Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of 3D Cultures from BT-474 Cells
2.2. RNA Isolation from BT-474 Cells in 2D Monolayer and 3D Cultures
2.3. Microarray Assays
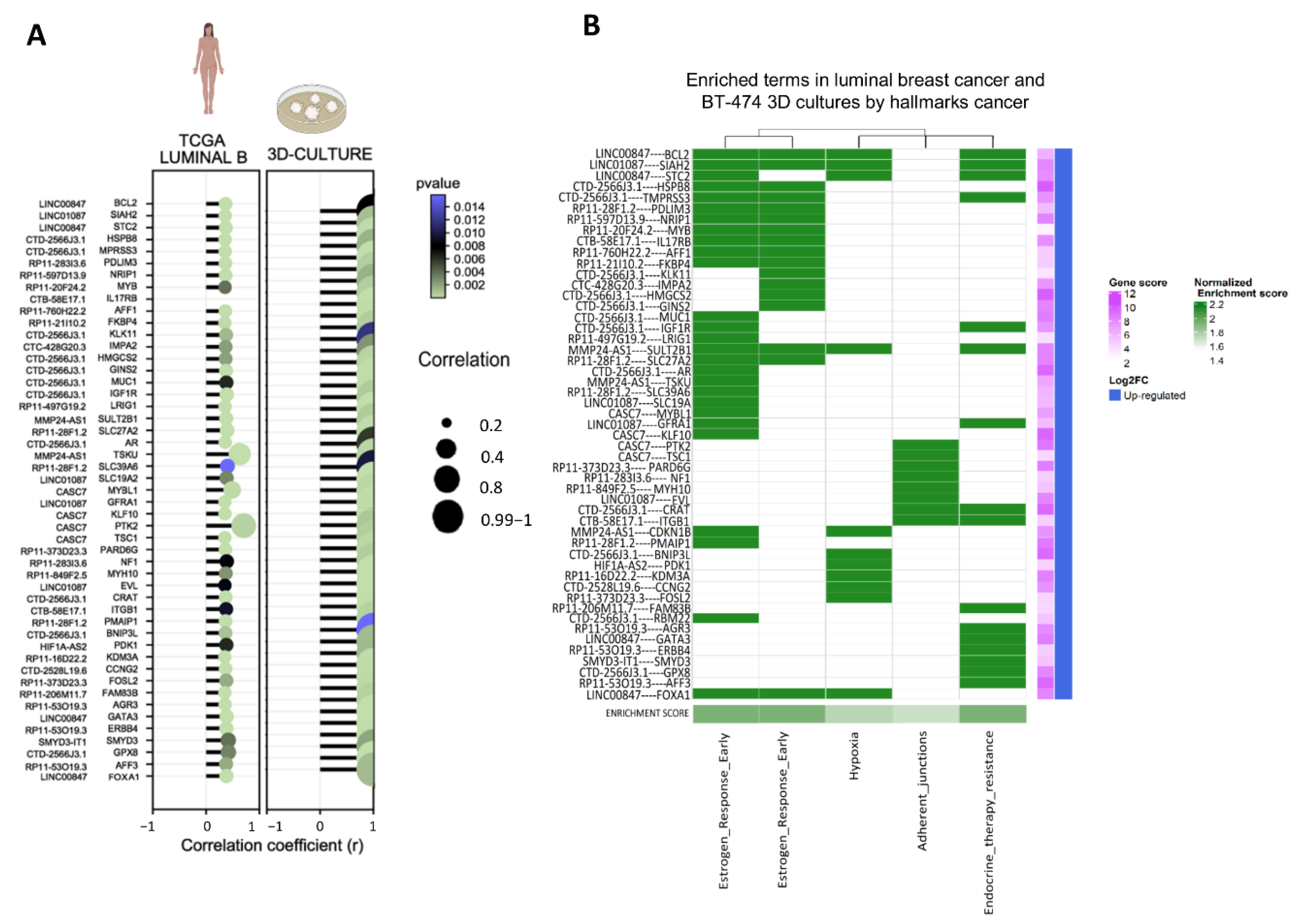
2.4. LncRNA/mRNA Correlation Analysis
2.5. GO and Pathway Analysis of DEGs
2.6. Gene Set Enrichment Analysis (GSEA)
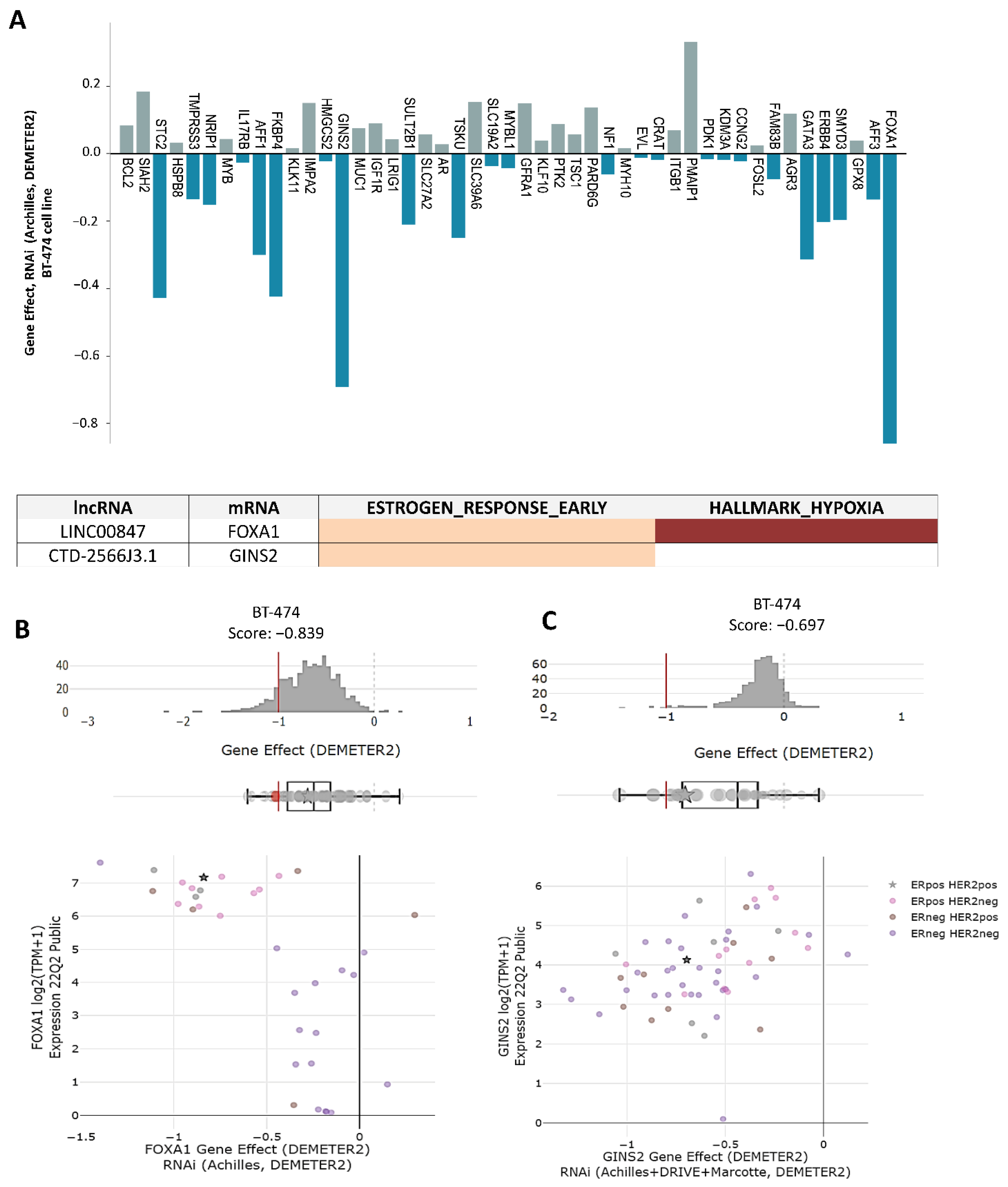
2.7. Analysis of Genetic Dependencies
2.8. Kaplan–Meier Analysis and ROC Curves
3. Results
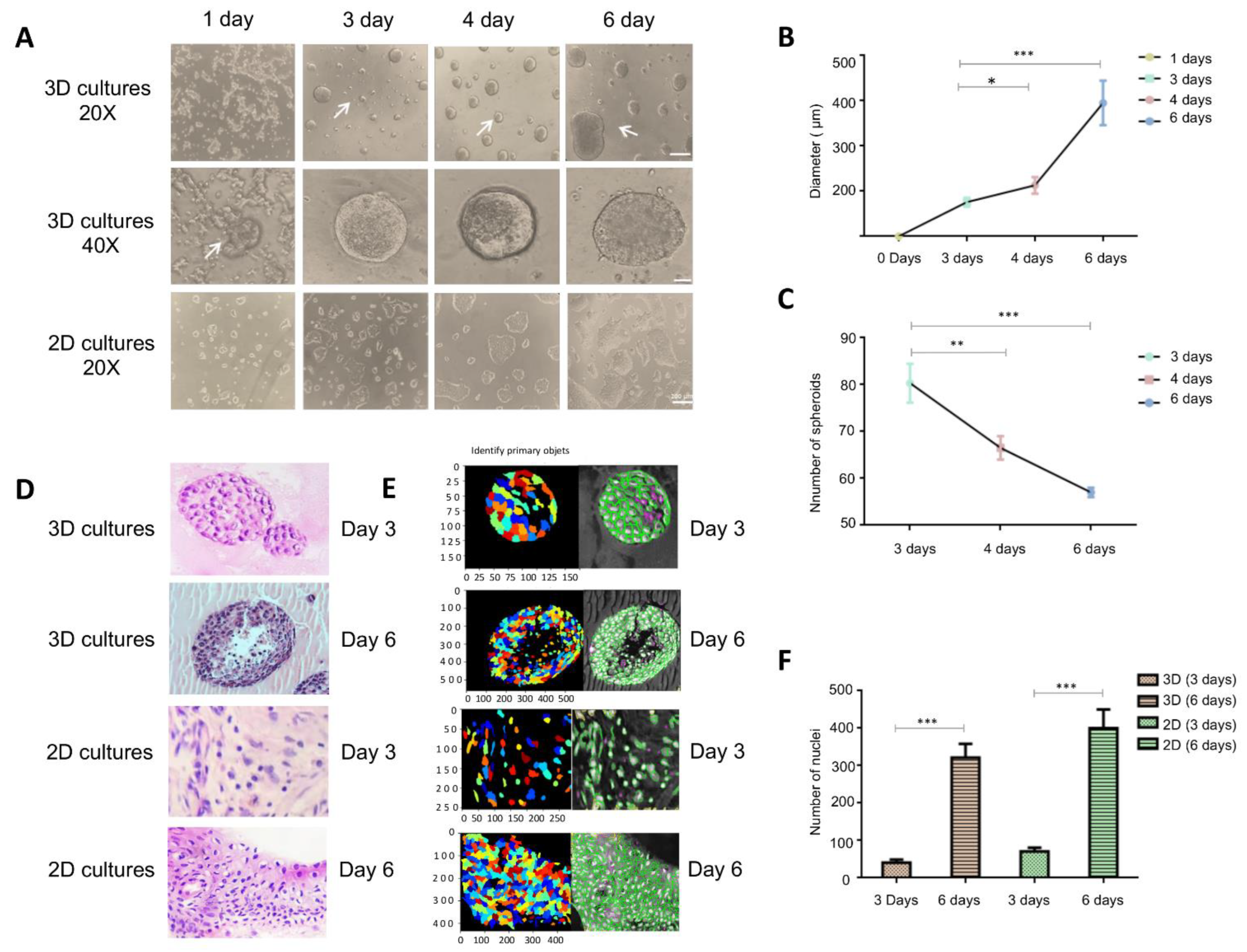
3.1. Generation and Morphological Characterization of 3D Organotypic Cultures of BT-474 Cells
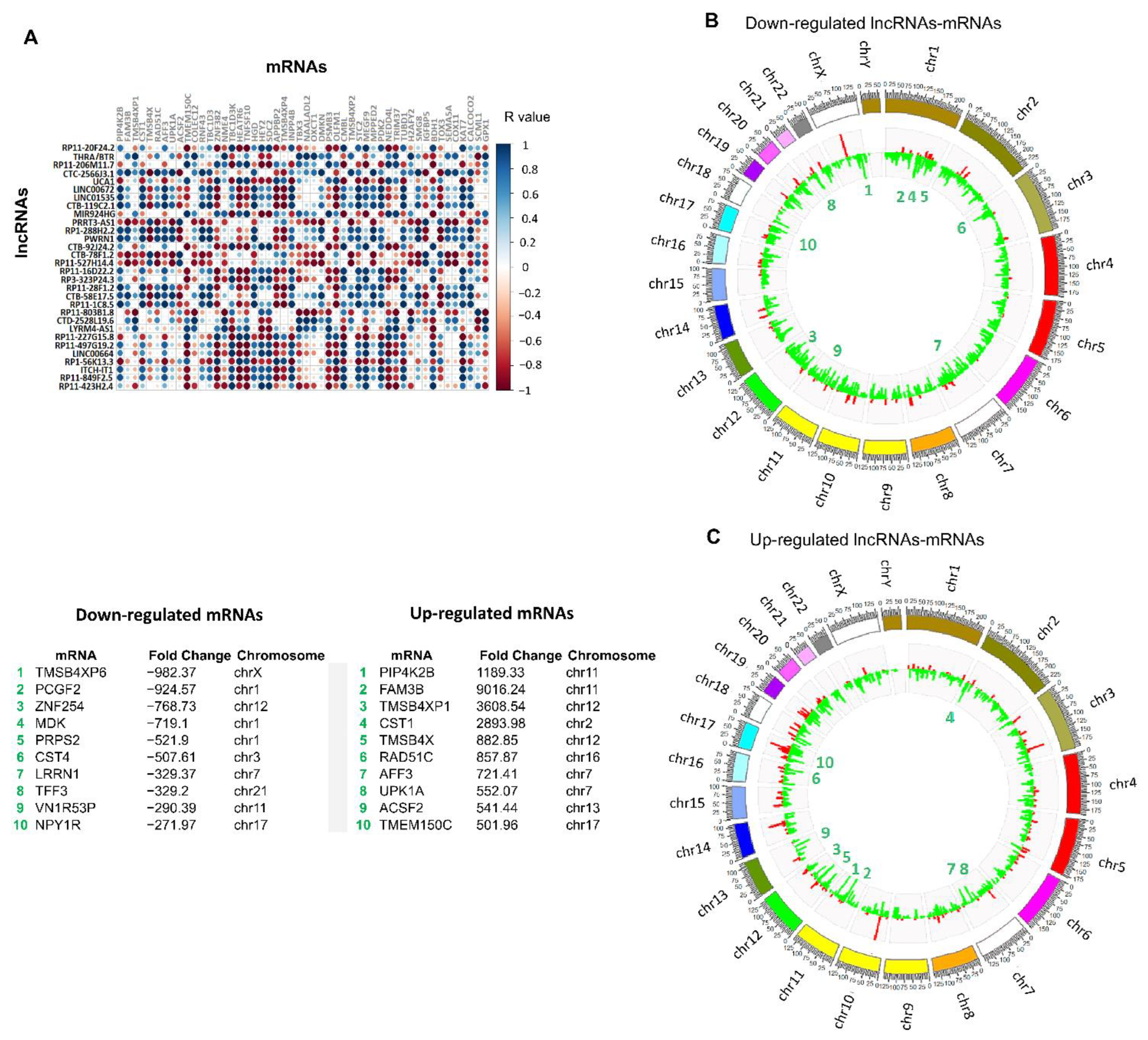
3.2. Identification of Differentially Expressed lncRNAs in 3D Cultures Derived from BT-474 Cell Line
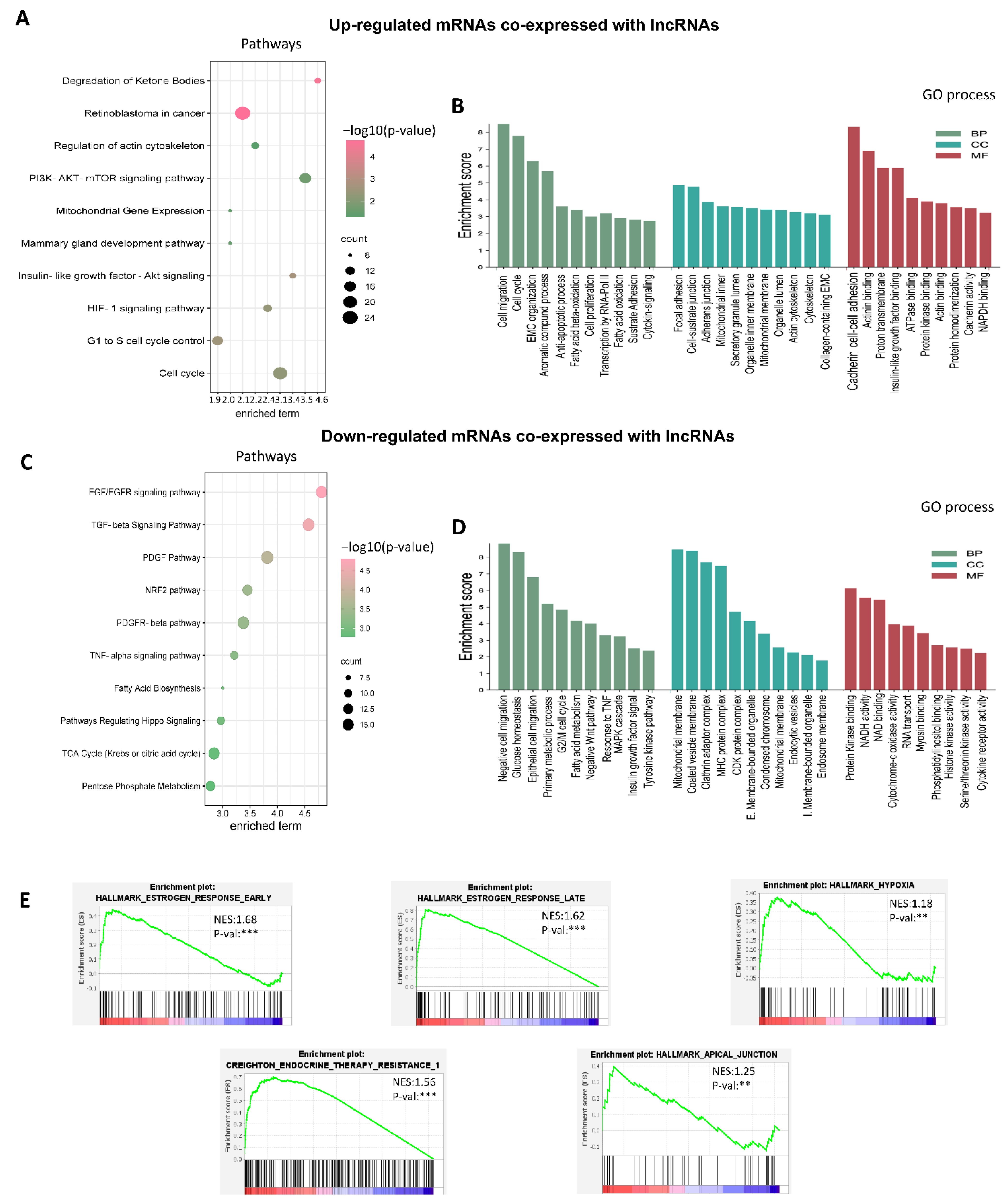
3.3. Establishment of Co-Expressed mRNAs/lncRNAs Pairs and Analysis of Biological Functions
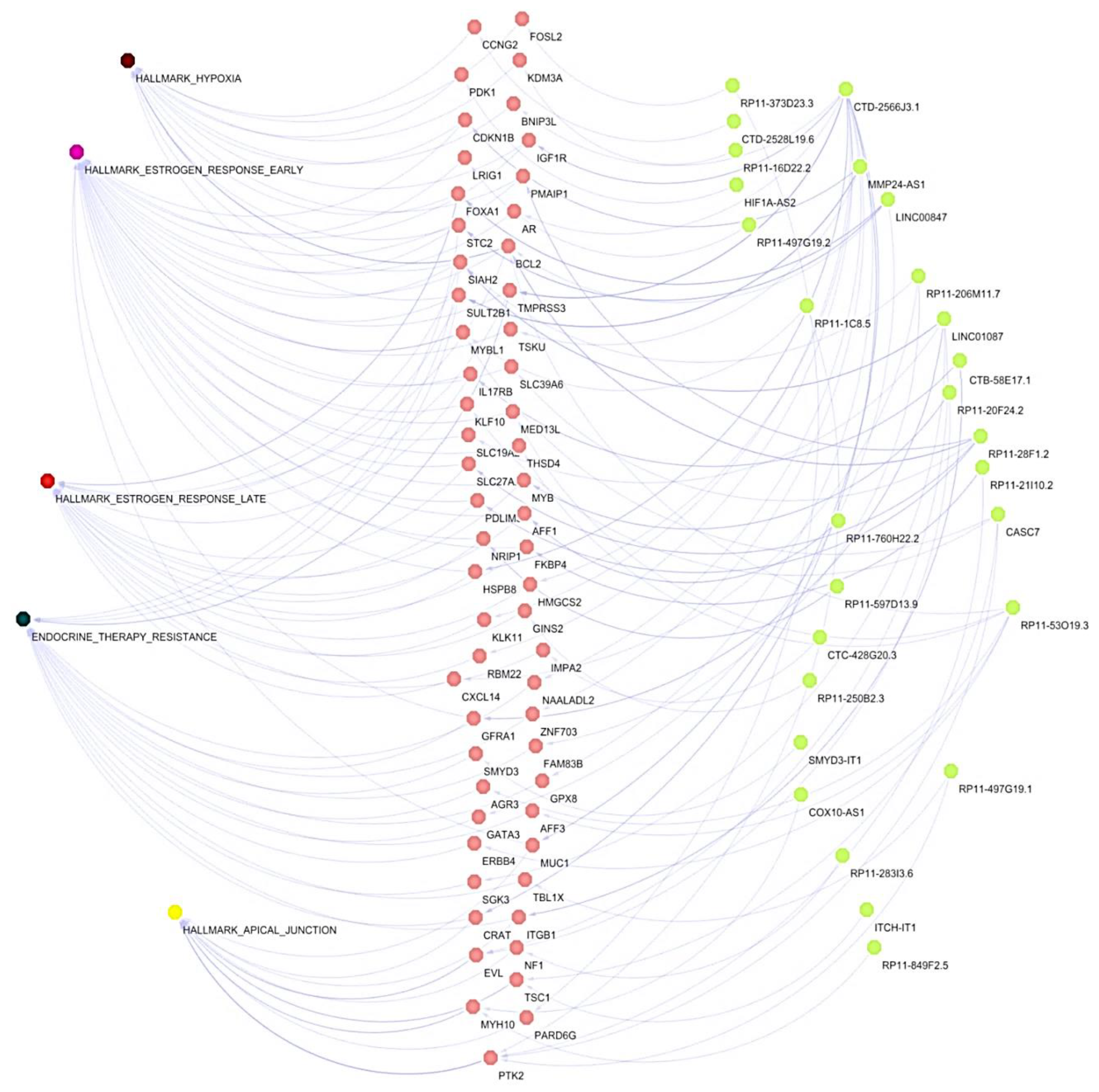
3.4. Co-Expression Networks between lncRNAs/mRNAs Could Regulate Key Processes for the Development and Maintenance of Cancer Hallmarks
3.5. lncRNAs/mRNAs Pairs Related to Genetic Dependency of Cancer
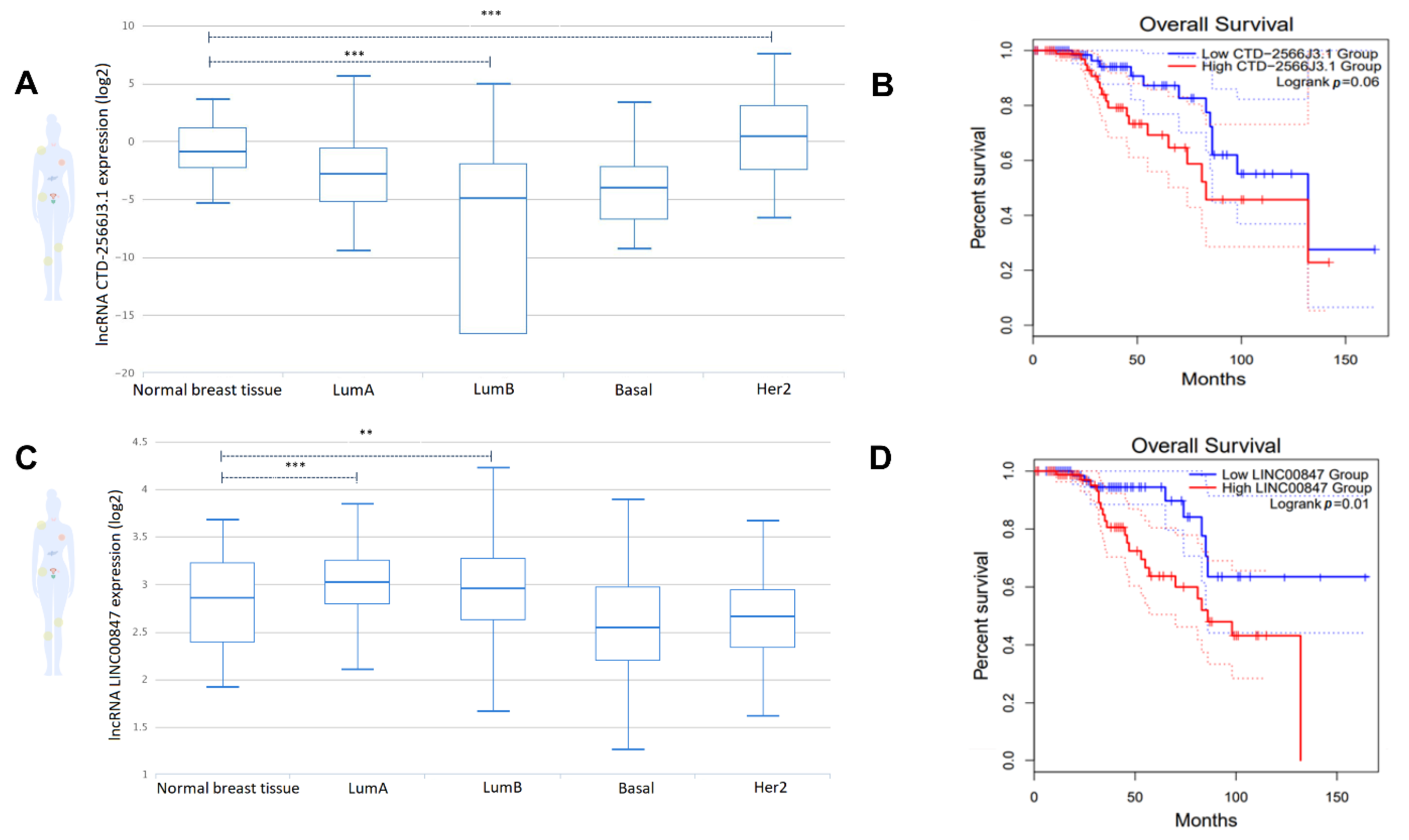
3.6. The lncRNAs, CTD-2566J3.1 and LINC00847, Are Associated with Overall Survival in Luminal B Breast Cancers
3.7. Several mRNAs Co-Expressed with CTD-2566J3.1, and LINC00847 May Be Involved in Pathological Complete Response to Chemotherapy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marusyk, A.; Polyak, K. Tumor heterogeneity: Causes and consequences. Biochim. Biophys. Acta 2010, 1805, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luce, E.; Messina, A.; Duclos-Vallée, J.C.; Dubart-Kupperschmitt, A. Advanced techniques and awaited clinical applications for human pluripotent stem cell differentiation into hepatocytes. Hepatology 2021, 74, 1101–1116. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A.; Kreutz, M.; Knuechel, R. Multicellular spheroids: A three-dimensional in vitro culture system to study tumour biology. Int. J. Exp. Pathol. 1998, 79, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, B.; Chen, H.; Gao, R.; Huang, K.; Guo, Q.; Li, F.; Chen, W.; He, J. Progress in the application of organoids to breast cancer research. J. Cell Mol. Med. 2020, 24, 5420–5427. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
- Ades, F.; Zardavas, D.; Bozovic-Spasojevic, I.; Pugliano, L.; Fumagalli, D.; de Azambuja, E.; Viale, G.; Sotiriou, C.; Piccart, M. Luminal B breast cancer: Molecular characterization, clinical management, and future perspectives. J. Clin. Oncol. 2014, 32, 2794–2803. [Google Scholar] [CrossRef] [Green Version]
- Perou, C.M.; Sørlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [Green Version]
- Tran, B.; Bedard, P.L. Luminal-B breast cancer and novel therapeutic targets. Breast Cancer Res. 2011, 13, 221. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, L.P.; Gaspar, V.M.; Mano, J.F. Design of spherically structured 3D in vitro tumor models—Advances and prospects. Acta Biomater. 2018, 75, 11–34. [Google Scholar] [CrossRef]
- Jensen, C.; Teng, Y. Is It Time to Start Transitioning From 2D to 3D Cell Culture? Front. Mol. Biosci. 2020, 7, 33. [Google Scholar] [CrossRef]
- Graf, B.W.; Boppart, S.A. Imaging and analysis of three-dimensional cell culture models. Methods Mol. Biol. 2010, 591, 211–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porter, R.J.; Murray, G.I.; McLean, M.H. Current concepts in tumour-derived organoids. Br. J. Cancer 2020, 123, 1209–1218. [Google Scholar] [CrossRef] [PubMed]
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinman, H.K.; Martin, G.R. Matrigel: Basement membrane matrix with biological activity. Semin. Cancer Biol. 2005, 15, 378–386. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.Z.; Chang, H.Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184. [Google Scholar] [CrossRef]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef]
- Boghaert, E.R.; Lu, X.; Hessler, P.E.; McGonigal, T.P.; Oleksijew, A.; Mitten, M.J.; Foster-Duke, K.; Hickson, J.A.; Santo, V.E.; Brito, C.; et al. The Volume of Three-Dimensional Cultures of Cancer Cells InVitro Influences Transcriptional Profile Differences and Similarities with Monolayer Cultures and Xenografted Tumors. Neoplasia 2017, 19, 695–706. [Google Scholar] [CrossRef]
- Fontoura, J.C.; Viezzer, C.; Dos Santos, F.G.; Ligabue, R.A.; Weinlich, R.; Puga, R.D.; Antonow, D.; Severino, P.; Bonorino, C. Comparison of 2D and 3D cell culture models for cell growth, gene expression and drug resistance. Mater. Sci. Eng. C Mater. Biol. Appl. 2020, 107, 110264. [Google Scholar] [CrossRef]
- Gamerith, G.; Rainer, J.; Huber, J.M.; Hackl, H.; Trajanoski, Z.; Koeck, S.; Lorenz, E.; Kern, J.; Kofler, R.; Kelm, J.M.; et al. 3D-cultivation of NSCLC cell lines induce gene expression alterations of key cancer-associated pathways and mimic. Oncotarget 2017, 8, 112647–112661. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, G.; Jiao, F.; Liao, Q.; Luo, H.; Li, H.; Sun, L.; Bu, D.; Yu, K.; Zhao, Y.; Chen, R. Genome-wide identification of cancer-related polyadenylated and non-polyadenylated RNAs in human breast and lung cell lines. Sci. China Life Sci. 2013, 56, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Peschansky, V.J.; Wahlestedt, C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics 2014, 9, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Mattick, J.S.; Rinn, J.L. Discovery and annotation of long noncoding RNAs. Nat. Struct. Mol. Biol. 2015, 22, 5–7. [Google Scholar] [CrossRef]
- Lim, L.J.; Jin, Y.; Yang, H.; Chung, A.Y.F.; Goh, B.K.P.; Chow, P.K.H.; Chan, C.Y.; Blanks, W.K.; Cheow, P.C.; Lee, S.Y.; et al. Network of clinically-relevant lncRNAs-mRNAs associated with prognosis of hepatocellular carcinoma patients. Sci. Rep. 2020, 10, 11124. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, Y.; Feng, L.; Li, F.; Sun, Z.; Wu, T.; Shi, X.; Li, J.; Li, X. Comprehensive characterization of lncRNA-mRNA related ceRNA network across 12 major cancers. Oncotarget 2016, 7, 64148–64167. [Google Scholar] [CrossRef] [Green Version]
- Cedro-Tanda, A.; Ríos-Romero, M.; Romero-Córdoba, S.; Cisneros-Villanueva, M.; Rebollar-Vega, R.G.; Alfaro-Ruiz, L.A.; Jiménez-Morales, S.; Domínguez-Reyes, C.; Villegas-Carlos, F.; Tenorio-Torres, A.; et al. A lncRNA landscape in breast cancer reveals a potential role for AC009283.1 in proliferation and apoptosis in HER2-enriched subtype. Sci. Rep. 2020, 10, 13146. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Melissaridou, S.; Wiechec, E.; Magan, M.; Jain, M.V.; Chung, M.K.; Farnebo, L.; Roberg, K. The effect of 2D and 3D cell cultures on treatment response, EMT profile and stem cell features in head and neck cancer. Cancer Cell Int. 2019, 19, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Shi, W.; Kuss, M.; Mirza, S.; Qi, D.; Krasnoslobodtsev, A.; Zeng, J.; Band, H.; Band, V.; Duan, B. 3D Bioprinting of Breast Cancer Models for Drug Resistance Study. ACS Biomater. Sci. Eng. 2018, 4, 4401–4411. [Google Scholar] [CrossRef]
- Koedoot, E.; Wolters, L.; Smid, M.; Stoilov, P.; Burger, G.A.; Herpers, B.; Yan, K.; Price, L.S.; Martens, J.W.M.; Le Dévédec, S.E.; et al. Differential reprogramming of breast cancer subtypes in 3D cultures and implications for sensitivity to targeted therapy. Sci. Rep. 2021, 11, 7259. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Han, L.; Roebuck, P.; Diao, L.; Liu, L.; Yuan, Y.; Weinstein, J.N.; Liang, H. TANRIC: An Interactive Open Platform to Explore the Function of lncRNAs in Cancer. Cancer Res. 2015, 75, 3728–3737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Zhang, J.; Gao, Y.; Li, Y.; Feng, C.; Song, C.; Ning, Z.; Zhou, X.; Zhao, J.; Feng, M.; et al. LncSEA: A platform for long non-coding RNA related sets and enrichment analysis. Nucleic Acids Res. 2021, 49, D969–D980. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M.; et al. Defining a Cancer Dependency Map. Cell 2017, 170, 564–576.e516. [Google Scholar] [CrossRef] [Green Version]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Xu, Q.; Liu, M.; Hu, H.; Xie, Y.; Zuo, Z.; Ren, J. lnCAR: A Comprehensive Resource for lncRNAs from Cancer Arrays. Cancer Res. 2019, 79, 2076–2083. [Google Scholar] [CrossRef] [Green Version]
- Fekete, J.T.; Győrffy, B. ROCplot.org: Validating predictive biomarkers of chemotherapy/hormonal therapy/anti-HER2 therapy using transcriptomic data of 3104 breast cancer patients. Int. J. Cancer 2019, 145, 3140–3151. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Xiu, J.; Liu, Y.; Zhang, T.; Pan, W.; Zheng, X.; Zhang, X. A 3D Printed Hanging Drop Dripper for Tumor Spheroids Analysis without Recovery. Sci. Rep. 2019, 9, 19717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondón-Lagos, M.; Verdun Di Cantogno, L.; Marchiò, C.; Rangel, N.; Payan-Gomez, C.; Gugliotta, P.; Botta, C.; Bussolati, G.; Ramírez-Clavijo, S.R.; Pasini, B.; et al. Differences and homologies of chromosomal alterations within and between breast cancer cell lines: A clustering analysis. Mol. Cytogenet. 2014, 7, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Prodduturi, N.; Sun, S.Y.; Thompson, E.A.; Kocher, J.P. Chromosome X genomic and epigenomic aberrations and clinical implications in breast cancer by base resolution profiling. Epigenomics 2015, 7, 1099–1110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cava, C.; Bertoli, G.; Castiglioni, I. Portrait of Tissue-Specific Coexpression Networks of Noncoding RNAs (miRNA and lncRNA) and mRNAs in Normal Tissues. Comput. Math. Methods Med. 2019, 2019, 9029351. [Google Scholar] [CrossRef]
- Choudhry, H. UCA1 Overexpression Promotes Hypoxic Breast Cancer Cell Proliferation and Inhibits Apoptosis via HIF-1. J. Oncol. 2021, 2021, 5512156. [Google Scholar] [CrossRef]
- Li, W.; Li, H.; Zhang, L.; Hu, M.; Li, F.; Deng, J.; An, M.; Wu, S.; Ma, R.; Lu, J.; et al. Long non-coding RNA LINC00672 contributes to p53 protein-mediated gene suppression and promotes endometrial cancer chemosensitivity. J. Biol. Chem. 2017, 292, 5801–5813. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Liu, Y.; Jin, X.; Yang, Y.; Li, L.; Wang, X.; Li, G. Long non-coding RNA LINC01535 promotes cervical cancer progression via targeting the miR-214/EZH2 feedback loop. J. Cell Mol. Med. 2019, 23, 6098–6111. [Google Scholar] [CrossRef]
- Fan, L.; Li, H.; Wang, W. Long non-coding RNA PRRT3-AS1 silencing inhibits prostate cancer cell proliferation and promotes apoptosis and autophagy. Exp. Physiol. 2020, 105, 793–808. [Google Scholar] [CrossRef]
- Huang, Y.S.; Chang, C.C.; Lee, S.S.; Jou, Y.S.; Shih, H.M. Xist reduction in breast cancer upregulates AKT phosphorylation via HDAC3-mediated repression of PHLPP1 expression. Oncotarget 2016, 7, 43256–43266. [Google Scholar] [CrossRef] [Green Version]
- Bian, Z.; Zhang, J.; Li, M.; Feng, Y.; Yao, S.; Song, M.; Qi, X.; Fei, B.; Yin, Y.; Hua, D.; et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis 2017, 6, 395. [Google Scholar] [CrossRef]
- Dong, X.; Yang, Z.; Yang, H.; Li, D.; Qiu, X. Long Non-coding RNA MIR4435-2HG Promotes Colorectal Cancer Proliferation and Metastasis through miR-206/YAP1 Axis. Front. Oncol. 2020, 10, 160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; He, Y.; Liu, W.; Bai, S.; Xiao, L.; Zhang, J.; Dhanasekaran, S.M.; Wang, Z.; Kalyana-Sundaram, S.; Balbin, O.A.; et al. Non-coding RNA LINC00857 is predictive of poor patient survival and promotes tumor progression via cell cycle regulation in lung cancer. Oncotarget 2016, 7, 11487–11499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Xie, Z.; Ma, M.; Duan, K.; Li, Y.; Ye, J. LncRNA and mRNA Expression Profiles in Methylprednisolone Stimulated Neural Stem Cells. Front. Neurosci. 2021, 15, 669224. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Li, H.; Hou, S.; Hu, B.; Liu, J.; Wang, J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS ONE 2013, 8, e65309. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Pei, W.; Zhu, L.; Nie, J.; Pei, H.; Zhang, J.; Li, B.; Hei, T.K.; Zhou, G. Microarray Profiling of TGF-β1-Induced Long Non-Coding RNA Expression Patterns in Human Lung Bronchial Epithelial BEAS-2B Cells. Cell Physiol. Biochem. 2018, 50, 2071–2085. [Google Scholar] [CrossRef]
- Hua, W.; Ten Dijke, P.; Kostidis, S.; Giera, M.; Hornsveld, M. TGFβ-induced metabolic reprogramming during epithelial-to-mesenchymal transition in cancer. Cell Mol. Life Sci. 2020, 77, 2103–2123. [Google Scholar] [CrossRef] [Green Version]
- Makinoshima, H.; Takita, M.; Matsumoto, S.; Yagishita, A.; Owada, S.; Esumi, H.; Tsuchihara, K. Epidermal growth factor receptor (EGFR) signaling regulates global metabolic pathways in EGFR-mutated lung adenocarcinoma. J. Biol. Chem. 2014, 289, 20813–20823. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Yang, Z.; Dong, D.L.; Jang, T.S.; Knowles, J.C.; Kim, H.W.; Jin, G.Z.; Xuan, Y. 3D culture technologies of cancer stem cells: Promising ex vivo tumor models. J. Tissue Eng. 2020, 11, 2041731420933407. [Google Scholar] [CrossRef]
- Kremheller, J.; Vuong, A.T.; Schrefler, B.A.; Wall, W.A. An approach for vascular tumor growth based on a hybrid embedded/homogenized treatment of the vasculature within a multiphase porous medium model. Int. J. Numer. Method. Biomed. Eng. 2019, 35, e3253. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Zhao, Y.; Zhang, Y.; AiErken, N.; Shao, N.; Ye, R.; Lin, Y.; Wang, S. AFF3 upregulation mediates tamoxifen resistance in breast cancers. J. Exp. Clin. Cancer Res. 2018, 37, 254. [Google Scholar] [CrossRef]
- Jansen, M.P.; Sas, L.; Sieuwerts, A.M.; Van Cauwenberghe, C.; Ramirez-Ardila, D.; Look, M.; Ruigrok-Ritstier, K.; Finetti, P.; Bertucci, F.; Timmermans, M.M.; et al. Decreased expression of ABAT and STC2 hallmarks ER-positive inflammatory breast cancer and endocrine therapy resistance in advanced disease. Mol. Oncol. 2015, 9, 1218–1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Aranda, M.; Pérez-Ruiz, E.; Redondo, M. Bcl-2 Inhibition to Overcome Resistance to Chemo- and Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, C.J.; Kohane, I.S.; Butte, A.J. Systematic survey reveals general applicability of “guilt-by-association” within gene coexpression networks. BMC Bioinform. 2005, 6, 227. [Google Scholar] [CrossRef] [Green Version]
- Santini, M.T.; Rainaldi, G.; Indovina, P.L. Apoptosis, cell adhesion and the extracellular matrix in the three-dimensional growth of multicellular tumor spheroids. Crit. Rev. Oncol. Hematol. 2000, 36, 75–87. [Google Scholar] [CrossRef]
- Pacheco-Marín, R.; Melendez-Zajgla, J.; Castillo-Rojas, G.; Mandujano-Tinoco, E.; Garcia-Venzor, A.; Uribe-Carvajal, S.; Cabrera-Orefice, A.; Gonzalez-Torres, C.; Gaytan-Cervantes, J.; Mitre-Aguilar, I.B.; et al. Transcriptome profile of the early stages of breast cancer tumoral spheroids. Sci. Rep. 2016, 6, 23373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, E. The role of oestrogen and progesterone receptors in human mammary development and tumorigenesis. Breast. Cancer Res. 2002, 4, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Bense, R.D.; de Vries, E.G.E.; Schröder, C.P.; Fehrmann, R.S.N. Higher expression of estrogen response genes in the primary tumor is associated with a greater risk for late recurrence in patients with ER+/HER2-breast cancer. Ann. Oncol. 2017, 28, v60. [Google Scholar] [CrossRef]
- Li, H.; Chen, Y.K.; Wan, Q.; Shi, A.Q.; Wang, M.; He, P.; Tang, L.X. Long Non-coding RNA LINC00847 Induced by E2F1 Accelerates Non-small Cell Lung Cancer Progression through Targeting miR-147a/IFITM1 Axis. Front. Med. 2021, 8, 663558. [Google Scholar] [CrossRef]
- Tu, L.R.; Li, W.; Liu, J.; Song, X.G.; Xu, H.W. LncRNA LINC00847 contributes to hepatocellular carcinoma progression by acting as a sponge of miR-99a to induce E2F2 expression. J. Biol. Regul. Homeost. Agents 2020, 34, 2195–2203. [Google Scholar] [CrossRef]
- Brantley, K.D.; Kjærsgaard, A.; Cronin-Fenton, D.; Yacoub, R.; Nielsen, A.S.; Lauridsen, K.L.; Hamilton-Dutoit, S.; Lash, T.L. Stanniocalcin Expression as a Predictor of Late Breast Cancer Recurrence. Cancer Epidemiol. Biomark. Prev. 2018, 27, 653–659. [Google Scholar] [CrossRef]
- Fu, X.; Pereira, R.; De Angelis, C.; Veeraraghavan, J.; Nanda, S.; Qin, L.; Cataldo, M.L.; Sethunath, V.; Mehravaran, S.; Gutierrez, C.; et al. FOXA1 upregulation promotes enhancer and transcriptional reprogramming in endocrine-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 26823–26834. [Google Scholar] [CrossRef] [PubMed]
- Theodorou, V.; Stark, R.; Menon, S.; Carroll, J.S. GATA3 acts upstream of FOXA1 in mediating ESR1 binding by shaping enhancer accessibility. Genome Res. 2013, 23, 12–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merikhian, P.; Ghadirian, R.; Farahmand, L.; Mansouri, S.; Majidzadeh-A, K. MUC1 induces tamoxifen resistance in estrogen receptor-positive breast cancer. Expert Rev. Anticancer 2017, 17, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wester, L.; He, J.; Geiger, T.; Moerkens, M.; Siddappa, R.; Helmijr, J.A.; Timmermans, M.M.; Look, M.P.; van Deurzen, C.H.M.; et al. IGF1R signaling drives antiestrogen resistance through PAK2/PIX activation in luminal breast cancer. Oncogene 2018, 37, 1869–1884. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.J.; Chen, S.M.; Guo, C.L.; Li, Y.X.; Ding, J.; Meng, L.H. The mTOR inhibitor AZD8055 overcomes tamoxifen resistance in breast cancer cells by down-regulating HSPB8. Acta Pharm. Sin. 2018, 39, 1338–1346. [Google Scholar] [CrossRef]


| LncRNA | Fold Change | Function | Targets | Cancer Type | Reference |
|---|---|---|---|---|---|
| RP11-20F24.2 | 632.4 | Metastasis | ANKRD30A | Normal breast tissues | [44] |
| THRA1/BTR | 243.7 | Unknown | Unknown | N/A | |
| RP11-206M11.7 | 138.68 | Unknown | Unknown | N/A | |
| CTD-2566J3.1 | 115.75 | Unknown | Unknown | N/A | |
| UCA1 | 28.37 | Hypoxia and apoptosis | HIF-1α, miR-206 | Breast cancer/2D cell lines | [45] |
| LINC00672 | 27.32 | Chemosensitivity | p53 | Endometrial cancer/Xenograft mouse model | [46] |
| LINC01535 | 23.34 | Proliferation, migration and invasion | miR-214/EZH2 | Cervical cancer/Tumor samples and 2D cell lines | [47] |
| CTB-119C2.1 | 20.69 | Unknown | Unknown | N/A | |
| MIR924HG | 19.66 | Unknown | Unknown | N/A | |
| PRRT3-AS1 | 19.41 | Proliferation, apoptosis and autophagy | PPARγ | Prostate cancer/Tumor samples and 2D cell lines | [48] |
| LncRNA | Fold Change | Function | Targets | Cancer Type | Reference |
|---|---|---|---|---|---|
| XIST | −2786.08 | Proliferation | AKT/HDAC3 PD-L1 | Breast cancer/2D cell lines | [49] |
| RP11-782C8.3 | −122.01 | Unknown | Unknown | N/A | |
| LINC00152 | −78.28 | Proliferation | miR-139-5p | Colorectal Cancer/Tumor samples and 2D cell lines | [50] |
| MIR4435-2HG | −35.98 | Proliferation and migration | miR-206/YAP1 | Colorectal Cancer/Tumor samples and 2D cell lines | [51] |
| CTD-2538C1.2 | −32.1 | Unknown | Unknown | N/A | |
| LINC00857 | −29.98 | Proliferation and invasion. | CCNE1/CDK2 | Lung Cancer/Tissues and 2D cell lines | [52] |
| RP11-425M5.7 | −29.61 | Unknown | Unknown | N/A | |
| BLCAP-AS1 | −25.26 | Unknown | Unknown | N/A | |
| RP11-383J24.6 | −24.71 | Unknown | Unknown | N/A | |
| CH17-360D5.2 | −24.61 | Unknown | Unknown | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nuñez-Olvera, S.I.; Aguilar-Arnal, L.; Cisneros-Villanueva, M.; Hidalgo-Miranda, A.; Marchat, L.A.; Salinas-Vera, Y.M.; Ramos-Payán, R.; Pérez-Plasencia, C.; Carlos-Reyes, Á.; Puente-Rivera, J.; et al. Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment. Cells 2022, 11, 3458. https://doi.org/10.3390/cells11213458
Nuñez-Olvera SI, Aguilar-Arnal L, Cisneros-Villanueva M, Hidalgo-Miranda A, Marchat LA, Salinas-Vera YM, Ramos-Payán R, Pérez-Plasencia C, Carlos-Reyes Á, Puente-Rivera J, et al. Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment. Cells. 2022; 11(21):3458. https://doi.org/10.3390/cells11213458
Chicago/Turabian StyleNuñez-Olvera, Stephanie I., Lorena Aguilar-Arnal, Mireya Cisneros-Villanueva, Alfredo Hidalgo-Miranda, Laurence A. Marchat, Yarely M. Salinas-Vera, Rosalio Ramos-Payán, Carlos Pérez-Plasencia, Ángeles Carlos-Reyes, Jonathan Puente-Rivera, and et al. 2022. "Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment" Cells 11, no. 21: 3458. https://doi.org/10.3390/cells11213458
APA StyleNuñez-Olvera, S. I., Aguilar-Arnal, L., Cisneros-Villanueva, M., Hidalgo-Miranda, A., Marchat, L. A., Salinas-Vera, Y. M., Ramos-Payán, R., Pérez-Plasencia, C., Carlos-Reyes, Á., Puente-Rivera, J., & López-Camarillo, C. (2022). Breast Cancer Cells Reprogram the Oncogenic lncRNAs/mRNAs Coexpression Networks in Three-Dimensional Microenvironment. Cells, 11(21), 3458. https://doi.org/10.3390/cells11213458








