The Potential Application of Human Gingival Fibroblast-Conditioned Media in Pulp Regeneration: An In Vitro Study
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primary Culture of DPSCs and hGF
2.2. Preparation of hGF-Conditioned Medium
2.3. Cell Viability and Proliferation Assays
2.4. Cell Migration Assay
2.5. Antioxidative Stress Assay
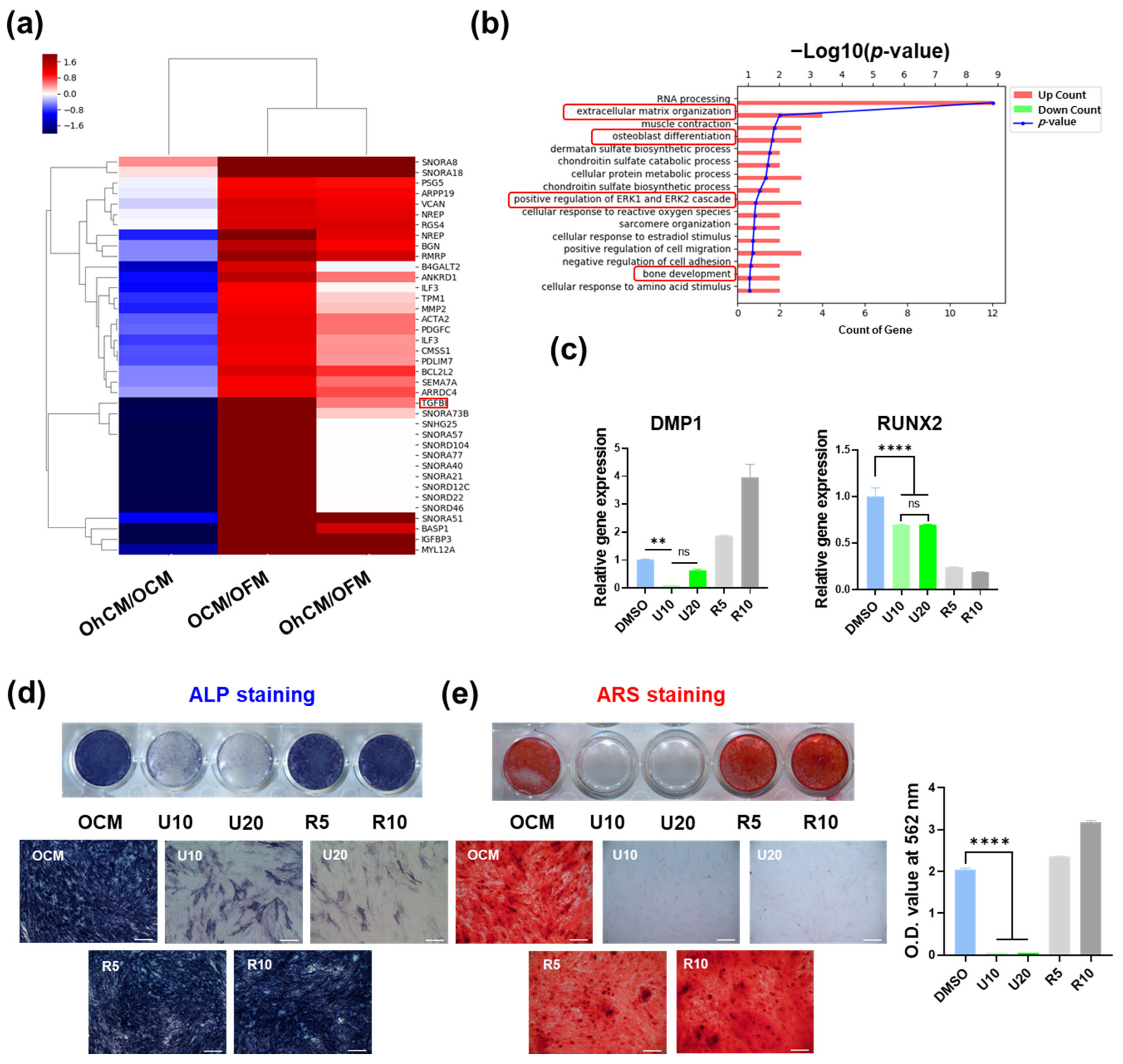
2.6. Osteogenic Differentiation Assays
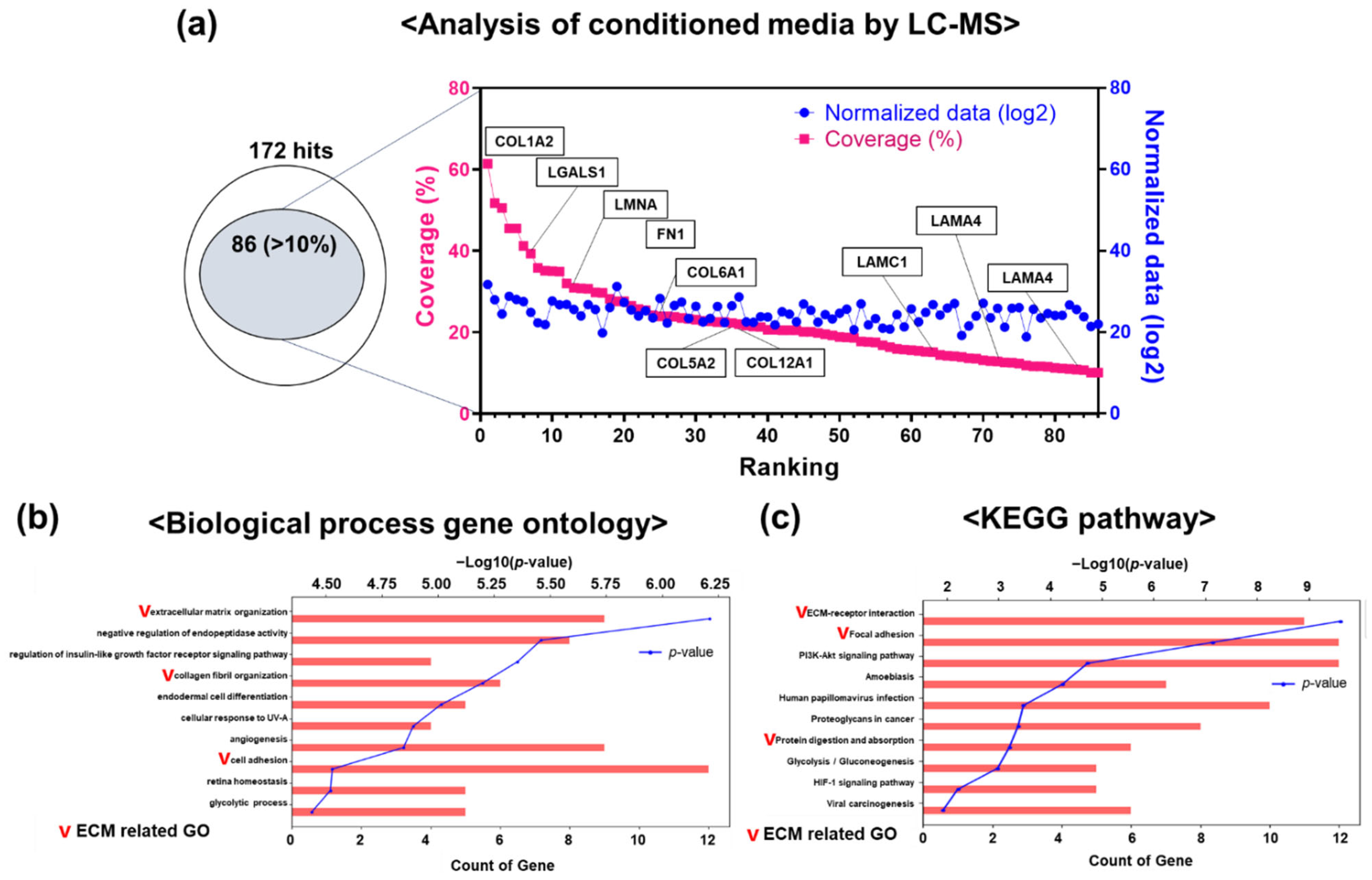
2.7. Proteomic Analysis
2.8. mRNA Sequencing
2.9. Inhibition Assay
2.10. Statistical Analyses
3. Results
3.1. Effect of hGF-CM on DPSC Proliferation
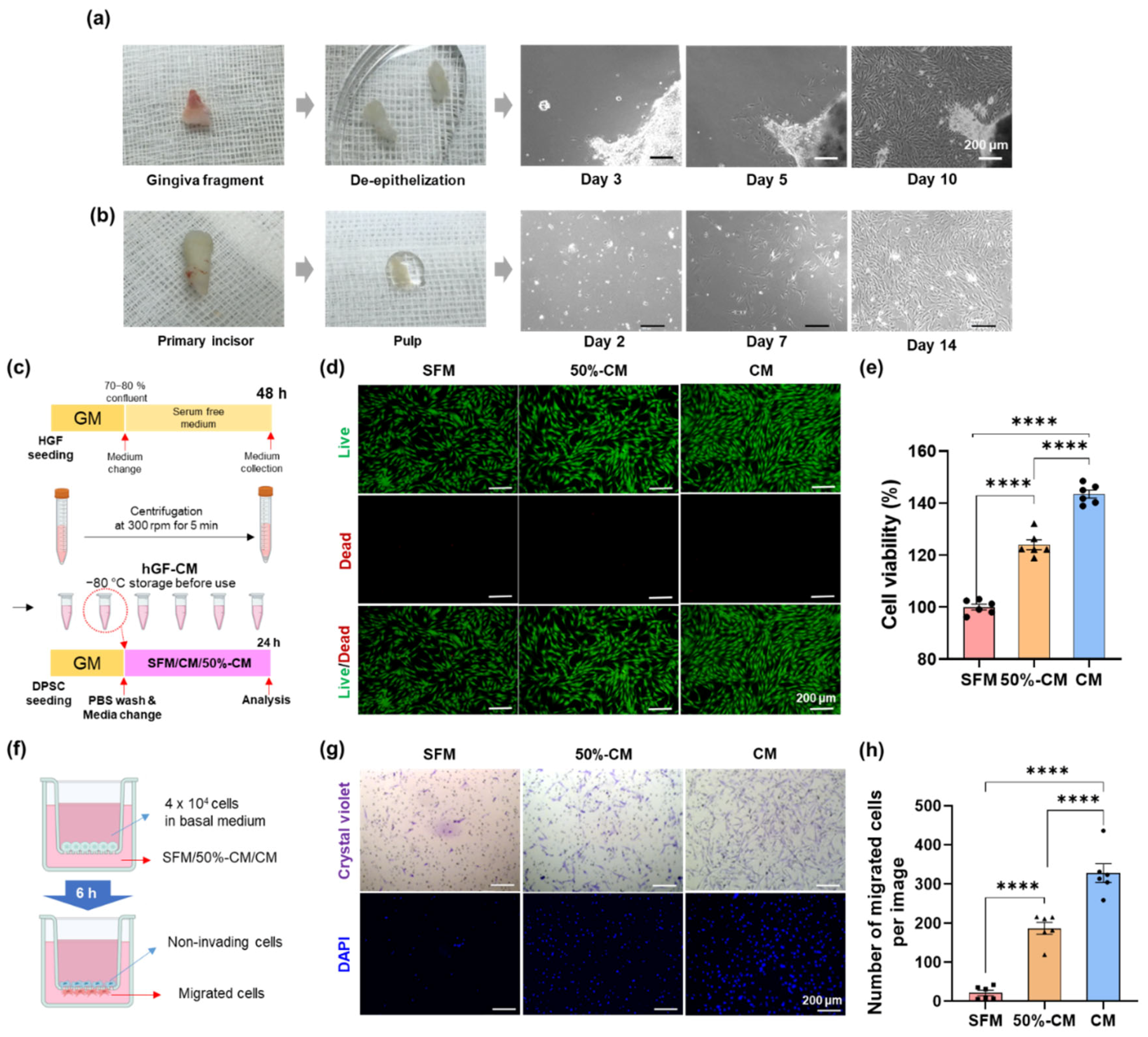
3.2. Effect of hGF-CM on DPSC Migration
3.3. Protective Effect of hGF-CM against H2O2-Induced Cell Death in DPSCs
3.4. hGF-CM Enhances DPSC Odontoblast Differentiation Capacity
3.5. Differential Protein Expression in hGF-CM
3.6. Signaling Pathway Involved in Enhancing the Capacity of hGF-CM in Odonto/Osteogenesis
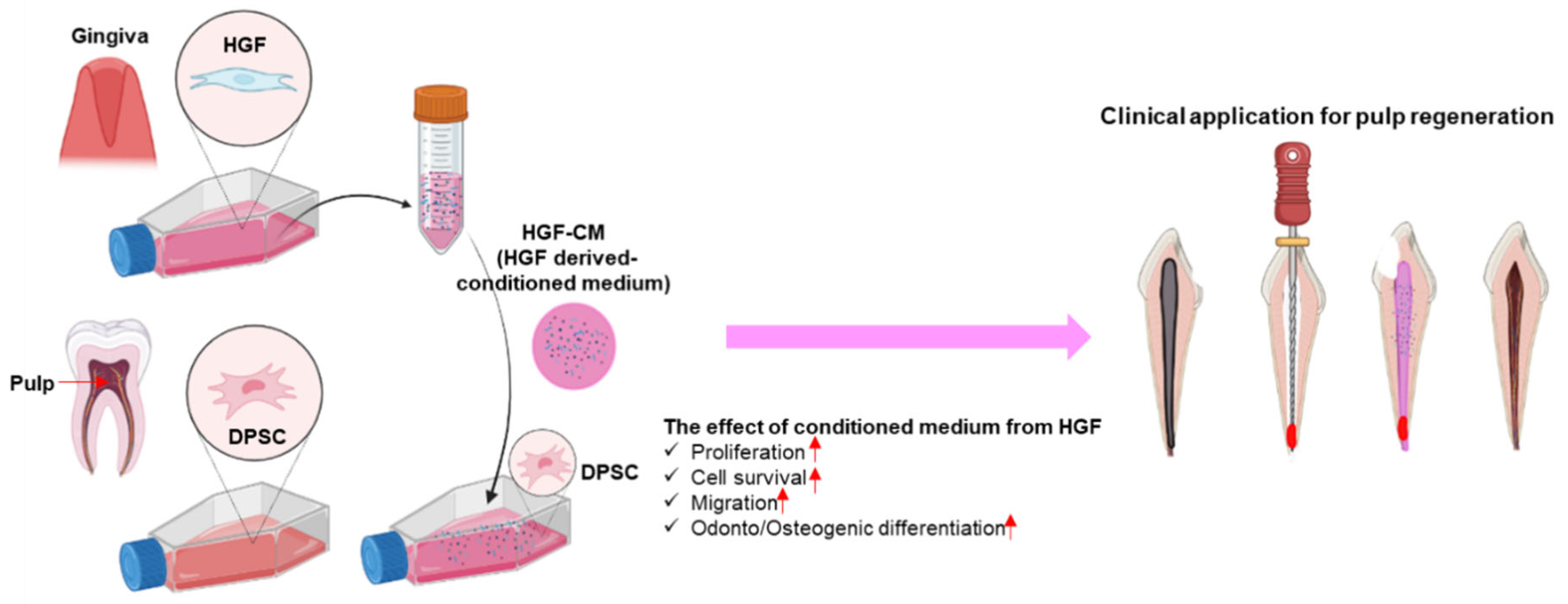
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hull, P.S.; Worthington, H.V.; Clerehugh, V.; Tsirba, R.; Davies, R.M.; Clarkson, J.E. The reasons for tooth extractions in adults and their validation. J. Dent. 1997, 25, 233–237. [Google Scholar] [CrossRef]
- Olatosi, O.O.; Sote, E.O. Causes and pattern of tooth loss in children and adolescents in a Nigerian tertiary hospital. Nig. Q. J. Hosp. Med. 2012, 22, 258–262. [Google Scholar] [PubMed]
- Stefanac, S.J. 4—Developing the treatment plan. In Diagnosis and Treatment Planning in Dentistry, 3rd ed.; Stefanac, S.J., Nesbit, S.P., Eds.; Mosby: St. Louis, MO, USA, 2017; pp. 104–120. [Google Scholar] [CrossRef]
- Kojima, K.; Inamoto, K.; Nagamatsu, K.; Hara, A.; Nakata, K.; Morita, I.; Nakagaki, H.; Nakamura, H. Success rate of endodontic treatment of teeth with vital and nonvital pulps. A meta-analysis. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2004, 97, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Oviir, T. Outcome of the root canal treatment on permanent teeth is related to the preoperative diagnosis and the accuracy of the treatment procedure. J. Evid. Based Dent. Pract. 2005, 5, 26–28. [Google Scholar] [CrossRef] [PubMed]
- American Academy on Pediatric Dentistry Clinical Affairs Committee—Pulp Therapy subcommittee; American Academy on Pediatric Dentistry Council on Clinical Affairs. Guideline on pulp therapy for primary and young permanent teeth. Pediatr. Dent. 2008, 30 (Suppl. S7), 170–174. [Google Scholar]
- Fuks, A.B.; Nuni, E. Pulp Therapy for the Young Permanent Dentition. In Pediatric Dentistry, 6th ed.; Nowak, A.J., Christensen, J.R., Mabry, T.R., Townsend, J.A., Wells, M.H., Eds.; Elsevier: Philadelphia, PA, USA, 2019; pp. 482–496. [Google Scholar] [CrossRef]
- Rafter, M. Apexification: A review. Dent. Traumatol. 2005, 21, 1–8. [Google Scholar] [CrossRef]
- Harlamb, S.C. Management of incompletely developed teeth requiring root canal treatment. Aust. Dent. J. 2016, 61 (Suppl. S1), 95–106. [Google Scholar] [CrossRef]
- Andreasen, J.O.; Farik, B.; Munksgaard, E.C. Long-term calcium hydroxide as a root canal dressing may increase risk of root fracture. Dent. Traumatol. 2002, 18, 134–137. [Google Scholar] [CrossRef]
- Możyńska, J.; Metlerski, M.; Lipski, M.; Nowicka, A. Tooth Discoloration Induced by Different Calcium Silicate-based Cements: A Systematic Review of In Vitro Studies. J. Endod. 2017, 43, 1593–1601. [Google Scholar] [CrossRef]
- Jeeruphan, T.; Jantarat, J.; Yanpiset, K.; Suwannapan, L.; Khewsawai, P.; Hargreaves, K.M. Mahidol study 1: Comparison of radiographic and survival outcomes of immature teeth treated with either regenerative endodontic or apexification methods: A retrospective study. J. Endod. 2012, 38, 1330–1336. [Google Scholar] [CrossRef]
- Banchs, F.; Trope, M. Revascularization of immature permanent teeth with apical periodontitis: New treatment protocol? J. Endod. 2004, 30, 196–200. [Google Scholar] [CrossRef] [PubMed]
- Becerra, P.; Ricucci, D.; Loghin, S.; Gibbs, J.L.; Lin, L.M. Histologic study of a human immature permanent premolar with chronic apical abscess after revascularization/revitalization. J. Endod. 2014, 40, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Ricucci, D.; Gibbs, J.L.; Lin, L.M. Histological findings of revascularized/revitalized immature permanent molar with apical periodontitis using platelet-rich plasma. J. Endod. 2013, 39, 138–144. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Liao, L.; Tian, W. Stem Cell-based Dental Pulp Regeneration: Insights From Signaling Pathways. Stem Cell Rev. Rep. 2021, 17, 1251–1263. [Google Scholar] [CrossRef]
- Miura, M.; Gronthos, S.; Zhao, M.; Lu, B.; Fisher, L.W.; Robey, P.G.; Shi, S. SHED: Stem cells from human exfoliated deciduous teeth. Proc. Natl. Acad. Sci. USA 2003, 100, 5807–5812. [Google Scholar] [CrossRef] [Green Version]
- Gronthos, S.; Brahim, J.; Li, W.; Fisher, L.W.; Cherman, N.; Boyde, A.; DenBesten, P.; Robey, P.G.; Shi, S. Stem cell properties of human dental pulp stem cells. J. Dent. Res. 2002, 81, 531–535. [Google Scholar] [CrossRef]
- Hussain, A.; Tebyaniyan, H.; Khayatan, D. The Role of Epigenetic in Dental and Oral Regenerative Medicine by Different Types of Dental Stem Cells: A Comprehensive Overview. Stem Cells Int. 2022, 2022, 1–15. [Google Scholar] [CrossRef]
- Ahangar, P.; Mills, S.J.; Smith, L.E.; Gronthos, S.; Cowin, A.J. Human gingival fibroblast secretome accelerates wound healing through anti-inflammatory and pro-angiogenic mechanisms. NPJ Regen. Med. 2020, 5, 24. [Google Scholar] [CrossRef]
- Kumar, P.; Kandoi, S.; Misra, R.; Vijayalakshmi, S.; Rajagopal, K.; Verma, R.S. The mesenchymal stem cell secretome: A new paradigm towards cell-free therapeutic mode in regenerative medicine. Cytokine Growth Factor Rev. 2019, 46, 1–9. [Google Scholar] [CrossRef]
- Hiraki, T.; Kunimatsu, R.; Nakajima, K.; Abe, T.; Yamada, S.; Rikitake, K.; Tanimoto, K. Stem cell-derived conditioned media from human exfoliated deciduous teeth promote bone regeneration. Oral Dis. 2020, 26, 381–390. [Google Scholar] [CrossRef]
- Md Fadilah, N.I.; Mohd Abdul Kader Jailani, M.S.; Badrul Hisham, M.A.I.; Sunthar Raj, N.; Shamsuddin, S.A.; Ng, M.H.; Fauzi, M.B.; Maarof, M. Cell secretomes for wound healing and tissue regeneration: Next generation acellular based tissue engineered products. J. Tissue Eng. 2022, 13, 20417314221114273. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Q.; Chen, S.; Zhang, F.; Li, Y.; Fan, W.; Mai, L.; He, H.; Huang, F. Extracellular vesicles delivering nuclear factor I/C for hard tissue engineering: Treatment of apical periodontitis and dentin regeneration. J. Tissue Eng. 2022, 13, 20417314221084095. [Google Scholar] [CrossRef]
- Jin, Q.; Li, P.; Yuan, K.; Zhao, F.; Zhu, X.; Zhang, P.; Huang, Z. Extracellular vesicles derived from human dental pulp stem cells promote osteogenesis of adipose-derived stem cells via the MAPK pathway. J. Tissue Eng. 2020, 11, 2041731420975569. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Yoon, J.Y.; Lee, J.H.; Lee, H.H.; Knowles, J.C.; Kim, H.W. Emerging biogenesis technologies of extracellular vesicles for tissue regenerative therapeutics. J. Tissue Eng. 2021, 12, 20417314211019015. [Google Scholar] [CrossRef] [PubMed]
- Chouaib, B.; Cuisinier, F.; Collart-Dutilleul, P.-Y. Dental stem cell-conditioned medium for tissue regeneration: Optimization of production and storage. World J. Stem Cells 2022, 14, 287. [Google Scholar] [CrossRef]
- de Cara, S.P.H.M.; Origassa, C.S.T.; de Sá Silva, F.; Moreira, M.S.N.; de Almeida, D.C.; Pedroni, A.C.F.; Carvalho, G.L.; Cury, D.P.; Câmara, N.O.S.; Marques, M.M. Angiogenic properties of dental pulp stem cells conditioned medium on endothelial cells in vitro and in rodent orthotopic dental pulp regeneration. Heliyon 2019, 5, e01560. [Google Scholar] [CrossRef] [Green Version]
- Xian, X.; Gong, Q.; Li, C.; Guo, B.; Jiang, H. Exosomes with highly angiogenic potential for possible use in pulp regeneration. J. Endod. 2018, 44, 751–758. [Google Scholar] [CrossRef]
- Hong, H.; Chen, X.; Li, K.; Wang, N.; Li, M.; Yang, B.; Yu, X.; Wei, X. Dental follicle stem cells rescue the regenerative capacity of inflamed rat dental pulp through a paracrine pathway. Stem Cell Res. Ther. 2020, 11, 1–16. [Google Scholar] [CrossRef]
- Kim, D.; Lee, A.E.; Xu, Q.; Zhang, Q.; Le, A.D. Gingiva-Derived Mesenchymal Stem Cells: Potential Application in Tissue Engineering and Regenerative Medicine—A Comprehensive Review. Front. Immunol. 2021, 12, 667221. [Google Scholar] [CrossRef]
- Grawish, M. Gingival-derived mesenchymal stem cells: An endless resource for regenerative dentistry. World J. Stem Cells. 2018, 10, 116–118. [Google Scholar] [CrossRef]
- Ge, S.; Mrozik, K.M.; Menicanin, D.; Gronthos, S.; Bartold, P.M. Isolation and characterization of mesenchymal stem cell-like cells from healthy and inflamed gingival tissue: Potential use for clinical therapy. Regen. Med. 2012, 7, 819–832. [Google Scholar] [CrossRef] [PubMed]
- Soudi, A.; Yazdanian, M.; Ranjbar, R.; Tebyanian, H.; Yazdanian, A.; Tahmasebi, E.; Keshvad, A.; Seifalian, A. Role and application of stem cells in dental regeneration: A comprehensive overview. Excli. J. 2021, 20, 454. [Google Scholar] [PubMed]
- Silvestro, S.; Chiricosta, L.; Gugliandolo, A.; Pizzicannella, J.; Diomede, F.; Bramanti, P.; Trubiani, O.; Mazzon, E. Extracellular Vesicles Derived from Human Gingival Mesenchymal Stem Cells: A Transcriptomic Analysis. Genes 2020, 11, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, J.; Wang, X.; Zhou, H.; Zhang, C.; Wang, Y.; Huang, J.; Liu, M.; Yang, P.; Song, A. Enhancement of periodontal tissue regeneration by conditioned media from gingiva-derived or periodontal ligament-derived mesenchymal stem cells: A comparative study in rats. Stem Cell Res. Ther. 2020, 11, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diomede, F.; Gugliandolo, A.; Scionti, D.; Merciaro, I.; Cavalcanti, M.F.; Mazzon, E.; Trubiani, O. Biotherapeutic Effect of Gingival Stem Cells Conditioned Medium in Bone Tissue Restoration. Int. J. Mol. Sci. 2018, 19, 329. [Google Scholar] [CrossRef] [Green Version]
- Ghuman, M.S.; Al-Masri, M.; Xavier, G.; Cobourne, M.T.; McKay, I.J.; Hughes, F.J. Gingival fibroblasts prevent BMP-mediated osteoblastic differentiation. J. Periodontal Res. 2019, 54, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Kang, K.J.; Ju, S.M.; Jang, Y.J.; Kim, J. Indirect co-culture of stem cells from human exfoliated deciduous teeth and oral cells in a microfluidic platform. Tissue Eng. Regen. Med. 2016, 13, 428–436. [Google Scholar] [CrossRef]
- Rajan, T.S.; Diomede, F.; Bramanti, P.; Trubiani, O.; Mazzon, E. Conditioned medium from human gingival mesenchymal stem cells protects motor-neuron-like NSC-34 cells against scratch-injury-induced cell death. Int. J. Immunopathol. Pharmacol. 2017, 30, 383–394. [Google Scholar] [CrossRef]
- Jin, S.; Yang, C.; Huang, J.; Liu, L.; Zhang, Y.; Li, S.; Zhang, L.; Sun, Q.; Yang, P. Conditioned medium derived from FGF-2-modified GMSCs enhances migration and angiogenesis of human umbilical vein endothelial cells. Stem Cell Res. Ther. 2020, 11, 68. [Google Scholar] [CrossRef]
- Cochran, D.L.; Rouse, C.A. The effect of conditioned medium from connective tissue fibroblasts and epithelium on calcium release from mouse calvarial organ culture. Arch. Oral Biol. 1993, 38, 61–65. [Google Scholar] [CrossRef]
- Kim, K.C.; Lee, E.J. Studies on Conditioned Media in Human Cells: Evaluation Using Various Cell and Culture Conditions, Animal Disease Models. J. Korean Soc. Anim. Biotechnol. 2018, 33, 41–48. [Google Scholar] [CrossRef]
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.G.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Shi, S.; Liu, Y.; Uyanne, J.; Shi, Y.; Shi, S.; Le, A.D. Mesenchymal stem cells derived from human gingiva are capable of immunomodulatory functions and ameliorate inflammation-related tissue destruction in experimental colitis. J. Immunol. 2009, 183, 7787–7798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.-Z.; Su, W.-R.; Shi, S.-H.; Wilder-Smith, P.; Xiang, A.P.; Wong, A.; Nguyen, A.L.; Kwon, C.W.; Le, A.D. Human gingiva-derived mesenchymal stem cells elicit polarization of m2 macrophages and enhance cutaneous wound healing. Stem Cells 2010, 28, 1856–1868. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Huang, Z.; Wu, D.; Kou, X.; Mao, X.; Shi, S. CD146 controls the quality of clinical grade mesenchymal stem cells from human dental pulp. Stem Cell Res. Ther. 2021, 12, 488. [Google Scholar] [CrossRef]
- Jung, Y.; Yoon, J.-Y.; Dev Patel, K.; Ma, L.; Lee, H.-H.; Kim, J.; Lee, J.-H.; Shin, J. Biological effects of tricalcium silicate nanoparticle-containing cement on stem cells from human exfoliated deciduous teeth. Nanomaterials 2020, 10, 1373. [Google Scholar] [CrossRef]
- Chalisserry, E.P.; Nam, S.Y.; Park, S.H.; Anil, S. Therapeutic potential of dental stem cells. J. Tissue Eng. 2017, 8, 2041731417702531. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.; Kim, S.; Sun, T.; Cho, Y.B.; Song, M. Pulp-dentin regeneration: Current approaches and challenges. J. Tissue Eng. 2019, 10, 2041731418819263. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Kim, B.; Bae, H.; Kim, W.; Baek, J.; Woo, K.; Lee, G.; Seol, Y.; Lee, Y.; Ku, Y.; et al. Direct Gingival Fibroblast/Osteoblast Transdifferentiation via Epigenetics. J. Dent. Res. 2017, 96, 555–561. [Google Scholar] [CrossRef]
- Sarra, G.; Machado, M.E.D.L.; Caballero-Flores, H.V.; Moreira, M.S.; Pedroni, A.C.F.; Marques, M.M. Effect of human dental pulp stem cell conditioned medium in the dentin-pulp complex regeneration: A pilot in vivo study. Tissue Cell 2021, 72, 101536. [Google Scholar] [CrossRef]
- Yu, S.; Zhao, Y.; Fang, T.J.; Ge, L. Effect of the Soluble Factors Released by Dental Apical Papilla-Derived Stem Cells on the Osteo/Odontogenic, Angiogenic, and Neurogenic Differentiation of Dental Pulp Cells. Stem Cells Dev. 2020, 29, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, C.; Huang, G.T.; Cheung, G.S.; Dissanayaka, W.L.; Zhu, W. Transplantation of dental pulp stem cells and platelet-rich plasma for pulp regeneration. J. Endod. 2012, 38, 1604–1609. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Kim, D.S.; Jang, H.; Kim, H.-R.; Kang, H.-W. Bioprinting of three-dimensional dentin–pulp complex with local differentiation of human dental pulp stem cells. J. Tissue Eng. 2019, 10, 2041731419845849. [Google Scholar] [CrossRef] [PubMed]
- Metzler, V.M.; Pritz, C.; Riml, A.; Romani, A.; Tuertscher, R.; Steinbichler, T.; Dejaco, D.; Riechelmann, H.; Dudás, J. Separation of cell survival, growth, migration, and mesenchymal transdifferentiation effects of fibroblast secretome on tumor cells of head and neck squamous cell carcinoma. Tumour Biol. 2017, 39, 1010428317705507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, N.S.; Yusop, N.; Ahmad, A. Importance of Stem Cell Migration and Angiogenesis Study for Regenerative Cell-based Therapy: A Review. Curr. Stem Cell Res. Ther. 2020, 15, 284–299. [Google Scholar] [CrossRef]
- Fu, X.; Liu, G.; Halim, A.; Ju, Y.; Luo, Q.; Song, A.G. Mesenchymal Stem Cell Migration and Tissue Repair. Cells 2019, 8, 784. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.; Xu, L. Exosomes from gingival mesenchymal stem cells enhance migration and osteogenic differentiation of pre-osteoblasts. Pharmazie 2020, 75, 576–580. [Google Scholar] [CrossRef]
- Lee, S.; Choi, E.; Cha, M.-J.; Hwang, K.-C. Cell Adhesion and Long-Term Survival of Transplanted Mesenchymal Stem Cells: A Prerequisite for Cell Therapy. Oxid. Med. Cell Longev. 2015, 2015, 632902. [Google Scholar] [CrossRef] [Green Version]
- Zhong, S.; He, X.; Li, Y.; Lou, X. Conditioned Medium Enhances Osteogenic Differentiation of Induced Pluripotent Stem Cell-Derived Mesenchymal Stem Cells. Tissue Eng. Regen. Med. 2019, 16, 141–150. [Google Scholar] [CrossRef]
- Wang, J.; Liu, B.; Gu, S.; Liang, J. Effects of Wnt/β-catenin signalling on proliferation and differentiation of apical papilla stem cells. Cell Prolif. 2012, 45, 121–131. [Google Scholar] [CrossRef]
- Jussila, M.; Thesleff, I. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 2012, 4, a008425. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, R.K.; Jaiswal, N.; Bruder, S.P.; Mbalaviele, G.; Marshak, D.R.; Pittenger, M.F. Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J. Biol. Chem. 2000, 275, 9645–9652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, G.; Jiang, D.; Thomas, P.; Benson, M.D.; Guan, K.; Karsenty, G.; Franceschi, R.T. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J. Biol. Chem. 2000, 275, 4453–4459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, J.; Lu, Y.; Song, M.; Yang, L.; Liu, J.; Chen, X.; Ma, Y.; Wang, Y. Effects of ERK/p38 MAPKs signaling pathways on MTA-mediated osteo/odontogenic differentiation of stem cells from apical papilla: A vitro study. BMC Oral Health 2020, 20, 1–9. [Google Scholar] [CrossRef]


| Forward Primer (5′-3′) | Reverse Primer (5′-3′) | |
|---|---|---|
| COL1A | 5′-CGTGACCAAAAACCAAAAGTGC-3′ | 5′-GGGTGGAGAAAGGAACAGAAA-3′ |
| ALP | 5′-ACACCTTGACTGTGGTTACT-3′ | 5′-CCTTGTAGCCAGGCCCGTTA-3′ |
| DMP1 | 5′-GGACGGCTCTGAGTTCGA-3′ | 5′-TGGGTTTCCCTGCTGTTG-3′ |
| OCN | 5′-AGACTCCGGCGCTACCTCAACAAT-3′ | 5′-CAGCTGTGCCGTCCATACT-3′ |
| RUNX2 | 5′GCTTCATTCGCCTCACAAAC-3′ | 5′-GTAGTGACCTGCGGAGATTAAG-3′ |
| GAPDH | 5′-GAGCATCTCCCTCACAATTT-3′ | 5′-GGGTGCAGCGAACTTTAT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vu, H.T.; Yoon, J.-Y.; Park, J.-H.; Lee, H.-H.; Dashnyam, K.; Kim, H.-W.; Lee, J.-H.; Shin, J.-S.; Kim, J.-B. The Potential Application of Human Gingival Fibroblast-Conditioned Media in Pulp Regeneration: An In Vitro Study. Cells 2022, 11, 3398. https://doi.org/10.3390/cells11213398
Vu HT, Yoon J-Y, Park J-H, Lee H-H, Dashnyam K, Kim H-W, Lee J-H, Shin J-S, Kim J-B. The Potential Application of Human Gingival Fibroblast-Conditioned Media in Pulp Regeneration: An In Vitro Study. Cells. 2022; 11(21):3398. https://doi.org/10.3390/cells11213398
Chicago/Turabian StyleVu, Huong Thu, Ji-Young Yoon, Jae-Hee Park, Hae-Hyoung Lee, Khandmaa Dashnyam, Hae-Won Kim, Jung-Hwan Lee, Ji-Sun Shin, and Jong-Bin Kim. 2022. "The Potential Application of Human Gingival Fibroblast-Conditioned Media in Pulp Regeneration: An In Vitro Study" Cells 11, no. 21: 3398. https://doi.org/10.3390/cells11213398
APA StyleVu, H. T., Yoon, J.-Y., Park, J.-H., Lee, H.-H., Dashnyam, K., Kim, H.-W., Lee, J.-H., Shin, J.-S., & Kim, J.-B. (2022). The Potential Application of Human Gingival Fibroblast-Conditioned Media in Pulp Regeneration: An In Vitro Study. Cells, 11(21), 3398. https://doi.org/10.3390/cells11213398







