Abstract
The impact of transposable elements (TEs) on the evolution of the eukaryote genome has been observed in a number of biological processes, such as the recruitment of the host’s gene expression network or the rearrangement of genome structure. However, TEs may also provide a substrate for the emergence of novel repetitive elements, which contribute to the generation of new genomic components during the course of the evolutionary process. In this review, we examine published descriptions of TEs that give rise to tandem sequences in an attempt to comprehend the relationship between TEs and the emergence of de novo satellite DNA families in eukaryotic organisms. We evaluated the intragenomic behavior of the TEs, the role of their molecular structure, and the chromosomal distribution of the paralogous copies that generate arrays of repeats as a substrate for the emergence of new repetitive elements in the genome. We highlight the involvement and importance of TEs in the eukaryote genome and its remodeling processes.
1. Introduction
The duplication and mobilization cycles of transposable elements (TEs) are evolutionary processes that enrich the paralogous copies that constitute the repetitive DNA content of the eukaryote genome. This repetitive DNA can be subdivided into two categories: tandem repeat sequences, which represent copies organized in juxtaposition to one another (e.g., DNA satellites, minisatellites, and microsatellites), or dispersed sequences, which include the transposable elements themselves [1,2].
In most cases, transposable elements and tandem repeats are studied independently, but evidence has been found that TEs may be involved in the origin of a library of tandem repeats that is typically dispersed throughout the eukaryote genome, where it plays a fundamental evolutionary role [3,4]. The sequences of homologies of satDNAs and transposons or retrotransposons that have been identified in many species indicate the existence of an intimate evolutionary relationship between the TEs and the emergence of tandem repeat sequences, which implies that the TEs are involved in the reshaping of the genomic architecture [3,5].
The conversion of one type of repetitive element into another has been reported in all the principal branches of the eukaryote tree of life [6,7,8]. Here, we review case studies of the emergence of repetitive DNA, focusing on the new satellite DNA (satDNA) families derived from TEs with the aim of understanding (i) the intragenomic behavior of the TEs, (ii) the role of their molecular structure, and (iii) the chromosomal distribution of the paralogous copies. We discuss the genetic mechanisms that produce the TE copies and contribute to the molecular co-option of these elements during the evolution of the genome and chromosomes, most of which are revealed by the interplay and turnover of the different repetitive classes in the genome.
There are two major categories of transposable elements: classes I and II. Class I elements, which are also known as retrotransposons, are dependent on the RNA for their transposition in the genome, while class II elements (DNA transposons) do not depend on any retrotranscription mechanism for their mobility [9].
The recruitment of copies of the TEs as a substrate for the evolutionary emergence of new tandem repeats and satDNA families is directly dependent on the behavior of these elements in the host genome. The intra-genomic behavior of the TEs in the host genome follows a four-phase life cycle (Figure 1), starting with (i) the birth or invasion phase, when the TEs are inserted into a new locus, followed by (ii) amplification, (iii) maturation, and finally, (iv) death or degeneration [10,11,12].
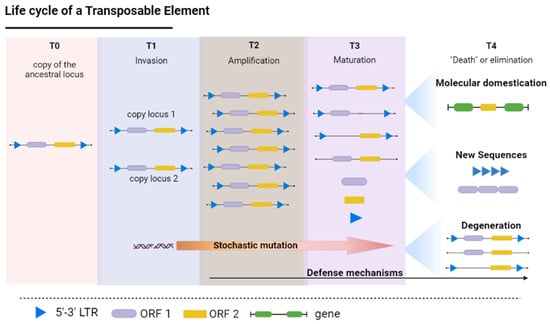
Figure 1.
Life cycle of a transposable element, using LTR transposons with their respective Open Reading Frames (ORFs) as an example. T0—simple copy of the element in the genome; T1—Invasion: the chromosomal locus acquires at least one repeat unit; T2—Amplification: an increase in the number of copies present in the genome, either by burst events (in the case of class I elements) or the repair or homologous recombination of double-stranded DNA (class II); T3—Maturation: the elements are inactivated or silenced through epigenetic silencing, piRNAs, DNA methylation or other mechanisms, such as mutations; T4—Death or degeneration: the elements may be either eliminated from the genome, undergo molecular domestication, begin to exercise new functions or give rise to new repetitive sequences, such as tandem repeats. LTR = Long Terminal Repeat; ORF = Open Reading Frame. (Source: the authors / created in Biorender.com, accessed on 5 September 2022).
The birth or invasion phase (Figure 1—T1) occurs when a new chromosomal locus acquires at least one copy of a TE. In most cases, the TE is transmitted vertically through the evolutionary lineage by splitting from the ancestral species, while invasion occurs through new transposition events, either by the recombination of sequences already present in the genome or events of horizontal transfer [13,14].
As a new repeat unit of a class I element (retrotransposon) is established in the genome, the number of copies begins to increase rapidly (amplification phase; T3 in Figure 1). These paralogous copies, generated by retrotransposition, are distributed randomly throughout the genome [15,16]. Genomes of all characterized higher eukaryotes possess examples of transposable element (TE) bursts [17]. Burst events can cause a local accumulation of TEs in the chromosomes, generating extensive clusters of TEs [18,19,20,21].
Events of this type are more frequent in germ cells due to the temporary relaxation of the epigenetic control of the TEs during the early development of the germline. This opens a temporal window that may allow these elements to escape from their constraints and propagate in the host genome [22,23]. Amplification occurs primarily in male germ cells due to the continuous spermatogenesis occurring throughout the life span of the animal, whereas female germlines are arrested in meiosis and do not undergo the temporary relaxation of epigenetic control observed in the male germline [24,25].
By contrast, the repair mechanism of double-strand (subclass I) or single-strand breaks (subclass II; see [9]) that occur following mobilization events is a central genetic process that results in the addition of a number of copies of the DNA transposons, given that the chromosomal gap generated by the excision of a transposon sequence is repaired by a homologous recombination mechanism, which results in the reintroduction of the transposon at the donor site [26]. If the mobilization event occurs during the S phase, the homologous recombination can use a sister chromatid as a template for the repair of the chromosome gap, which also restores the excised element and results in an increase in the number of copies of the class II elements [26,27].
The maturation phase (T4 in Figure 1) is the most prolonged stage of the process, and is the most subject to the action of mechanisms of molecular evolution. The host genome can evolve defenses to stop the spread of the TEs by temporary epigenetic inactivation, irrespective of the mobilization mechanism involved in the insertion of the new TE [28]. During this phase, the silencing of transposition activity and the relaxation of selection pressures on the paralogous copies contribute to the accumulation of random mutations in the molecular structure of the TE sequences [29,30].
One of these pathways is through the silencing of the RNA by small RNA molecules—endogenous siRNAs (endo-siRNAs) or PIWI-associated RNAs (piRNAs)—which guide the process by modulating chromatin states or targeting the degradation of the RNA [31]. The piRNA pathway suppresses transposon activity in the metazoan germline, whereas the endo-siRNAs repress TE activity in the somatic tissue [31,32].
The transposition of the TEs may also be suppressed by DNA methylation, in which a methyl group is added covalently to the C’5 position of the cytosine (5-methylcytosine) [31,33]. This methylation is associated with repression of the TE, as first observed in the retrotransposon Activator (Ac), Suppressor-mutator (Spm), and Mutator (Mu) of maize [34,35,36]. This epigenetic suppression by DNA methylation is thought to contribute not only to the silencing of transcription, but also to the formation of the heterochromatic regions of the genome [37,38]. Epigenetic histone modifications, such as acetylation, phosphorylation, and methylation, may also be involved in the repression of the TEs [39]. In the genus Arabidopsis, for example, the histone methylation of H3K9me2 and H3K27me1 appears to contribute to the transcriptional silencing of some TEs [40], while histone deacetylase triggers the transcriptional activity of the different TEs [41]. The long-time prevalence of TEs under epigenetic control results in the progressive accumulation of mutations (substitutions or indels), which leads, in turn, to the degeneration of the sequence and a loss of identity, which either reduces or eliminates its capacity for amplification in the genome (Figure 1—T3). Recombination is often suppressed in the heterochromatic regions [42,43], which appears to be one of the reasons why these portions of the chromosome are prone to the accumulation of TEs, maintained by unequal crossing-over or genetic drift [44,45].
Stochastic mutations that interfere with the mobilization capacity of the silenced TE sequences are either fixed or lost primarily through genetic drift, which may represent the degeneration or death phase of the respective locus (Figure 1—T4). In this case, the heterochromatin is often the “final resting place” of dysfunctional TEs, due to the low frequency of recombination in these regions [44,45], leading to the formation of a “TE cemetery” [45,46], where the TEs may play an important role in the structure of the heterochromatin itself [47]. Given the high level of degeneration of the TE sequences, the recognition of genomes by searching for homologies often recovers contigs that are not very similar to the active copies. This hampers the understanding of their evolutionary history in the host genome.
The insertion and accumulation of some TEs in the vicinity of genes, for example, or their degenerate copies, represent an opportunity for molecular evolution or domestication [48,49,50]. Numerous studies have revealed protein motifs or repetitive regions of TEs being recruited for new genomic functions, providing an important source of novel sequences for the evolution of the genome [51,52,53]. New regulatory sequences and new protein-coding or even non-coding RNAs may play a beneficial role in the host genome [11,54,55], and may also be involved in a number of different processes of the regulation of gene expression. These processes include the regulation of transcription by the domesticated LTR retrotransposons and the action of the Amniota SINE1 element, which enhances the genes encoding the fibroblast growth factor of the inhibitor of an apoptosis protein family [56,57].
Given this, the ability of the TEs to provide a substrate for evolutionary innovations has attracted considerable interest over the years, and has been the focus of numerous studies of eukaryote species, which have found evidence of their involvement in the evolution of the genome through their reorganization.
2. Transposons as a Source of Repetitive Units for the Emergence of Tandem Repeats
A number of studies have provided examples of TE sequences that give rise to new repetitive classes, such as microsatellites, minisatellites, and satellite DNA [3,5,11,54,58]. Transposable elements represent an important substrate for genomic remodeling, and the emergence of new repetitive sequences, which generate a library of short repeat arrays that will subsequently be dispersed through the genome, to eventually become novel tandem repeats [59] (Figure 1—T4).
Transposable elements may contain sites predisposed for the formation of microsatellite DNA, which favors the dispersal of these repetitive units in the genome [5,58,60]. In the human genome, for example, approximately 23% of all tandem arrays (satellite, mini- and microsatellite sequences) are derived from TEs [61].
2.1. Classes and Mobility of the TEs
Studies of a range of different organisms have revealed a subtle prevalence of class I elements as a source of repetitions in the genome. Retrotransposons of the SINE (Short Interspersed Nuclear Element) superfamily, in particular the Alu elements, are a compelling example of this process. The non-autonomous SINEs, which range in size from 100 to 600 base pairs (bps), are widely distributed in the eukaryotes, where they play an important role in the organization of the genome, given their involvement in cell survival during many different types of physiological stress, for example [62]. The Alu elements, which are primate-specific, are considered to be the most widespread of the transposable elements, representing approximately 11% of the human genome [63,64,65]. These elements are considered to be the origin of the pλg3 satDNA and (GAA)n-type repeats, which are both found in the human genome [66,67,68], and the A-rich microsatellites in the primates [69]. Approximately 7276 minisatellites in the human genome have been derived from transposable elements, and 2663 are associated with Alu elements [61].
Non-autonomous TEs prevail as a source of the sequences for the formation of novel repetitive elements (Table 1), and the MITE (Miniature Transposable Elements with inverted repetitions) elements may be an especially interesting group for the understanding of this process. The MITE-type elements are non-autonomous, class II elements of approximately 400 bps, which are characterized by inverted repetitions flanking a variable region [9,70,71]. The MITE repeats are known to give rise to (GTCY)n repeats in lepidopterans [72], Xstir satDNA in Xenopus leavis [73], miDNA4 in Xenopus tropicalis [74], D1100 satDNA in rye [75], a number of different types of satellite DNA in Messor ants [76], and HindIII satDNA in bivalves [77].

Table 1.
Recorded cases of transposable elements giving rise to satellite, mini- and microsatellite DNA (satDNA), according to their mobility and the regions of the transposable elements that have given rise to the tandem repeat sequences. ORF (Open Reading Frame); TR (Tandem Repeat); UTR (Untranslated Region); LTR (Long Terminal Repeat), TIR (Terminal Inverted Repeats), IR (Inverted Repeat).
The MITE sequences experience burst events, which may lead to a dramatic increase in the number of copies, resulting in the rapid accumulation of arranged units with highly similar sequences in the chromosomes [100,101,102], as seen in the amphibian Xenopus tropicalis [74]. In the genome of X. tropicalis, the presence of a miDNA4-MITE that contains a satellite DNA motif, demonstrates the synergic interplay between the MITE structure and genomic context, which contributes to the emergence of the repetitive arrays of the new satDNA. In this model, the birth of the satellite monomer within a MITE is followed by the amplification of tandem repeats, with the higher recombination rates of the tandem arrays of paralogous copies leading to the rapid homogenization of the repetitive arrays and, over time, the generation of satDNA following the putative concerted evolution model [74].
Helitron elements (subclass II of the DNA transposons) also represent a source of the spread of satDNA-like arrays in the genome. The transposition of these Class II elements occurs through semi-replicative transposition, in which only one strand of the transposon is transferred between genomic sites without duplicating the target site (TSD) upon insertion, a process known as Rolling Circle Transposition, or RCT [2,9,103,104]. Analysis of the RCT mechanism has revealed cases in which the rolling-circle transposases are unable to recognize the termination sites located at the extremity of each TE, which results in the transposition of fragments of the genomic DNA located in the immediate vicinity of the sequence. This makes the TEs prone to the capture and propagation of a range of different genomic sequences [2,105].
As satDNA may be formed by the tandem amplification of a whole TE, or only a part of it, where fractions of short satDNA-like arrays would be expected to be found dispersed throughout the genome as an intermediate stage of the emergence of satDNA from the TEs, which are normally distributed in euchromatic regions [2,8,74,106]. In Crassostrea gigas, for example, the genome assembly presents 13 clusters of satDNA-like tandem repeats, which represent the central repeats of 11 non-autonomous elements belonging to the Helentron superfamily of DNA transposons known as the CgHINE [2]. The genome-wide distribution of this element in this species indicates that Helentrons are able to propagate tandem repeats.
2.2. Are Certain Portions of a TE More Prone to the Generation of Tandem Repeat Sequences?
Although the entire sequence of a transposable element has the potential to act as the substrate for the generation of new repetitive elements [61], we observed a prevalence of the repetitive portions of these elements as the source of the monomeric units of tandem repeats. The greater similarities between the monomeric units and the Terminal Inverted Repeats (TIRs), Long Terminal Repeats (LTRs), and other non-coding regions of the transposable elements highlights the importance of these naturally repetitive segments for the emergence of new classes of repetitive DNA (Table 1; Figure 2).
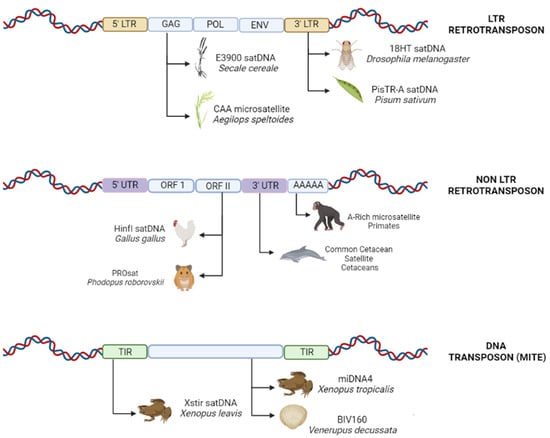
Figure 2.
Schematic diagram of the different regions of transposable elements that may provide the starting point of origin for new micro-, mini- or satellite DNA. LTR (Long Terminal Repeat); GAG (GAG domain); POL (Reverse Transcriptase); ENV (Envelope Protein); UTR (Untranslated Region); ORF (Open Reading Frame); TIR (Terminal Inverted Repeats). (Source: the authors/created in Biorender.com, accessed on 5 September 2022).
The untranslated 5′ and 3′ (5′UTR and 3′UTR) extremities also serve as a source of satellite DNA. The evidence available for the pea, Pisum sativum, indicates that the satDNA PisTR-A originated through the amplification and homogenization of tandem repeats present in the hypervariable 3′UTR of the Ty3/gypsy-like Ogre elements [59]. The 3′ terminal may also play a key role in this process, as observed in Drosophila melanogaster, in which the emergence of new satDNA corresponded to the 3′ non-coding region of the transposable element HeT-A [87], and in the cetaceans, the DNA of the common satellite monomer is similar to the 3’-terminal portion of the mammalian L1 (LINE-1) retrotransposon [82] (Figure 2).
Open Reading Frames (ORFs) of TEs have also been found to be related to the satDNA in Gallus gallus, with similarities in the sequences being observed among the regions of the satDNA HinfI and the CR1 retrotransposon, which contains the partial ORF II of the CR1 element [7] (Figure 2). In the rodent Phodopus roborovskii, the ORF II of a LINE-1 retrotransposon is 88% similar to a small region of the PROsat present in the genome [81]. In rye, Secale cereale, the E900 satDNA contains a fragment of a retrotransposon which encodes a partial reading frame for the GAG-like protein of an LTR retrotransposon [75]. The GAG-like gene was also related to the CAA microsatellite in wheat, Aegilops speltoides, and presented similarities with the upstream region of the Ty3/gypsy-like retroelement [98] (Figure 2). Overall, then, the sum of the evidence indicates that any part of a transposable element may provide a substrate for the generation of new sequences, and there does not appear to be any conclusive evidence that specific portions of these elements are involved preferentially in the generation of satellites or other types of tandem repeat.
3. Is the Centromeric Region a Hotspot of the Emergence of de novo satDNA Derived from TEs?
Centromeric regions are favorable to the emergence and establishment of new families of satellite DNA and are ideal models for studying the TEs as an evolutionary substrate in chromosomal evolution [107,108]. Centromeres are gene-poor, and the majority of the transcriptional activity observed in this region involves non-coding RNAs that interact in the organization of the kinetochores [109].
Meiotic recombination is avoided in centromeric regions, which appears to be an evolutionary strategy to avoid chromosome aneuploidy, given that crossovers in the vicinity of the centromere make chromosomes more prone to mis-segregation [109,110]. The centromeres are thus a cold spot for crossovers, making this chromosomal region prone to the emergence of de novo satDNA from TEs. The mobile genetic elements inserted in these regions cannot be deleted easily by crossovers, and thus accumulate in the centromeric and pericentromeric regions [111].
Plant and animal centromeres are rich in TEs, and these sequences may sometimes be highly specific and/or involved directly in the architecture of this chromosomal region. Two species of beetle, Dichotomius schiffleri and Tribolium castaneum, have unusually extended centromeres, which appear to be related to the prevalence of DsGypsy6, LINE-1, and Helitron-like sequences in these structures [112,113]. In Poaceae species, the centromere is enriched with a specific retrotransposon family known as the centromeric retrotransposon (CR), which has highly conserved motifs, that are also found in their B chromosomes, when present (see [114]). The CR elements and centromere connection may contribute to the maintenance of the centromere/kinetochore complex, given that these sequences interact with the kinetochore protein CENH3 [115].
Recent studies of neo-centromeres have established a new perspective for the understanding of the role of TEs in the emergence of new satDNA families. Talbot and Hennikof [111] proposed a model which indicates an active role of the TEs in the evolutionary emergence of neo-centromeres. De novo centromeres appear to arise from the deposition of epigenetic markers in a particular region of the chromosome, followed by the enrichment of repetitive sequences at this new site [116]. The transposons appear to play an important role in the maintenance of the size of the region and the increase in the repeat content of the neo-centromeres that do not have many tandem repeats during this initial period [117].
Large numbers of CENP-A are detected flanking centromeres, but low levels of CENP-A may spread to the non-canonical centromere regions, predisposing them to acquire a potential centromeric function [118,119], and the insertion of TEs in an ectopic CENP-A site may create favorable conditions for the evolution of new a centromeric complex [118].
New cycles of transposition activity increase the density of TEs and may result in insertions of copies adjacent to the original, resulting in tandem duplications (tandem dimers, trimers, or more repetitions), as observed in the Rice8 satDNA in centromere of rice [120], for example. Retrotransposons comprise approximately 70% of the functional centromeres of D. melanogaster, and are composed of complex DNA rich in non-LTR retroelements inserted within large blocks of tandem repeats [121]. Some plant centromeres are composed of long arrays of satDNA interspersed with Gypsy LTR-retrotransposons [122,123,124,125], which highlights the interplay of the different classes of repetitive DNA in the establishment and maintenance of the centromere region.
Satellite DNA repeats derived from TEs in the centromere region have been identified in a number of different species [3,61,121,126]. The centromeres of Zea mays are a good example because it is composed of CRM1TR and CRM4TR satDNA, two tandem arrays that were derived from an LTR-retrotransposon in at least two separate events [96]. The repeat arrays of satellites in the centromeres of chromosomes 1, 2, 3, 5, 7, and 8 of the potato, Solanum tuberosus, were also amplified from retrotransposon sequences [95]. In the chicken karyotype, several centromere-specific types of satDNA are highly similar to retrotransposons [127]. In the centromeres of Prunus species, a highly conserved monomer unit of 166 bps has been identified from assembled genomes and sequencing reads, with varying signal intensities in fluorescence in situ hybridization (FISH) experiments, which indicates that the centromeric regions of this genus are enriched with this sequence [128].
The centromeric region of the Y chromosome of Drosophila melanogaster is composed of 18HT satDNA, a satellite DNA derived from HeT-A and TART non-LTR retrotransposons [87], the same elements that were co-opted to the telomeric function in Drosophila. In the Drosophila obscura species group (Drosophila subobscura, D. guanche, and D. madeirensis), a DNA transposon (SGM elements) was the substrate for the emergence of a species-specific SGM satDNA that makes up the centromeric heterochromatin in D. guanche, in which the SGM element appears to be inactive [85].
The subtelomeric heterochromatin also appears to be prone to the emergence of tandem copies of transposable elements. Drosophila melanogaster, for example, has three telomere-specialized retrotransposons (HeT-A, TART, and TAHRE) that have been co-opted functionally to the organization of the chromatin and the maintenance of the telomere, and resemble telomere extensions containing telomerase [129,130]. The subtelomeric region of the D. melanogaster chromosomes presents a complex combination of potentially active elements and truncated TEs arranged in a long array [129]. In Drosophila biarmipes, by contrast, Helitron transposons may play an important role in the structure of the telomere, and while full-length Helitrons can be observed in the telomeres, fragments are interspersed within the abundant satellite sequences [129]. The richness of the repetitive arrays derived from the TEs in the subtelomeric heterochromatin of Drosophila represents a promising model for the study of the emergence of satellite DNA from TE elements and another potential chromosomal hotspot for the formation of de novo satDNA from TEs.
4. Mechanisms of the Production of Repeats from TEs
Unequal crossovers are an important mechanism of the expansion of tandem arrays and the homogenization of the satDNA by concerted evolution [131], although the absence of canonical meiotic crossovers in the centromeric region would require a mechanism that is independent of the emergence and evolution of the satDNA in this region. The transposition process may cause a Double Strand Break (DBS) of the DNA, which the DNA repair mechanism will attempt to fix, through alternative pathways, such as Non-Homologous End Joining (NHEJ), Non-Allelic Homologous Recombination (NAHR), or by Homologous Recombination (HR).
The NHEJ may contribute to the expansion of tandem arrays as a result of a single transposition event. Microhomology-mediated NHEJ has the potential to give rise to a wide range of chromosome events including duplications of sequences if it happens between sister chromatids [132] This mechanism also plays an important role in finalizing some TE-related instability events [133] and has been found with a dominant role in gene duplications [134]. TSD sequences are identical; therefore, it provides microhomology making it possible for NHEJ to restore the genome back to its original state before the insertion of a transportable element [135].
For example, Target Site Duplications (TSD) may be brought together by the NHEJ mechanism following a “cut and paste” transposition event (Figure 3), which rejoins the broken ends without the use of extensive homology [136,137]. As each TSD normally has fewer than 10 bps, these dimers of low complexity can arise easily through polymerase slippage and the subsequent expansion of the number of copies (Figure 3).
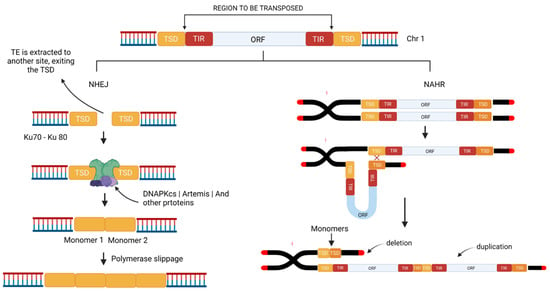
Figure 3.
Schematic diagram showing how the Non-Homologous End-Joining (NHEJ) and Non-Allelic Homologous Recombination (NAHR) DNA repair mechanisms contribute to the expansion of tandem repeats from a Transposable Element. TSD (Target Site Duplication); TIR (Terminal Inverted Repeats); ORF (Open Reading Frame). (Source: Authors/created in Biorender.com, accessed on 5 September 2022).
Studies on TSD and TE insertions were performed in D. melanogaster [1], where it was hypothesized that TEs organized in tandem could be generated through multiple insertions at the same chromosomal site, which also presents a TSD. The authors concluded, analyzing reads of the element known as P-element, that the junctions of the tandem repeats had the same size as the TSD of the said element, and the consensus motif was found similar to the TSD from other P-element insertions.
On the other hand, NAHR may also result in recurrent genome rearrangements, such as inversions, translocations, deletions, or duplications, in sequences that are not in intra- or inter-chromatid allelic positions, i.e., they are paralogous [138,139]. This mechanism provides valuable insights into the expansion of repeat arrays to the emergence of new satDNA, given that it depends on the position of the paralogous TE copies that will used as the template for the repair of the chromatid that has suffered the DBS, which may result in tandem duplications of the repetitive segments of the TE (Figure 3). Given this, intra- or inter-chromatid NAHR may contribute to the expansion of the initial repeat arrays in a manner similar to that of the unequal crossovers performed in the concerted evolution model.
One other especially interesting pathway of double-strand DNA repair is the use of a strand from a sister chromatid through Homologous Recombination (HR). Given either the existence of the repetitive elements that make up the molecular structure of the TEs (such as LTRs and ITRs) or the greater similarities of the TSD regions, the repair of DBSs using intrachromosomal HR mechanisms has the potential to generate tandem duplications in these regions [140] which may cause repeat arrays of the TE to accumulate in this region, if the Holliday junctions are processed to yield a crossover.
Similar results may also be obtained when a DBS is repaired by ectopic recombination, in which the paralogous copies used as the template are from non-homologous chromosomes [141]. In this case, the resolution of the Holliday junctions in the crossover events between dispersed repeats [142] will also result in the tandem duplications of the repetitive portions of the TEs in the region of the DBS. The outcome of these mechanisms of DBS repair is the formation of repeat arrays in the region of the DNA damage, which will be a source of other mechanisms of sequence expansion, such as polymerase slippage, that contribute to an increase in the number of monomers, leading to the emergence of satDNA from TE segments.
Finally, the intrinsic mechanisms involved in the transposition events are prone to the production of repeats derived from TEs. For example, the genesis of the Bari1 (Tc1-mariner superfamily) repeat clusters in D. melanogaster was provoked by anomalous rolling circle mechanisms and the subsequent reintegration within the Stalker LTR-retrotransposon, which generated an 80-repeat organized in heterochromatic clusters [143].
5. Conclusions
Repetitive DNA sequences have long been studied because of their structural role and impact on the genome of a range of different organisms. Here, however, we highlight the importance of also focusing on the relationships among the different elements. The possible presence of pre-existing transposable elements that serve as a substrate for the emergence of new families of repetitive sequences reinforces the potential importance of these elements in genome and karyotype remodeling during the evolutionary process. Given this, a better comprehension of the evolutionary relationships of these elements may be extremely valuable for a better understanding of the evolution of the eukaryote genome.
Author Contributions
M.L.Z. drafted and revised the manuscript. D.P.B. coordinated the study and revised the text. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Brazilian Coordination for Higher Education Personnel Training (CAPES/PROAP—Finance Code 001) and the Brazilian National Council for Scientific and Technological Development (CNPq #303646/2021-7; CNPq/MCTI/FNDCT Nº 18/2021 #405041/2021-7).
Acknowledgments
We thank the Brazilian Coordination for Higher Education Personnel Training (CAPES/PROAP—Finance Code 001) for the scholarships provided to MLZ. DPB thanks the Brazilian National Council for Scientific and Technological Development (CNPq #303646/2021-7; CNPq/MCTI/FNDCT Nº 18/2021 #405041/2021-7), for its financial support.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- McGurk, M.P.; Barbash, D.A. Double Insertion of Transposable Elements Provides a Substrate for the Evolution of Satellite DNA. Genome Res. 2018, 28, 714–725. [Google Scholar] [CrossRef]
- Zeljko, V.T.; Pavlek, M.; Meštrović, N.; Plohl, M. Satellite DNA-like Repeats Are Dispersed throughout the Genome of the Pacific Oyster Crassostrea Gigas Carried by Helentron Non-Autonomous Mobile Elements. Sci. Rep. 2020, 10, 15107. [Google Scholar] [CrossRef] [PubMed]
- Meštrović, N.; Mravinac, B.; Pavlek, M.; Vojvoda-Zeljko, T.; Šatović, E.; Plohl, M. Structural and Functional Liaisons between Transposable Elements and Satellite DNAs. Chromosome Res. 2015, 23, 583–596. [Google Scholar] [CrossRef] [PubMed]
- Belyayev, A.; Josefiová, J.; Jandová, M.; Mahelka, V.; Krak, K.; Mandák, B. Transposons and Satellite DNA: On the Origin of the Major Satellite DNA Family in the Chenopodium Genome. Mob. DNA 2020, 11, 20. [Google Scholar] [CrossRef] [PubMed]
- Paço, A.; Freitas, R.; Vieira-da-Silva, A. Conversion of DNA Sequences: From a Transposable Element to a Tandem Repeat or to a Gene. Genes 2019, 10, 1014. [Google Scholar] [CrossRef]
- Tek, A.L.; Song, J.; Macas, J.; Jiang, J. Sobo, a Recently Amplified Satellite Repeat of Potato, and Its Implications for the Origin of Tandemly Repeated Sequences. Genetics 2005, 170, 1231–1238. [Google Scholar] [CrossRef]
- Li, J.; Leung, F.C. A CR1 Element Is Embedded in a Novel Tandem Repeat (Hin FI Repeat) within the Chicken Genome. Genome 2006, 49, 97–103. [Google Scholar] [CrossRef]
- Dias, G.B.; Heringer, P.; Svartman, M.; Kuhn, G.C.S. Helitrons Shaping the Genomic Architecture of Drosophila: Enrichment of DINE-TR1 in α- and β-Heterochromatin, Satellite DNA Emergence, and PiRNA Expression. Chromosome Res. 2015, 23, 597–613. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A Unified Classification System for Eukaryotic Transposable Elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef]
- Kidwell, M.G.; Lisch, D.R. Perspective: Transposable elements, parasitic dna, and genome evolution. Evolution 2001, 55, 1–24. [Google Scholar] [CrossRef]
- Fernández-Medina, R.D.; Ribeiro, J.M.C.; Carareto, C.M.A.; Velasque, L.; Struchiner, C.J. Losing Identity: Structural Diversity of Transposable Elements Belonging to Different Classes in the Genome of Anopheles Gambiae. BMC Genom. 2012, 13, 272. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.L.; Lozovskaya, E.R.; Nurminsky, D.I.; Lohe, A.R. What Restricts the Activity of Mariner-like Transposable Elements. Trends Genet. 1997, 13, 197–201. [Google Scholar] [CrossRef]
- Le Rouzic, A.; Capy, P. The First Steps of Transposable Elements Invasion. Genetics 2005, 169, 1033–1043. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, S.G.; Bao, W.; Martins, C.; Jurka, J. Horizontal Transfers of Mariner Transposons between Mammals and Insects. Mob. DNA 2012, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Pace, J.K.; Feschotte, C. The Evolutionary History of Human DNA Transposons: Evidence for Intense Activity in the Primate Lineage. Genome Res. 2007, 17, 422–432. [Google Scholar] [CrossRef]
- Pritham, E.J.; Feschotte, C. Massive Amplification of Rolling-Circle Transposons in the Lineage of the Bat Myotis Lucifugus. Proc. Natl. Acad. Sci. USA 2007, 104, 1895–1900. [Google Scholar] [CrossRef]
- Chen, J.; Lu, L.; Robb, S.M.C.; Collin, M.; Okumoto, Y.; Stajich, J.E.; Wessler, S.R. Genomic Diversity Generated by a Transposable Element Burst in a Rice Recombinant Inbred Population. Proc. Natl. Acad. Sci. USA 2020, 117, 26288–26297. [Google Scholar] [CrossRef]
- Nitta, N.; Farman, M.L.; Leong, S.A. Genome Organization of Magnaporthe Grisea: Integration of Genetic Maps, Clustering of Transposable Elements and Identification of Genome Duplications and Rearrangements. Theor. Appl. Genet. 1997, 95, 20–32. [Google Scholar] [CrossRef]
- Dasilva, C.; Hadji, H.; Ozouf-Costaz, C.; Nicaud, S.; Jaillon, O.; Weissenbach, J.; Crollius, H.R. Remarkable Compartmentalization of Transposable Elements and Pseudogenes in the Heterochromatin of the Tetraodon Nigroviridis Genome. Proc. Natl. Acad. Sci. USA 2002, 99, 13636–13641. [Google Scholar] [CrossRef]
- Thon, M.R.; Martin, S.L.; Goff, S.; Wing, R.A.; Dean, R.A. BAC End Sequences and a Physical Map Reveal Transposable Element Content and Clustering Patterns in the Genome of Magnaporthe Grisea. Fungal Genet. Biol. 2004, 41, 657–666. [Google Scholar] [CrossRef]
- Thon, M.; Pan, H.; Diener, S.; Papalas, J.; Taro, A.; Mitchell, T.; Dean, R. BAC End Sequences and a Physical Map Reveal Transposable Element Content and Clustering Patterns in the Genome of Magnaporthe Grisea. Genome Biol. 2006, 7, R16. [Google Scholar] [CrossRef] [PubMed]
- Zamudio, N.; Bourc’his, D. Transposable Elements in the Mammalian Germline: A Comfortable Niche or a Deadly Trap? Heredity 2010, 105, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Yan, W. Male Germline Control of Transposable Elements. Biol. Reprod. 2012, 86, 162-1. [Google Scholar] [CrossRef]
- Shi, X.; Seluanov, A.; Gorbunova, V. Cell Divisions Are Required for L1 Retrotransposition. Mol. Cell. Biol. 2007, 27, 1264–1270. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.; Odom, D.T.; Kutter, C. The Emergence of PiRNAs against Transposon Invasion to Preserve Mammalian Genome Integrity. Nat. Commun. 2017, 8, 1411. [Google Scholar] [CrossRef]
- Feschotte, C.; Pritham, E.J. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef]
- Engels, W.R.; Johnson-Schlitz, D.M.; Eggleston, W.B.; Sved, J. High-Frequency P Element Loss in Drosophila Is Homolog Dependent. Cell 1990, 62, 515–525. [Google Scholar] [CrossRef]
- Maupetit-Mehouas, S.; Vaury, C. Transposon Reactivation in the Germline May Be Useful for Both Transposons and Their Host Genomes. Cells 2020, 9, 1172. [Google Scholar] [CrossRef]
- Sigman, M.J.; Slotkin, R.K. The First Rule of Plant Transposable Element Silencing: Location, Location, Location. Plant Cell 2016, 28, 304–313. [Google Scholar] [CrossRef]
- Ward, M.C.; Zhao, S.; Luo, K.; Pavlovic, B.J.; Karimi, M.M.; Stephens, M.; Gilad, Y. Silencing of Transposable Elements May Not Be a Major Driver of Regulatory Evolution in Primate IPSCs. eLife 2018, 7, e33084. [Google Scholar] [CrossRef]
- Rigal, M.; Mathieu, O. A “Mille-Feuille” of Silencing: Epigenetic Control of Transposable Elements. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2011, 1809, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xi, R. Silencing Transposable Elements in the Drosophila Germline. Cell. Mol. Life Sci. 2017, 74, 435–448. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.; Rivin, C.; Walbot, V. Stable non-mutator stocks of maize have sequences homologous to the Mu1 transposable element. Genetics 1986, 114, 1007–1021. [Google Scholar] [CrossRef]
- Chomet, P.S.; Wessler, S.; Dellaporta, S.L. Inactivation of the Maize Transposable Element Activator (Ac) Is Associated with Its DNA Modification. EMBO J. 1987, 6, 295–302. [Google Scholar] [CrossRef]
- Banks, J.A.; Masson, P.; Fedoroff, N. Molecular Mechanisms in the Developmental Regulation of the Maize Suppressor-Mutator Transposable Element. Genes Dev. 1988, 2, 1364–1380. [Google Scholar] [CrossRef]
- Collings, C.K.; Anderson, J.N. Links between DNA Methylation and Nucleosome Occupancy in the Human Genome. Epigenetics Chromatin 2017, 10, 18. [Google Scholar] [CrossRef]
- Rose, N.R.; Klose, R.J. Understanding the Relationship between DNA Methylation and Histone Lysine Methylation. Biochim. Biophys. Acta Gene Regul. Mech. 2014, 1839, 1362–1372. [Google Scholar] [CrossRef]
- Walter, M.; Teissandier, A.; Pérez-Palacios, R.; Bourc’his, D. An Epigenetic Switch Ensures Transposon Repression upon Dynamic Loss of DNA Methylation in Embryonic Stem Cells. eLife 2016, 5, e11418. [Google Scholar] [CrossRef]
- Ebbs, M.L.; Bender, J. Locus-Specific Control of DNA Methylation by the Arabidopsis SUVH5 Histone Methyltransferase. Plant Cell 2006, 18, 1166–1176. [Google Scholar] [CrossRef]
- Lippman, Z.; May, B.; Yordan, C.; Singer, T.; Martienssen, R. Distinct Mechanisms Determine Transposon Inheritance and Methylation via Small Interfering RNA and Histone Modification. PLoS Biol. 2003, 1, E67. [Google Scholar] [CrossRef] [PubMed]
- Janssen, A.; Colmenares, S.U.; Karpen, G.H. Heterochromatin: Guardian of the Genome. Annu. Rev. Cell Dev. Biol. 2018, 34, 265–288. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ali, M.; Zhou, Q. Establishment and Evolution of Heterochromatin. Ann. N. Y. Acad. Sci. 2020, 1476, 59–77. [Google Scholar] [CrossRef] [PubMed]
- Kidwell, M.G. Transposable Elements and the Evolution of Genome Size in Eukaryotes. Genetica 2002, 115, 49–63. [Google Scholar] [CrossRef]
- Sirijovski, N.; Woolnough, C.; Rock, J.; Joss, J.M.P. NfCR1, the First Non-LTR Retrotransposon Characterized in the Australian Lungfish Genome, Neoceratodus Forsteri, Shows Similarities to CR1-like Elements. J. Exp. Zool. 2005, 304, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, J.; Zhu, C.; Yang, L.; Ren, Y.; Ruan, J.; Fan, G.; Hu, J.; Xu, W.; Bi, X.; et al. African Lungfish Genome Sheds Light on the Vertebrate Water-to-Land Transition. Cell 2021, 184, 1362–1376.e18. [Google Scholar] [CrossRef] [PubMed]
- Marsano, R.M.; Dimitri, P. Constitutive Heterochromatin in Eukaryotic Genomes: A Mine of Transposable Elements. Cells 2022, 11, 761. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. RAG1 Core and V(D)J Recombination Signal Sequences Were Derived from Transib Transposons. PLoS Biol. 2005, 3, e181. [Google Scholar] [CrossRef]
- Feschotte, C. Transposable Elements and the Evolution of Regulatory Networks. Nat. Rev. Genet. 2008, 9, 397–405. [Google Scholar] [CrossRef]
- Alzohairy, A.M.; Gyulai, G.; Jansen, R.K.; Bahieldin, A. Transposable Elements Domesticated and Neofunctionalized by Eukaryotic Genomes. Plasmid 2013, 69, 1–15. [Google Scholar] [CrossRef]
- Qiu, Y.; Köhler, C. Mobility Connects: Transposable Elements Wire New Transcriptional Networks by Transferring Transcription Factor Binding Motifs. Biochem. Soc. Trans. 2020, 48, 1005–1017. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Han, K.; Liang, P. Role of Transposable Elements in Gene Regulation in the Human Genome. Life 2021, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Fueyo, R.; Judd, J.; Feschotte, C.; Wysocka, J. Roles of Transposable Elements in the Regulation of Mammalian Transcription. Nat. Rev. Mol. Cell Biol. 2022, 23, 481–497. [Google Scholar] [CrossRef] [PubMed]
- Kelkar, Y.D.; Eckert, K.A.; Chiaromonte, F.; Makova, K.D. A Matter of Life or Death: How Microsatellites Emerge in and Vanish from the Human Genome. Genome Res. 2011, 21, 2038–2048. [Google Scholar] [CrossRef]
- Etchegaray, E.; Naville, M.; Volff, J.-N.; Haftek-Terreau, Z. Transposable Element-Derived Sequences in Vertebrate Development. Mob. DNA 2021, 12, 1. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable Elements and the Epigenetic Regulation of the Genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as Regulators of Gene Expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef]
- Buschiazzo, E.; Gemmell, N.J. The Rise, Fall and Renaissance of Microsatellites in Eukaryotic Genomes—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/16998838/ (accessed on 15 May 2022).
- Macas, J.; Koblízková, A.; Navrátilová, A.; Neumann, P. Hypervariable 3’ UTR Region of Plant LTR-Retrotransposons as a Source of Novel Satellite Repeats. Gene 2009, 448, 198–206. [Google Scholar] [CrossRef]
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef]
- Ahmed, M.; Liang, P. Transposable Elements Are a Significant Contributor to Tandem Repeats in the Human Genome. Comp. Funct. Genom. 2012, 2012, 947089. [Google Scholar] [CrossRef]
- Kanhayuwa, L.; Coutts, R.H.A. Short Interspersed Nuclear Element (SINE) Sequences in the Genome of the Human Pathogenic Fungus Aspergillus Fumigatus Af293. PLoS ONE 2016, 11, e0163215. [Google Scholar] [CrossRef]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. International Human Genome Sequencing Consortium. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Dewannieux, M.; Esnault, C.; Heidmann, T. LINE-Mediated Retrotransposition of Marked Alu Sequences. Nat. Genet. 2003, 35, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Deininger, P. Alu Elements: Know the SINEs. Genome Biol. 2011, 12, 236. [Google Scholar] [CrossRef]
- Armour, J.A.L.; Wong, Z.; Wilson, V.; Royle, N.J.; Jeffreys, A.J. Sequences Flanking the Repeat Arrays of Human Minlsatellites: Association with Tandem and Dispersed Repeat Elements. Nucl. Acids Res. 1989, 17, 4925–4936. [Google Scholar] [CrossRef] [PubMed]
- Clark, R.M.; Dalgliesh, G.L.; Endres, D.; Gomez, M.; Taylor, J.; Bidichandani, S.I. Expansion of GAA Triplet Repeats in the Human Genome: Unique Origin of the FRDA Mutation at the Center of an Alu. Genomics 2004, 83, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Jurka, J.; Gentles, A.J. Origin and Diversification of Minisatellites Derived from Human Alu Sequences. Gene 2006, 365, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Arcot, S.S.; Wang, Z.; Weber, J.L.; Deininger, P.L.; Batzer, M.A. Alu Repeats: A Source for the Genesis of Primate Microsatellites. Genomics 1995, 29, 136–144. [Google Scholar] [CrossRef]
- Bureau, T.E.; Wessler, S.R. Mobile Inverted-Repeat Elements of the Tourist Familyare Associated with the Genes of Many Cereal Grasses. Proc. Natl. Acad. Sci. USA 1994, 91, 1411–1415. [Google Scholar] [CrossRef]
- Feschotte, C.; Swamy, L.; Wessler, S.R. Genome-Wide Analysis of Mariner -Like Transposable Elements in Rice Reveals Complex Relationships with Stowaway Miniature Inverted Repeat Transposable Elements (MITEs). Genetics 2003, 163, 747–758. [Google Scholar] [CrossRef]
- Coates, B.S.; Kroemer, J.A.; Sumerford, D.V.; Hellmich, R.L. A Novel Class of Miniature Inverted Repeat Transposable Elements (MITEs) That Contain Hitchhiking (GTCY)n Microsatellites: Lepidopteran Mobile Microsatellites. Insect Mol. Biol. 2011, 20, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Hikosaka, A.; Kawahara, A. Lineage-Specific Tandem Repeats Riding on a Transposable Element of MITE in Xenopus Evolution: A New Mechanism for Creating Simple Sequence Repeats. J. Mol. Evol. 2004, 59, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Scalvenzi, T.; Pollet, N. Insights on Genome Size Evolution from a Miniature Inverted Repeat Transposon Driving a Satellite DNA. Mol. Phylogenet. Evol. 2014, 81, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Langdon, T.; Seago, C.; Jones, R.N.; Ougham, H.; Thomas, H.; Forster, J.W.; Jenkins, G. De Novo Evolution of Satellite DNA on the Rye B Chromosome. Genetics 2000, 154, 869–884. [Google Scholar] [CrossRef] [PubMed]
- López-Flores, I.; Garrido-Ramos, M.A. The Repetitive DNA Content of Eukaryotic Genomes. In Genome Dynamics; Garrido-Ramos, M.A., Ed.; S. Karger AG: Basel, Switzerland, 2012; Volume 7, pp. 1–28. [Google Scholar] [CrossRef]
- López-Flores, I.; de la Herrán, R.; Garrido-Ramos, M.A.; Boudry, P.; Ruiz-Rejón, C.; Ruiz-Rejón, M. The Molecular Phylogeny of Oysters Based on a Satellite DNA Related to Transposons. Gene 2004, 339, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Bois, P.; Williamson, J.; Brown, J.; Dubrova, Y.E.; Jeffreys, A.J. A Novel Unstable Mouse VNTR Family Expanded from SINE B1 Elements. Genomics 1998, 49, 122–128. [Google Scholar] [CrossRef]
- Kelly, R.G. Similar Origins of Two Mouse Minisatellites within Transposon-like LTRs. Genomics 1994, 24, 509–515. [Google Scholar] [CrossRef]
- Rossi, M.S.; Pesce, C.G.; Reig, O.A.; Kornblihtt, A.R.; Zorzópulos, J. Retroviral-like Features in the Monomer of the Major Satellite DNA from the South American Rodents of the Genus Ctenomys. DNA Sequence 1993, 3, 379–381. [Google Scholar] [CrossRef]
- Paço, A.; Adega, F.; Meštrović, N.; Plohl, M.; Chaves, R. The Puzzling Character of Repetitive DNA in Phodopus Genomes (Cricetidae, Rodentia). Chromosome Res. 2015, 23, 427–440. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Holmquist, G.P.; Jurka, J. L1 Repeat Is a Basic Unit of Heterochromatin Satellites in Cetaceans. Mol. Biol. Evol. 1998, 15, 611–612. [Google Scholar] [CrossRef]
- Duffy, A.J.; Coltman, D.W.; Wright, J.M. Microsatellites at a Common Site in the Second ORF of L1 Elements in Mammalian Genomes. Mamm. Genome 1996, 7, 386–387. [Google Scholar] [CrossRef] [PubMed]
- Palomeque, T.; Antonio Carrillo, J.; Muñoz-López, M.; Lorite, P. Detection of a Mariner-like Element and a Miniature Inverted-Repeat Transposable Element (MITE) Associated with the Heterochromatin from Ants of the Genus Messor and Their Possible Involvement for Satellite DNA Evolution. Gene 2006, 371, 194–205. [Google Scholar] [CrossRef] [PubMed]
- Miller, W.J.; Nagel, A.; Bachmann, J.; Bachmann, L. Evolutionary Dynamics of the SGM Transposon Family in the Drosophila Obscura Species Group. Mol. Biol. Evol. 2000, 17, 1597–1609. [Google Scholar] [CrossRef][Green Version]
- Heikkinen, E.; Launonen, V.; Müller, E.; Bachmann, L. The PvB370 BamHI Satellite DNA Family of the Drosophila Virilis Group and Its Evolutionary Relation to Mobile Dispersed Genetic PDv Elements. J. Mol. Evol. 1995, 41, 604–614. [Google Scholar] [CrossRef] [PubMed]
- Agudo, M.; Losada, A.; Abad, J.P.; Pimpinelli, S.; Ripoll, P.; Villasante, A. Centromeres from Telomeres? The Centromeric Region of the Y Chromosome of Drosophila Melanogaster Contains a Tandem Array of Telomeric HeT-A- and TART-Related Sequences. Nucleic Acids Res. 1999, 27, 3318–3324. [Google Scholar] [CrossRef] [PubMed]
- Dias, G.B.; Svartman, M.; Delprat, A.; Ruiz, A.; Kuhn, G.C.S. Tetris Is a Foldback Transposon That Provided the Building Blocks for an Emerging Satellite DNA of Drosophila Virilis. Genome Biol. Evol. 2014, 6, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Batistoni, R.; Pesole, G.; Marracci, S.; Nardi, I. A Tandemly Repeated DNA Family Originated from SINE-Related Elements in the European Plethodontid Salamanders (Amphibia, Urodela). J. Mol. Evol. 1995, 40, 608–615. [Google Scholar] [CrossRef]
- Suntronpong, A.; Singchat, W.; Kruasuwan, W.; Prakhongcheep, O.; Sillapaprayoon, S.; Muangmai, N.; Somyong, S.; Indananda, C.; Kraichak, E.; Peyachoknagul, S.; et al. Characterization of Centromeric Satellite DNAs (MALREP) in the Asian Swamp Eel (Monopterus Albus) Suggests the Possible Origin of Repeats from Transposable Elements. Genomics 2020, 112, 3097–3107. [Google Scholar] [CrossRef]
- Pasero, P.; Sjakste, N.; Blettry, C.; Got, C.; Marilley, M. Long-Range Organization and Sequence-Directed Curvature of Xenopus Laevis Satellite 1 DNA. Nucleic Acids Res. 1993, 21, 4703–4710. [Google Scholar] [CrossRef]
- Plohl, M.; Petrović, V.; Luchetti, A.; Ricci, A.; Šatović, E.; Passamonti, M.; Mantovani, B. Long-Term Conservation vs High Sequence Divergence: The Case of an Extraordinarily Old Satellite DNA in Bivalve Mollusks. Heredity 2010, 104, 543–551. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Molecular Paleontology of Transposable Elements from Arabidopsis Thaliana. In Transposable Elements and Genome Evolution; McDonald, J.F., Ed.; Springer: Dordrecht, The Netherlands, 2000; pp. 27–37. [Google Scholar] [CrossRef]
- Mogil, L.S.; Slowikowski, K.; Laten, H.M. Computational and Experimental Analyses of Retrotransposon-Associated Minisatellite DNAs in the Soybean Genome. BMC Bioinform. 2012, 13, S13. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Wu, Y.; Koblížková, A.; Torres, G.A.; Wang, K.; Iovene, M.; Neumann, P.; Zhang, W.; Novák, P.; Buell, C.R.; et al. Repeatless and Repeat-Based Centromeres in Potato: Implications for Centromere Evolution. Plant Cell 2012, 24, 3559–3574. [Google Scholar] [CrossRef]
- Sharma, A.; Wolfgruber, T.K.; Presting, G.G. Tandem Repeats Derived from Centromeric Retrotransposons. BMC Genom. 2013, 14, 142. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, L.; Macaulay, M.; Cardle, L.; Morgante, M.; degli Ivanissevich, S.; Maestri, E.; Powell, W.; Waugh, R. Intimate Association of Microsatellite Repeats with Retrotransposons and Other Dispersed Repetitive Elements in Barley. Plant J. 1999, 17, 415–425. [Google Scholar] [CrossRef]
- Cheng, Z.-J.; Murata, M. A Centromeric Tandem Repeat Family Originating From a Part of Ty3/ Gypsy -Retroelement in Wheat and Its Relatives. Genetics 2003, 164, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Vondrak, T.; Ávila Robledillo, L.; Novák, P.; Koblížková, A.; Neumann, P.; Macas, J. Characterization of Repeat Arrays in Ultra-Long Nanopore Reads Reveals Frequent Origin of Satellite DNA from Retrotransposon-Derived Tandem Repeats. Plant J. 2020, 101, 484–500. [Google Scholar] [CrossRef] [PubMed]
- Han, M.-J.; Shen, Y.-H.; Gao, Y.-H.; Chen, L.-Y.; Xiang, Z.-H.; Zhang, Z. Burst Expansion, Distribution and Diversification of MITEs in the Silkworm Genome. BMC Genom. 2010, 11, 520. [Google Scholar] [CrossRef]
- Deprá, M.; Ludwig, A.; Valente, V.L.; Loreto, E.L. Mar, a MITE Family of HAT Transposons in Drosophila. Mob. DNA 2012, 3, 13. [Google Scholar] [CrossRef]
- Lu, C.; Chen, J.; Zhang, Y.; Hu, Q.; Su, W.; Kuang, H. Miniature Inverted-Repeat Transposable Elements (MITEs) Have Been Accumulated through Amplification Bursts and Play Important Roles in Gene Expression and Species Diversity in Oryza Sativa. Mol. Biol. Evol. 2012, 29, 1005–1017. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. Helitrons on a Roll: Eukaryotic Rolling-Circle Transposons. Trends Genet. 2007, 23, 521–529. [Google Scholar] [CrossRef]
- Bennetzen, J.L.; Wang, H. The Contributions of Transposable Elements to the Structure, Function, and Evolution of Plant Genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef] [PubMed]
- Wawrzyniak, P.; Płucienniczak, G.; Bartosik, D. The Different Faces of Rolling-Circle Replication and Its Multifunctional Initiator Proteins. Front. Microbiol. 2017, 8, 2353. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Choo, K.H.A. Evolutionary Dynamics of Transposable Elements at the Centromere. Trends Genet. 2004, 20, 611–616. [Google Scholar] [CrossRef]
- Garrido-Ramos, M. Satellite DNA: An Evolving Topic. Genes 2017, 8, 230. [Google Scholar] [CrossRef]
- Thakur, J.; Packiaraj, J.; Henikoff, S. Sequence, Chromatin and Evolution of Satellite DNA. Int. J. Mol. Sci. 2021, 22, 4309. [Google Scholar] [CrossRef] [PubMed]
- Perea-Resa, C.; Blower, M.D. Centromere Biology: Transcription Goes on Stage. Mol. Cell Biol. 2018, 38, e00263-18. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, M.; Chuang, Y.-C.; Smith, G.R. Distributing Meiotic Crossovers for Optimal Fertility and Evolution. DNA Repair 2019, 81, 102648. [Google Scholar] [CrossRef] [PubMed]
- Talbert, P.B.; Henikoff, S. What Makes a Centromere? Exp. Cell Res. 2020, 389, 111895. [Google Scholar] [CrossRef]
- Gržan, T.; Despot-Slade, E.; Meštrović, N.; Plohl, M.; Mravinac, B. CenH3 Distribution Reveals Extended Centromeres in the Model Beetle Tribolium Castaneum. PLoS Genet. 2020, 16, e1009115. [Google Scholar] [CrossRef]
- Amorim, I.C.; Sotero-Caio, C.G.; Costa, R.G.C.; Xavier, C.; de Moura, R.d.C. Comprehensive Mapping of Transposable Elements Reveals Distinct Patterns of Element Accumulation on Chromosomes of Wild Beetles. Chromosome Res. 2021, 29, 203–218. [Google Scholar] [CrossRef]
- Jiang, J.; Birchler, J.A.; Parrott, W.A.; Kelly Dawe, R. A Molecular View of Plant Centromeres. Trends Plant Sci. 2003, 8, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Mroczek, R.J.; Dawe, R.K. Distribution of Retroelements in Centromeres and Neocentromeres of Maize. Genetics 2003, 165, 809–819. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Su, H.; Shi, Q.; Fu, S.; Wang, J.; Zhang, X.; Hu, Z.; Han, F. De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and Its Wide Hybrids. PLoS Genet. 2016, 12, e1005997. [Google Scholar] [CrossRef] [PubMed]
- Presting, G.G. Centromeric Retrotransposons and Centromere Function. Curr. Opin. Genet. Dev. 2018, 49, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Leo, L.; Marchetti, M.; Giunta, S.; Fanti, L. Epigenetics as an Evolutionary Tool for Centromere Flexibility. Genes 2020, 11, 809. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.-H.; Hori, T.; Martins, N.M.C.; Toyoda, A.; Misu, S.; Monma, N.; Hiratani, I.; Maeshima, K.; Ikeo, K.; Fujiyama, A.; et al. Chromosome Engineering Allows the Efficient Isolation of Vertebrate Neocentromeres. Dev. Cell 2013, 24, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Gao, D.; Jiang, N.; Wing, R.A.; Jiang, J.; Jackson, S.A. Transposons Play an Important Role in the Evolution and Diversification of Centromeres among Closely Related Species. Front. Plant Sci. 2015, 6, 216. [Google Scholar] [CrossRef]
- Chang, C.-H.; Chavan, A.; Palladino, J.; Wei, X.; Martins, N.M.C.; Santinello, B.; Chen, C.-C.; Erceg, J.; Beliveau, B.J.; Wu, C.-T.; et al. Islands of Retroelements Are Major Components of Drosophila Centromeres. PLoS Biol. 2019, 17, e3000241. [Google Scholar] [CrossRef]
- Wolfgruber, T.K.; Sharma, A.; Schneider, K.L.; Albert, P.S.; Koo, D.-H.; Shi, J.; Gao, Z.; Han, F.; Lee, H.; Xu, R.; et al. Maize Centromere Structure and Evolution: Sequence Analysis of Centromeres 2 and 5 Reveals Dynamic Loci Shaped Primarily by Retrotransposons. PLoS Genet. 2009, 5, e1000743. [Google Scholar] [CrossRef]
- Zhang, H.-H.; Shen, Y.-H.; Xu, H.-E.; Liang, H.-Y.; Han, M.-J.; Zhang, Z. A Novel HAT Element in Bombyx Mori and Rhodnius Prolixus: Its Relationship with Miniature Inverted Repeat Transposable Elements (MITEs) and Horizontal Transfer. Insect Mol. Biol. 2013, 22, 584–596. [Google Scholar] [CrossRef]
- Sharma, A.; Presting, G.G. Evolution of Centromeric Retrotransposons in Grasses. Genome Biol. Evol. 2014, 6, 1335–1352. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, S.; Ishii, T.; Brown, C.T.; Houben, A.; Comai, L. Centromere Location in Arabidopsis Is Unaltered by Extreme Divergence in CENH3 Protein Sequence. Genome Res. 2017, 27, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Satović, E.; Vojvoda Zeljko, T.; Luchetti, A.; Mantovani, B.; Plohl, M. Adjacent Sequences Disclose Potential for Intra-Genomic Dispersal of Satellite DNA Repeats and Suggest a Complex Network with Transposable Elements. BMC Genom. 2016, 17, 997. [Google Scholar] [CrossRef] [PubMed]
- Shang, W.-H.; Hori, T.; Toyoda, A.; Kato, J.; Popendorf, K.; Sakakibara, Y.; Fujiyama, A.; Fukagawa, T. Chickens Possess Centromeres with Both Extended Tandem Repeats and Short Non-Tandem-Repetitive Sequences. Genome Res. 2010, 20, 1219–1228. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Zhang, J.; Feng, Y.; Chen, Q.; Liu, Z.-S.; Liu, C.-L.; He, W.; Wang, H.; Yang, S.-F.; et al. Comparative Analysis of Transposable Elements and the Identification of Candidate Centromeric Elements in the Prunus Subgenus Cerasus and Its Relatives. Genes 2022, 13, 641. [Google Scholar] [CrossRef]
- Saint-Leandre, B.; Nguyen, S.C.; Levine, M.T. Diversification and Collapse of a Telomere Elongation Mechanism. Genome Res. 2019, 29, 920–931. [Google Scholar] [CrossRef]
- George, J.A.; DeBaryshe, P.G.; Traverse, K.L.; Celniker, S.E.; Pardue, M.-L. Genomic Organization of the Drosophila Telomere Retrotransposable Elements. Genome Res. 2006, 16, 1231–1240. [Google Scholar] [CrossRef]
- Dover, G. Molecular Drive: A Cohesive Mode of Species Evolution. Nature 1982, 299, 111–117. [Google Scholar] [CrossRef]
- Elliott, B.; Richardson, C.; Jasin, M. Chromosomal Translocation Mechanisms at Intronic Alu Elements in Mammalian Cells. Mol. Cell 2005, 17, 885–894. [Google Scholar] [CrossRef]
- Hedges, D.J.; Deininger, P.L. Inviting Instability: Transposable Elements, Double-Strand Breaks, and the Maintenance of Genome Integrity. Mutat. Res. 2007, 616, 46–59. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Q.; Gao, D.; Wang, J.; Lang, Y.; Liu, T.; Li, B.; Bai, Z.; Luis Goicoechea, J.; Liang, C.; et al. Whole-Genome Sequencing of Oryza Brachyantha Reveals Mechanisms Underlying Oryza Genome Evolution. Nat. Commun. 2013, 4, 1595. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, D.M.; Bridges, M.C.; Strother, A.E.; Burckhalter, C.E.; Burnette, J.M.; Hancock, C.N. Precise Repair of MPing Excision Sites Is Facilitated by Target Site Duplication Derived Microhomology. Mob. DNA 2015, 6, 15. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, K.; McVey, M. Error-Prone Repair of DNA Double-Strand Breaks. J. Cell Physiol. 2016, 231, 15–24. [Google Scholar] [CrossRef]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-Homologous DNA End Joining and Alternative Pathways to Double-Strand Break Repair. Nat. Rev. Mol. Cell. Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Liu, P.; Lacaria, M.; Zhang, F.; Withers, M.; Hastings, P.J.; Lupski, J.R. Frequency of Nonallelic Homologous Recombination Is Correlated with Length of Homology: Evidence That Ectopic Synapsis Precedes Ectopic Crossing-Over. Am. J. Hum. Genet. 2011, 89, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Parks, M.M.; Lawrence, C.E.; Raphael, B.J. Detecting Non-Allelic Homologous Recombination from High-Throughput Sequencing Data. Genome Biol. 2015, 16, 72. [Google Scholar] [CrossRef] [PubMed]
- Al-Zain, A.M.; Symington, L.S. The Dark Side of Homology-Directed Repair. DNA Repair 2021, 106, 103181. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.T.; Drouin, G. Ectopic Gene Conversions in the Genome of Ten Hemiascomycete Yeast Species. Int. J. Evol. Biol. 2010, 2011, 970768. [Google Scholar] [CrossRef]
- Mazón, G.; Lam, A.F.; Ho, C.K.; Kupiec, M.; Symington, L.S. The Rad1–Rad10 Nuclease Promotes Chromosome Translocations between Dispersed Repeats. Nat. Struct. Mol. Biol. 2012, 19, 964–971. [Google Scholar] [CrossRef]
- Marsano, R.M.; Milano, R.; Minervini, C.; Moschetti, R.; Caggese, C.; Barsanti, P.; Caizzi, R. Organization and Possible Origin of the Bari-1 Cluster in the Heterochromatic H39 Region of Drosophila Melanogaster. Genetica 2003, 117, 281–289. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).