin. Cell Dev. Biol. 2017, 63, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.D.; Gagne, J.P.; Poirier, G.G.; Goodlett, D.R. Mapping PARP-1 auto-ADP-ribosylation sites by liquid chro-matography-tandem mass spectrometry. J. Proteome Res. 2013, 12, 1868–1880. [Google Scholar] [CrossRef] [PubMed]
- Muthurajan, U.M.; Hepler, M.R.D.; Hieb, A.R.; Clark, N.J.; Kramer, M.; Yao, T.; Luger, K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA 2014, 111, 12752–12757. [Google Scholar] [CrossRef] [Green Version]
- Ummarino, S.; Hausman, C.; Di Ruscio, A. The PARP Way to Epigenetic Changes. Genes 2021, 12, 446. [Google Scholar] [CrossRef] [PubMed]
- Kurgina, T.A.; Moor, N.A.; Kutuzov, M.M.; Naumenko, K.N.; Ukraintsev, A.A.; Lavrik, O.I. Dual function of HPF1 in the modulation of PARP1 and PARP2 activities. Commun. Biol. 2021, 4, 1259. [Google Scholar] [CrossRef]
- Kutuzov, M.M.; Belousova, E.A.; Kurgina, T.A.; Ukraintsev, A.A.; Vasil’Eva, I.A.; Khodyreva, S.N.; Lavrik, O.I. The contribution of PARP1, PARP2 and poly(ADP-ribosyl)ation to base excision repair in the nucleosomal context. Sci. Rep. 2021, 11, 1–17. [Google Scholar] [CrossRef]
- Kotova, E.P.A.; Tulin, A.V. Small-molecule collection and high-throughput colorimetric assay to identify PARP-1 inhibitors. Methods Mol. Biol. 2011, 780, 491–516. [Google Scholar]
- Thomas, C.J.; Kotova, E.; Andrake, M.; Adolf-Bryfogle, J.; Glaser, R.; Regnard, C.; Tulin, A.V. Kinase-Mediated Changes in Nucleosome Conformation Trigger Chromatin Decondensation via Poly(ADP-Ribosyl)ation. Mol. Cell 2014, 53, 831–842. [Google Scholar] [CrossRef] [Green Version]
- Kotova, E.; Tulin, A.V. High-Throughput Colorimetric Assay for Identifying PARP-1 Inhibitors Using a Large Small-Molecule Collection. Methods Mol. Biol. 2017, 1608, 299–312. [Google Scholar] [CrossRef]
- Steffen, J.D.; Brody, J.R.; Armen, R.S.; Pascal, J.M. Structural Implications for Selective Targeting of PARPs. Front. Oncol. 2013, 3, 301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nilov, D.K.; Pushkarev, S.V.; Gushchina, I.V.; Manasaryan, G.A.; Kirsanov, K.I.; Svedas, V.K. Modeling of the En-zyme-Substrate Complexes of Human Poly(ADP-Ribose) Polymerase 1. Biochemistry. Biokhimiia 2020, 85, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tokuda, J.M.; Topping, T.; Meisburger, S.P.; Pabit, S.A.; Gloss, L.M.; Pollack, L. Asymmetric unwrapping of nucleosomal DNA propagates asymmetric opening and dissociation of the histone core. Proc. Natl. Acad. Sci. USA 2016, 114, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Gansen, A.; Felekyan, S.; Kühnemuth, R.; Lehmann, K.; Tóth, K.; Seidel, C.A.M.; Langowski, J. High precision FRET studies reveal reversible transitions in nucleosomes between microseconds and minutes. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudolph, J.; Mahadevan, J.; Dyer, P.; Luger, K. Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mecha-nism. eLife 2018, 7, e37818. [Google Scholar] [CrossRef] [PubMed]
- Eustermann, S.V.H.; Yang, J.C.; Cole, P.T.; Gruszka, D.; Veprintsev, D.; Neuhaus, D. The DNA-binding domain of humanPARP-1 interacts with DNAsingle-strand breaks as a monomer through its second zinc finger. J. Mol. Biol. 2011, 407, 149–170. [Google Scholar] [CrossRef] [Green Version]
- Alvarez-Gonzalez, R.; Althaus, F.R. Poly(ADP-ribose) catabolism in mammalian cells exposed to DNA-damaging agents. Mutat. Res. Repair 1989, 218, 67–74. [Google Scholar] [CrossRef]
- Sukhanova, M.V.; Hamon, L.; Kutuzov, M.M.; Joshi, V.; Abrakhi, S.; Dobra, I.; Curmi, P.A.; Pastre, D.; Lavrik, O.I. A Single-Molecule Atomic Force Microscopy Study of PARP1 and PARP2 Recognition of Base Excision Repair DNA Interme-diates. J. Mol. Biol. 2019, 431, 2655–2673. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maluchenko, N.; Koshkina, D.; Korovina, A.; Studitsky, V.; Feofanov, A. Interactions of PARP1 Inhibitors with PARP1-Nucleosome Complexes. Cells 2022, 11, 3343. https://doi.org/10.3390/cells11213343
Maluchenko N, Koshkina D, Korovina A, Studitsky V, Feofanov A. Interactions of PARP1 Inhibitors with PARP1-Nucleosome Complexes. Cells. 2022; 11(21):3343. https://doi.org/10.3390/cells11213343
Chicago/Turabian StyleMaluchenko, Natalya, Darya Koshkina, Anna Korovina, Vasily Studitsky, and Alexey Feofanov. 2022. "Interactions of PARP1 Inhibitors with PARP1-Nucleosome Complexes" Cells 11, no. 21: 3343. https://doi.org/10.3390/cells11213343
APA StyleMaluchenko, N., Koshkina, D., Korovina, A., Studitsky, V., & Feofanov, A. (2022). Interactions of PARP1 Inhibitors with PARP1-Nucleosome Complexes. Cells, 11(21), 3343. https://doi.org/10.3390/cells11213343






