Transcriptomic Landscape and Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Limbal Epithelial Progenitor Cells
Abstract
1. Introduction
2. Material and Methods
2.1. Tissue and Cells
2.2. Differentiation of hiPSC Using the SEAM Method
2.3. Cell Sorting
2.4. Total RNA Extraction
2.5. RNA Sequencing
2.6. Bioinformatics
2.7. CFU-E Assay
2.8. Flow Cytometry
2.9. Co-Culture of LMs with hiPSC-LEPC
2.10. Wound Healing Assays
2.11. Angiogenesis-Related Assays
2.12. Immunomodulatory Assays
2.13. Repopulation of Decellularized Corneal/Limbal Scaffolds with hiPSC-LEPC
2.14. Histology and Immunohistochemistry—Paraffin
2.15. Immunohistochemistry of Frozen Sections and Immunocytochemistry
2.16. Transmission Electron Microscopy
2.17. Statistical Analysis
3. Results
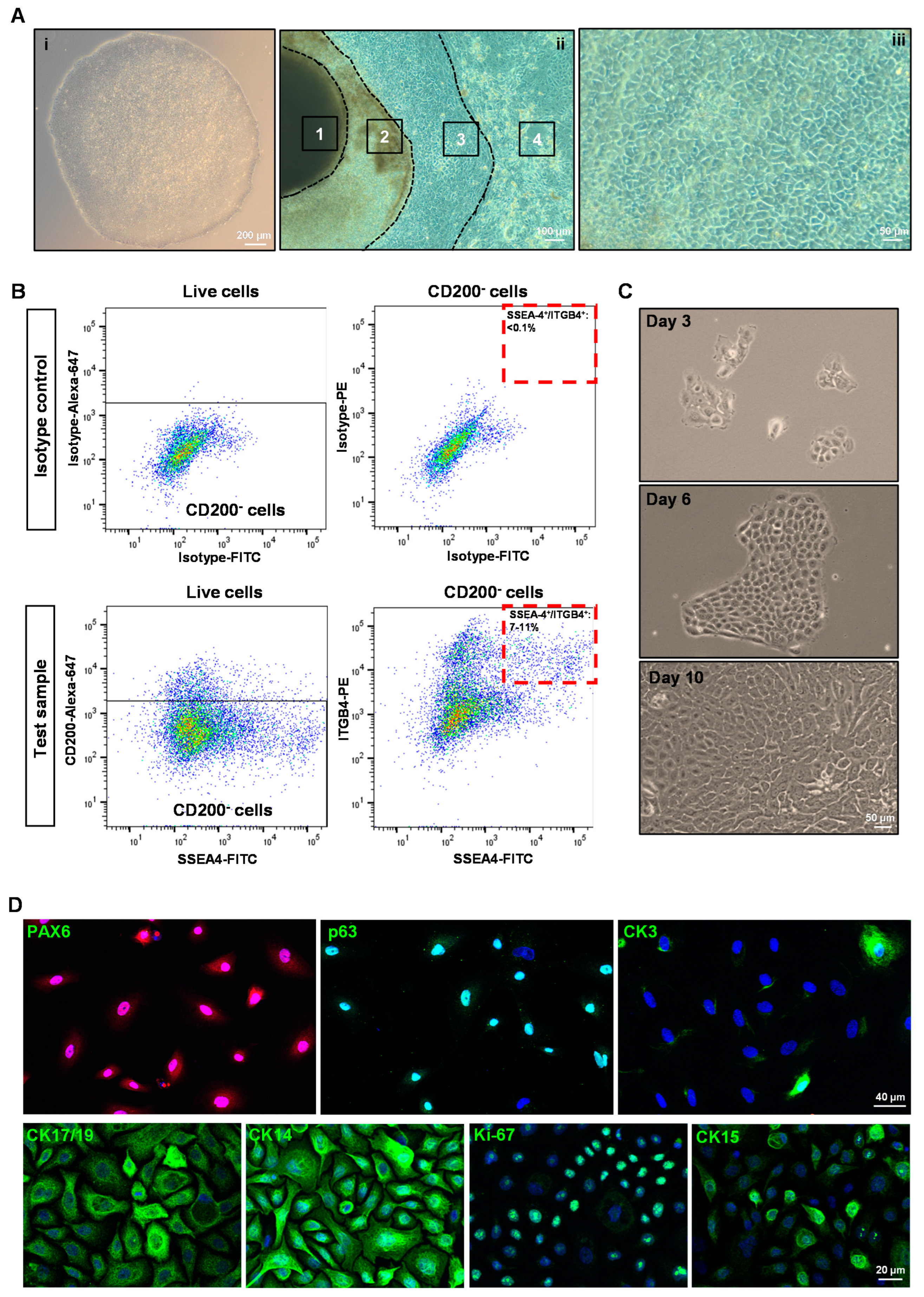
3.1. Isolation and Characterization of hiPSC-LEPC
3.2. Transcriptional Profiling of hiPSC-LEPC
3.3. Immunogenic Potential of hiPSC-LEPC
3.4. Angiogenic Potential of hiPSC-LEPC
3.5. Repopulation of Decellularized Scaffolds
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cotsarelis, G.; Cheng, S.Z.; Dong, G.; Sun, T.T.; Lavker, R.M. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell 1989, 57, 201–209. [Google Scholar] [CrossRef]
- Deng, S.X.; Borderie, V.; Chan, C.C.; Dana, R.; Figueiredo, F.C.; Gomes, J.A.P.; Pellegrini, G.; Shimmura, S.; Kruse, F.E. Global Consensus on Definition, Classification, Diagnosis, and Staging of Limbal Stem Cell Deficiency. Cornea 2019, 38, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Reinhard, T.; Spelsberg, H.; Henke, L.; Kontopoulos, T.; Enczmann, J.; Wernet, P.; Berschick, P.; Sundmacher, R.; Böhringer, D. Long-term results of allogeneic penetrating limbo-keratoplasty in total limbal stem cell deficiency. Ophthalmology 2004, 111, 775–782. [Google Scholar] [CrossRef]
- Shortt, A.J.; Secker, G.A.; Notara, M.D.; Limb, G.A.; Khaw, P.T.; Tuft, S.J.; Daniels, J.T. Transplantation of ex vivo cultured limbal epithelial stem cells: A review of techniques and clinical results. Surv. Ophthalmol. 2007, 52, 483–502. [Google Scholar] [CrossRef] [PubMed]
- Rama, P.; Matuska, S.; Paganoni, G.; Spinelli, A.; De Luca, M.; Pellegrini, G. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010, 363, 147–155. [Google Scholar] [CrossRef]
- Ghareeb, A.E.; Lako, M.; Figueiredo, F.C. Recent Advances in Stem Cell Therapy for Limbal Stem Cell Deficiency: A Narrative Review. Ophthalmol. Ther. 2020, 9, 809–831. [Google Scholar] [CrossRef] [PubMed]
- Kate, A.; Basu, S. A Review of the Diagnosis and Treatment of Limbal Stem Cell Deficiency. Front. Med. 2022, 9, 836009. [Google Scholar] [CrossRef] [PubMed]
- Dua, H.S.; Saini, J.S.; Azuara-Blanco, A.; Gupta, P. Limbal stem cell deficiency: Concept, aetiology, clinical presentation, diagnosis and management. Indian J. Ophthalmol. 2000, 48, 83–92. [Google Scholar]
- Shalom-Feuerstein, R.; Serror, L.; De La Forest Divonne, S.; Petit, I.; Aberdam, E.; Camargo, L.; Damour, O.; Vigouroux, C.; Solomon, A.; Gaggioli, C.; et al. Pluripotent stem cell model reveals essential roles for miR-450b-5p and miR-184 in embryonic corneal lineage specification. Stem Cells 2012, 30, 898–909. [Google Scholar] [CrossRef]
- Mikhailova, A.; Ilmarinen, T.; Uusitalo, H.; Skottman, H. Small-molecule induction promotes corneal epithelial cell differentiation from human induced pluripotent stem cells. Stem Cell Rep. 2014, 2, 219–231. [Google Scholar] [CrossRef]
- Mahmood, N.; Suh, T.C.; Ali, K.M.; Sefat, E.; Jahan, U.M.; Huang, Y.; Gilger, B.C.; Gluck, J.M. Induced Pluripotent Stem Cell-Derived Corneal Cells: Current Status and Application. Stem Cell Rev. Rep. 2022, 18, 2817–2832. [Google Scholar] [CrossRef]
- Hongisto, H.; Ilmarinen, T.; Vattulainen, M.; Mikhailova, A.; Skottman, H. Xeno- and feeder-free differentiation of human pluripotent stem cells to two distinct ocular epithelial cell types using simple modifications of one method. Stem Cell Res. Ther. 2017, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, R.; Ishikawa, Y.; Sasamoto, Y.; Katori, R.; Nomura, N.; Ichikawa, T.; Araki, S.; Soma, T.; Kawasaki, S.; Sekiguchi, K.; et al. Co-ordinated ocular development from human iPS cells and recovery of corneal function. Nature 2016, 531, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Sharaf, L.; Martin, G.; Schlunck, G.; Reinhard, T. P-Cadherin Is Expressed by Epithelial Progenitor Cells and Melanocytes in the Human Corneal Limbus. Cells 2022, 11, 1975. [Google Scholar] [CrossRef]
- Tang, C.; Drukker, M. Potential barriers to therapeutics utilizing pluripotent cell derivatives: Intrinsic immunogenicity of in vitro maintained and matured populations. Semin. Immunopathol. 2011, 33, 563–572. [Google Scholar] [CrossRef]
- Wang, Z.; Zhou, Q.; Duan, H.; Wang, Y.; Dong, M.; Shi, W. Immunological Properties of Corneal Epithelial-Like Cells Derived from Human Embryonic Stem Cells. PLoS ONE 2016, 11, e0150731. [Google Scholar] [CrossRef] [PubMed]
- Yoshinaga, Y.; Soma, T.; Azuma, S.; Maruyama, K.; Hashikawa, Y.; Katayama, T.; Sasamoto, Y.; Takayanagi, H.; Hosen, N.; Shiina, T.; et al. Long-term survival in non-human primates of stem cell-derived, MHC-unmatched corneal epithelial cell sheets. Stem Cell Rep. 2022, 17, 1714–1729. [Google Scholar] [CrossRef]
- Tseng, S.C.G.; He, H.; Zhang, S.; Chen, S.Y. Niche Regulation of Limbal Epithelial Stem Cells: Relationship between Inflammation and Regeneration. Ocul. Surf. 2016, 14, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Tsai, R.J.F.; Tsai, R.Y.N. From stem cell niche environments to engineering of corneal epithelium tissue. Jpn. J. Ophthalmol. 2014, 58, 111–119. [Google Scholar] [CrossRef]
- Chen, S.Y.; Mahabole, M.; Tseng, S.C.G. Optimization of Ex Vivo Expansion of Limbal Epithelial Progenitors by Maintaining Native Niche Cells on Denuded Amniotic Membrane. Transl. Vis. Sci. Technol. 2013, 2, 1. [Google Scholar] [CrossRef][Green Version]
- Eberwein, P.; Reinhard, T. Concise reviews: The role of biomechanics in the limbal stem cell niche: New insights for our understanding of this structure. Stem Cells 2015, 33, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Schmid, A.; Schlötzer-Schrehardt, U.; Maier, P.; Lang, S.J.; Steinberg, T.; Schlunck, G.; Reinhard, T. A decellularized human corneal scaffold for anterior corneal surface reconstruction. Sci. Rep. 2021, 11, 2992. [Google Scholar] [CrossRef]
- Polisetti, N.; Roschinski, B.; Schlötzer-Schrehardt, U.; Maier, P.; Schlunck, G.; Reinhard, T. A Decellularized Human Limbal Scaffold for Limbal Stem Cell Niche Reconstruction. Int. J. Mol. Sci. 2021, 22, 10067. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Zenkel, M.; Menzel-Severing, J.; Kruse, F.E.; Schlötzer-Schrehardt, U. Cell Adhesion Molecules and Stem Cell-Niche-Interactions in the Limbal Stem Cell Niche. Stem Cells 2016, 34, 203–219. [Google Scholar] [CrossRef] [PubMed]
- Polisetti, N.; Gießl, A.; Li, S.; Sorokin, L.; Kruse, F.E.; Schlötzer-Schrehardt, U. Laminin-511-E8 promotes efficient in vitro expansion of human limbal melanocytes. Sci. Rep. 2020, 10, 11074. [Google Scholar] [CrossRef]
- Polisetti, N.; Gießl, A.; Zenkel, M.; Heger, L.; Dudziak, D.; Naschberger, E.; Stich, L.; Steinkasserer, A.; Kruse, F.E.; Schlötzer-Schrehardt, U. Melanocytes as emerging key players in niche regulation of limbal epithelial stem cells. Ocul. Surf. 2021, 22, 172–189. [Google Scholar] [CrossRef]
- Polisetti, N.; Schlötzer-Schrehardt, U.; Reinhard, T.; Schlunck, G. Isolation and enrichment of melanocytes from human corneal limbus using CD117 (c-Kit) as selection marker. Sci. Rep. 2020, 10, 17588. [Google Scholar] [CrossRef]
- Huang, H.W.; Hu, F.R.; Wang, I.J.; Hou, Y.C.; Chen, W.L. Migration of limbal melanocytes onto the central cornea after ocular surface reconstruction: An in vivo confocal microscopic case report. Cornea 2010, 29, 204–206. [Google Scholar] [CrossRef]
- Polisetti, N.; Sharaf, L.; Schlötzer-Schrehardt, U.; Schlunck, G.; Reinhard, T. Efficient Isolation and Functional Characterization of Niche Cells from Human Corneal Limbus. Int. J. Mol. Sci. 2022, 23, 2750. [Google Scholar] [CrossRef]
- Wolf, J.; Boneva, S.; Schlecht, A.; Lapp, T.; Auw-Haedrich, C.; Lagrèze, W.; Agostini, H.; Reinhard, T.; Schlunck, G.; Lange, C. The Human Eye Transcriptome Atlas: A searchable comparative transcriptome database for healthy and diseased human eye tissue. Genomics 2022, 114, 110286. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. BibSonomy. Available online: https://www.bibsonomy.org/person/1f230a919c34360709aa298734d63dca3/author/0 (accessed on 5 October 2022).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Moreau, Y.; Kasprzyk, A.; Davis, S.; De Moor, B.; Brazma, A.; Huber, W. BioMart and Bioconductor: A powerful link between biological databases and microarray data analysis. Bioinformatics 2005, 21, 3439–3440. [Google Scholar] [CrossRef] [PubMed]
- Ggplot2. Available online: https://link.springer.com/book/10.1007/978-3-319-24277-4 (accessed on 4 October 2022).
- Sergushichev, A.A.; Loboda, A.A.; Jha, A.K.; Vincent, E.; Driggers, E.M.; Jones, R.G.; Pearce, E.J.; Artyomov, M.N. GAM: A web-service for integrated transcriptional and metabolic network analysis. Nucleic Acids Res. 2016, 44, W194–W200. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Bucher, F.; Aguilar, E.; Marra, K.V.; Rapp, J.; Arnold, J.; Diaz-Aguilar, S.; Lange, C.; Agostini, H.; Schlunck, G.; Stahl, A.; et al. CNTF Prevents Development of Outer Retinal Neovascularization Through Upregulation of CxCl10. Investig. Ophthalmol. Vis. Sci. 2020, 61, 20. [Google Scholar] [CrossRef] [PubMed]
- Dettmer, V.; Bloom, K.; Gross, M.; Weissert, K.; Aichele, P.; Ehl, S.; Cathomen, T. Retroviral UNC13D Gene Transfer Restores Cytotoxic Activity of T Cells Derived from Familial Hemophagocytic Lymphohistiocytosis Type 3 Patients In Vitro. Hum. Gene Ther. 2019, 30, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Wolf, J.; Auw-Haedrich, C.; Schlecht, A.; Boneva, S.; Lapp, T.; Horres, R.; Agostini, H.; Martin, G.; Reinhard, T.; et al. Expression of the COVID-19 receptor ACE2 in the human conjunctiva. J. Med. Virol. 2020, 92, 2081–2086. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, R.; Ishikawa, Y.; Katayama, T.; Quantock, A.J.; Nishida, K. CD200 facilitates the isolation of corneal epithelial cells derived from human pluripotent stem cells. Sci. Rep. 2018, 8, 16550. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Takeda, K.; Inatomi, T.; Sotozono, C.; Kinoshita, S. Long-term results of autologous cultivated oral mucosal epithelial transplantation in the scar phase of severe ocular surface disorders. Br. J. Ophthalmol. 2011, 95, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Sekiyama, E.; Nakamura, T.; Kawasaki, S.; Sogabe, H.; Kinoshita, S. Different expression of angiogenesis-related factors between human cultivated corneal and oral epithelial sheets. Exp. Eye Res. 2006, 83, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Kanayama, S.; Nishida, K.; Yamato, M.; Hayashi, R.; Sugiyama, H.; Soma, T.; Maeda, N.; Okano, T.; Tano, Y. Analysis of angiogenesis induced by cultured corneal and oral mucosal epithelial cell sheets in vitro. Exp. Eye Res. 2007, 85, 772–781. [Google Scholar] [CrossRef]
- Watanabe, S.; Hayashi, R.; Sasamoto, Y.; Tsujikawa, M.; Ksander, B.R.; Frank, M.H.; Quantock, A.J.; Frank, N.Y.; Nishida, K. Human iPS cells engender corneal epithelial stem cells with holoclone-forming capabilities. iScience 2021, 24, 102688. [Google Scholar] [CrossRef] [PubMed]
- Pepper, M.S. Positive and Negative Regulation of Angiogenesis: From Cell Biology to the Clinic. Vasc. Med. 1996, 1, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Satake, Y.; Higa, K.; Tsubota, K.; Shimazaki, J. Long-term outcome of cultivated oral mucosal epithelial sheet transplantation in treatment of total limbal stem cell deficiency. Ophthalmology 2011, 118, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
- Ljubimov, A.V.; Saghizadeh, M. Progress in corneal wound healing. Prog. Retin. Eye Res. 2015, 49, 17–45. [Google Scholar] [CrossRef] [PubMed]
- Saghizadeh, M.; Kramerov, A.A.; Svendsen, C.N.; Ljubimov, A.V. Concise Review: Stem Cells for Corneal Wound Healing. Stem Cells 2017, 35, 2105–2114. [Google Scholar] [CrossRef] [PubMed]
- Vattulainen, M.; Ilmarinen, T.; Viheriälä, T.; Jokinen, V.; Skottman, H. Corneal epithelial differentiation of human pluripotent stem cells generates ABCB5+ and ∆Np63α+ cells with limbal cell characteristics and high wound healing capacity. Stem Cell Res. Ther. 2021, 12, 609. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, H.; Ma, Q.; Chen, C.; Yue, J.; Li, B.; Zhang, X. Time-course single-cell RNA sequencing reveals transcriptional dynamics and heterogeneity of limbal stem cells derived from human pluripotent stem cells. Cell Biosci. 2021, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- Shibata, S.; Hayashi, R.; Kudo, Y.; Okubo, T.; Imaizumi, T.; Katayama, T.; Ishikawa, Y.; Kobayashi, Y.; Toga, J.; Taniguchi, Y.; et al. Cell-Type-Specific Adhesiveness and Proliferation Propensity on Laminin Isoforms Enable Purification of iPSC-Derived Corneal Epithelium. Stem Cell Rep. 2020, 14, 663–676. [Google Scholar] [CrossRef]
- Kulkarni, B.B.; Tighe, P.J.; Mohammed, I.; Yeung, A.M.; Powe, D.G.; Hopkinson, A.; A Shanmuganathan, V.; Dua, H.S. Comparative transcriptional profiling of the limbal epithelial crypt demonstrates its putative stem cell niche characteristics. BMC Genom. 2010, 11, 526. [Google Scholar] [CrossRef]
- Ramirez-Miranda, A.; Nakatsu, M.N.; Zarei-Ghanavati, S.; Nguyen, C.V.; Deng, S.X. Keratin 13 is a more specific marker of conjunctival epithelium than keratin 19. Mol. Vis. 2011, 17, 1652–1661. [Google Scholar]
- Ligocki, A.J.; Fury, W.; Gutierrez, C.; Adler, C.; Yang, T.; Ni, M.; Bai, Y.; Wei, Y.; Lehmann, G.L.; Romano, C. Molecular characteristics and spatial distribution of adult human corneal cell subtypes. Sci. Rep. 2021, 11, 16323. [Google Scholar] [CrossRef]
- Raicevic, G.; Rouas, R.; Najar, M.; Stordeur, P.; Boufker, H.I.; Bron, D.; Martiat, P.; Goldman, M.; Nevessignsky, M.T.; Lagneaux, L. Inflammation modifies the pattern and the function of Toll-like receptors expressed by human mesenchymal stromal cells. Hum. Immunol. 2010, 71, 235–244. [Google Scholar] [CrossRef]
- Araki, R.; Uda, M.; Hoki, Y.; Sunayama, M.; Nakamura, M.; Ando, S.; Sugiura, M.; Ideno, H.; Shimada, A.; Nifuji, A.; et al. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature 2013, 494, 100–104. [Google Scholar] [CrossRef]
- Drukker, M.; Benvenisty, N. The immunogenicity of human embryonic stem-derived cells. Trends Biotechnol. 2004, 22, 136–141. [Google Scholar] [CrossRef]
- Hori, J.; Wang, M.; Miyashita, M.; Tanemoto, K.; Takahashi, H.; Takemori, T.; Okumura, K.; Yagita, H.; Azuma, M. B7-H1-induced apoptosis as a mechanism of immune privilege of corneal allografts. J. Immunol. 2006, 177, 5928–5935. [Google Scholar] [CrossRef]
- Watson, M.P.; George, A.J.T.; Larkin, D.F.P. Differential effects of costimulatory pathway modulation on corneal allograft survival. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3417–3422. [Google Scholar] [CrossRef]
- Shen, L.; Jin, Y.; Freeman, G.J.; Sharpe, A.H.; Dana, M.R. The function of donor versus recipient programmed death-ligand 1 in corneal allograft survival. J. Immunol. 2007, 179, 3672–3679. [Google Scholar] [CrossRef]
- Collin, J.; Queen, R.; Zerti, D.; Bojic, S.; Dorgau, B.; Moyse, N.; Molina, M.M.; Yang, C.; Dey, S.; Reynolds, G.; et al. A single cell atlas of human cornea that defines its development, limbal progenitor cells and their interactions with the immune cells. Ocul. Surface 2021, 21, 279–298. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.H.; Tsai, R.J.; Chu, W.K.; Kao, C.H.; Chen, J.K. Inhibition of vascular endothelial cell morphogenesis in cultures by limbal epithelial cells. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1822–1828. [Google Scholar]
- Allen, C.L.; Clare, G.; Stewart, E.A.; Branch, M.J.; McIntosh, O.D.; Dadhwal, M.; Dua, H.S.; Hopkinson, A. Augmented dried versus cryopreserved amniotic membrane as an ocular surface dressing. PLoS ONE 2013, 8, e78441. [Google Scholar] [CrossRef]
- Shortt, A.J.; Secker, G.A.; Lomas, R.J.; Wilshaw, S.P.; Kearney, J.N.; Tuft, S.J.; Daniels, J.T. The effect of amniotic membrane preparation method on its ability to serve as a substrate for the ex-vivo expansion of limbal epithelial cells. Biomaterials 2009, 30, 1056–1065. [Google Scholar] [CrossRef]
- Badylak, S.F. The extracellular matrix as a scaffold for tissue reconstruction. Semin. Cell Dev. Biol. 2002, 13, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Pérez, J.; Ahearne, M. Decellularization and recellularization of cornea: Progress towards a donor alternative. Methods 2020, 171, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Schlötzer-Schrehardt, U.; Naumann, G.O.H. Ocular and Systemic Pseudoexfoliation Syndrome. Am. J. Ophthalmol. 2006, 141, 921–937.e2. [Google Scholar] [CrossRef]
- Pasutto, F.; Zenkel, M.; Hoja, U.; Berner, D.; Uebe, S.; Ferrazzi, F.; Schödel, J.; Liravi, P.; Ozaki, M.; Paoli, D.; et al. Pseudoexfoliation syndrome-associated genetic variants affect transcription factor binding and alternative splicing of LOXL1. Nat. Commun. 2017, 8, 15466. [Google Scholar] [CrossRef] [PubMed]
- Brzeszczynska, J.; Samuel, K.; Greenhough, S.; Ramaesh, K.; Dhillon, B.; Hay, D.; Ross, J. Differentiation and molecular profiling of human embryonic stem cell-derived corneal epithelial cells. Int. J. Mol. Med. 2014, 33, 1597–1606. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Polisetti, N.; Rapp, J.; Liang, P.; Dettmer-Monaco, V.; Bucher, F.; Pruszak, J.; Schlötzer-Schrehardt, U.; Cathomen, T.; Schlunck, G.; Reinhard, T. Transcriptomic Landscape and Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Limbal Epithelial Progenitor Cells. Cells 2022, 11, 3752. https://doi.org/10.3390/cells11233752
Polisetti N, Rapp J, Liang P, Dettmer-Monaco V, Bucher F, Pruszak J, Schlötzer-Schrehardt U, Cathomen T, Schlunck G, Reinhard T. Transcriptomic Landscape and Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Limbal Epithelial Progenitor Cells. Cells. 2022; 11(23):3752. https://doi.org/10.3390/cells11233752
Chicago/Turabian StylePolisetti, Naresh, Julian Rapp, Paula Liang, Viviane Dettmer-Monaco, Felicitas Bucher, Jan Pruszak, Ursula Schlötzer-Schrehardt, Toni Cathomen, Günther Schlunck, and Thomas Reinhard. 2022. "Transcriptomic Landscape and Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Limbal Epithelial Progenitor Cells" Cells 11, no. 23: 3752. https://doi.org/10.3390/cells11233752
APA StylePolisetti, N., Rapp, J., Liang, P., Dettmer-Monaco, V., Bucher, F., Pruszak, J., Schlötzer-Schrehardt, U., Cathomen, T., Schlunck, G., & Reinhard, T. (2022). Transcriptomic Landscape and Functional Characterization of Human Induced Pluripotent Stem Cell-Derived Limbal Epithelial Progenitor Cells. Cells, 11(23), 3752. https://doi.org/10.3390/cells11233752








