Histone Deacetylase HDA15 Restrains PHYB-Dependent Seed Germination via Directly Repressing GA20ox1/2 Gene Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. PHYB-Dependent Seed Germination Assays
2.3. RNA Isolation and qRT-PCR Analysis
2.4. ChIP Assays
2.5. Western Blot Assays
2.6. Statistical Analysis
3. Results
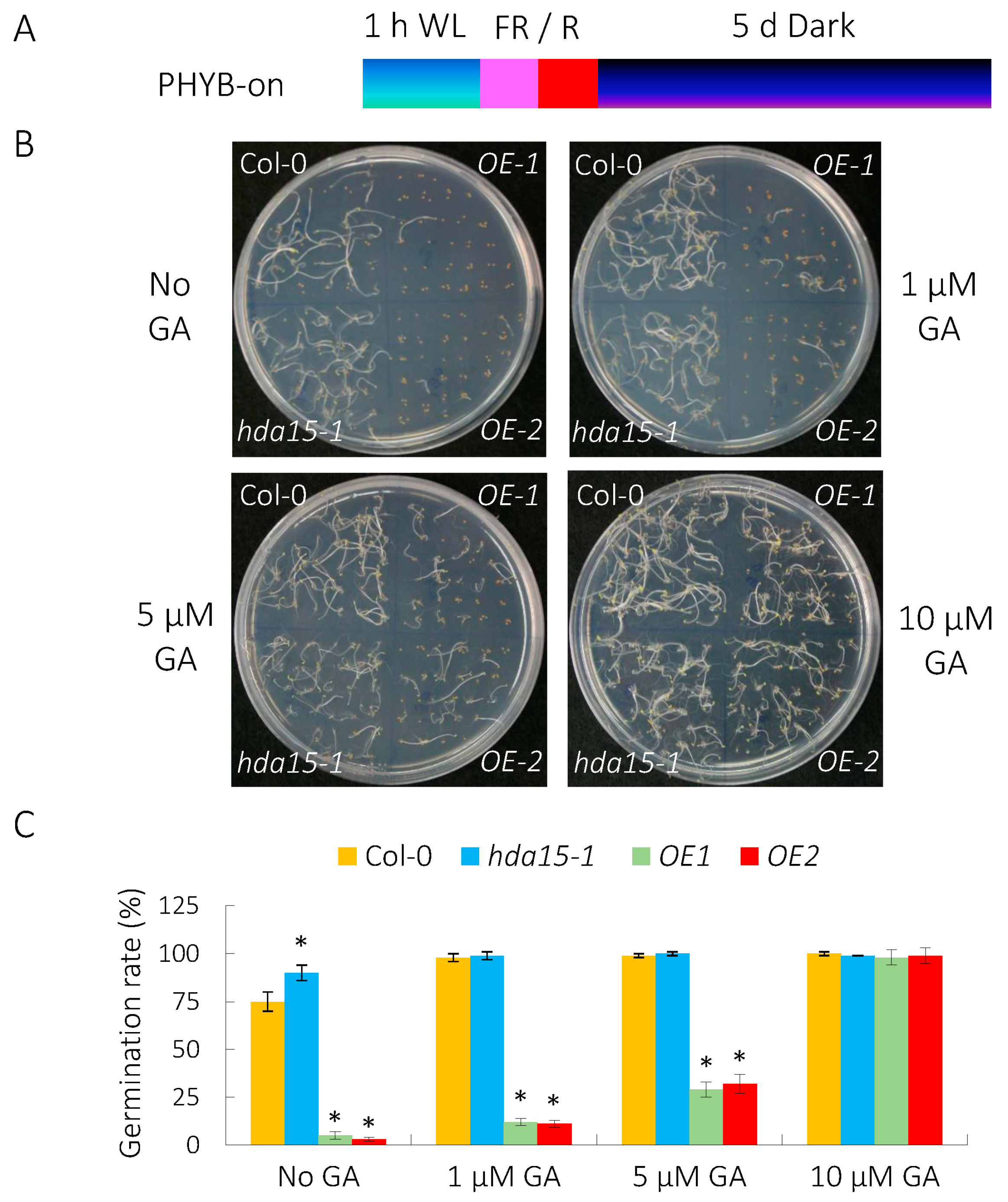
3.1. HDA15 Restrains PHYB-Dependent Seed Germination While GA Relieves Its Repressive Role
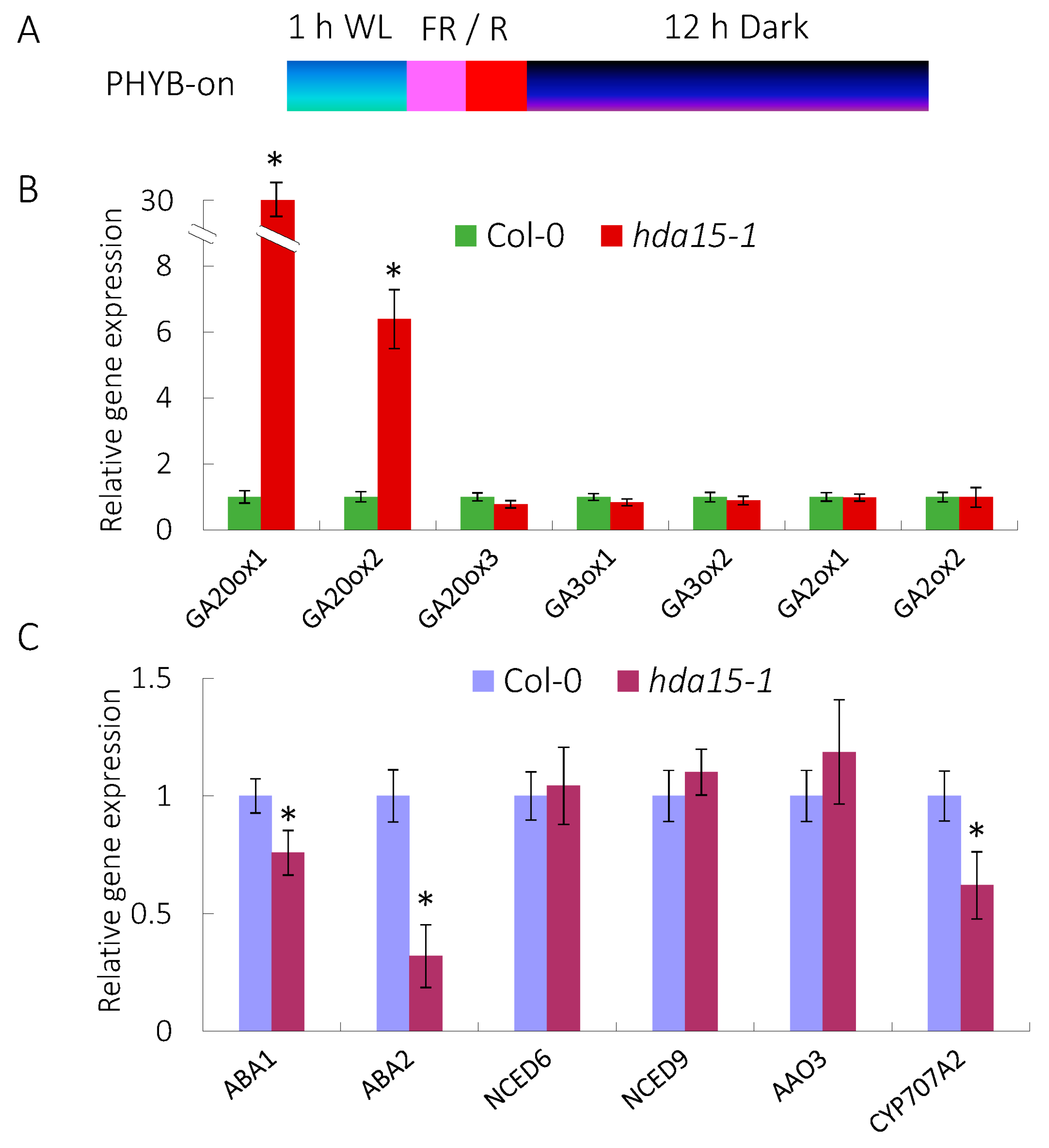
3.2. HDA15 Regulates the Expression of GA and ABA Metablism-Related Genes
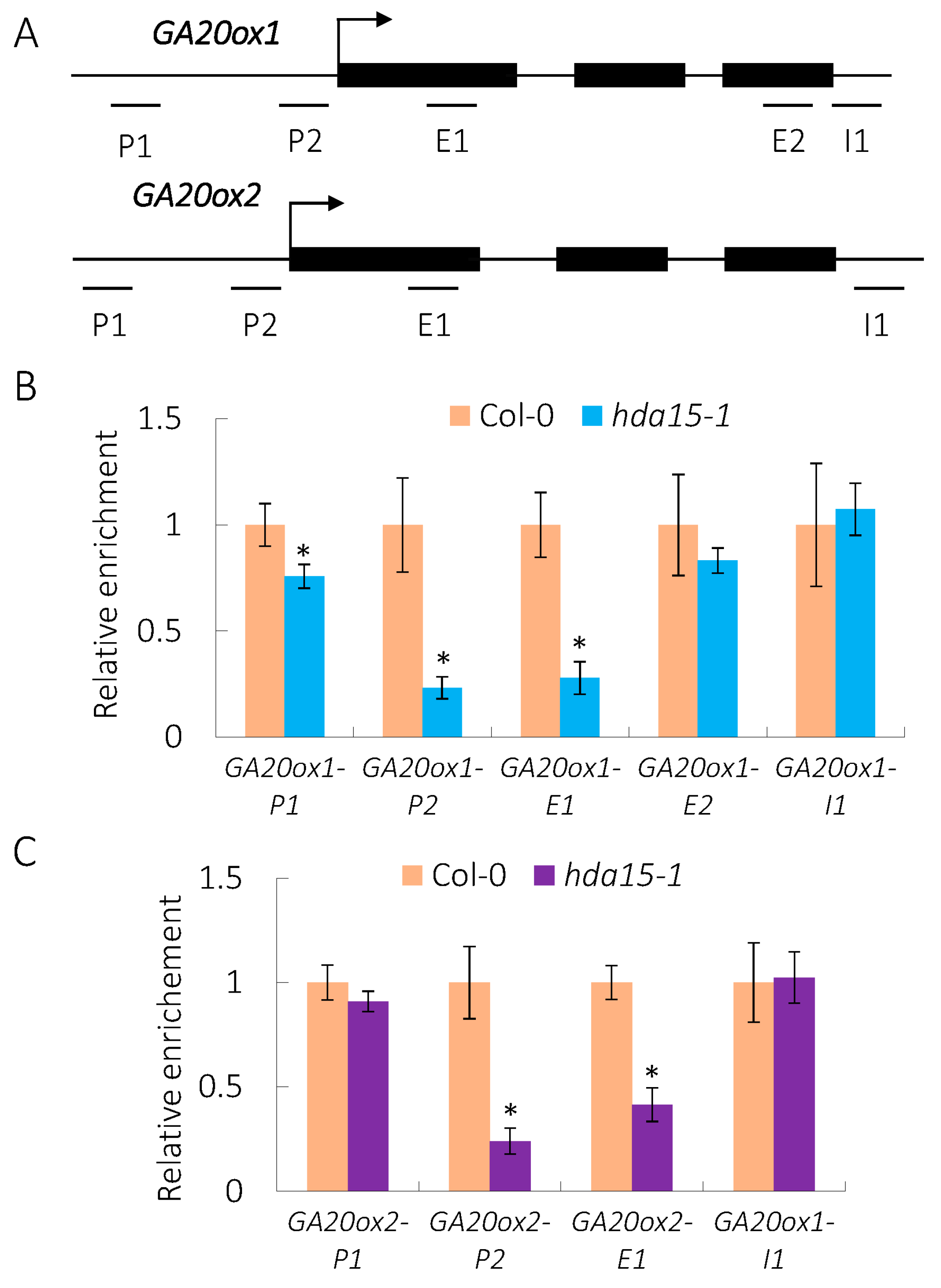
3.3. HDA15 Directly Targets GA20ox1 and GA20ox2 in Imbibed Seeds
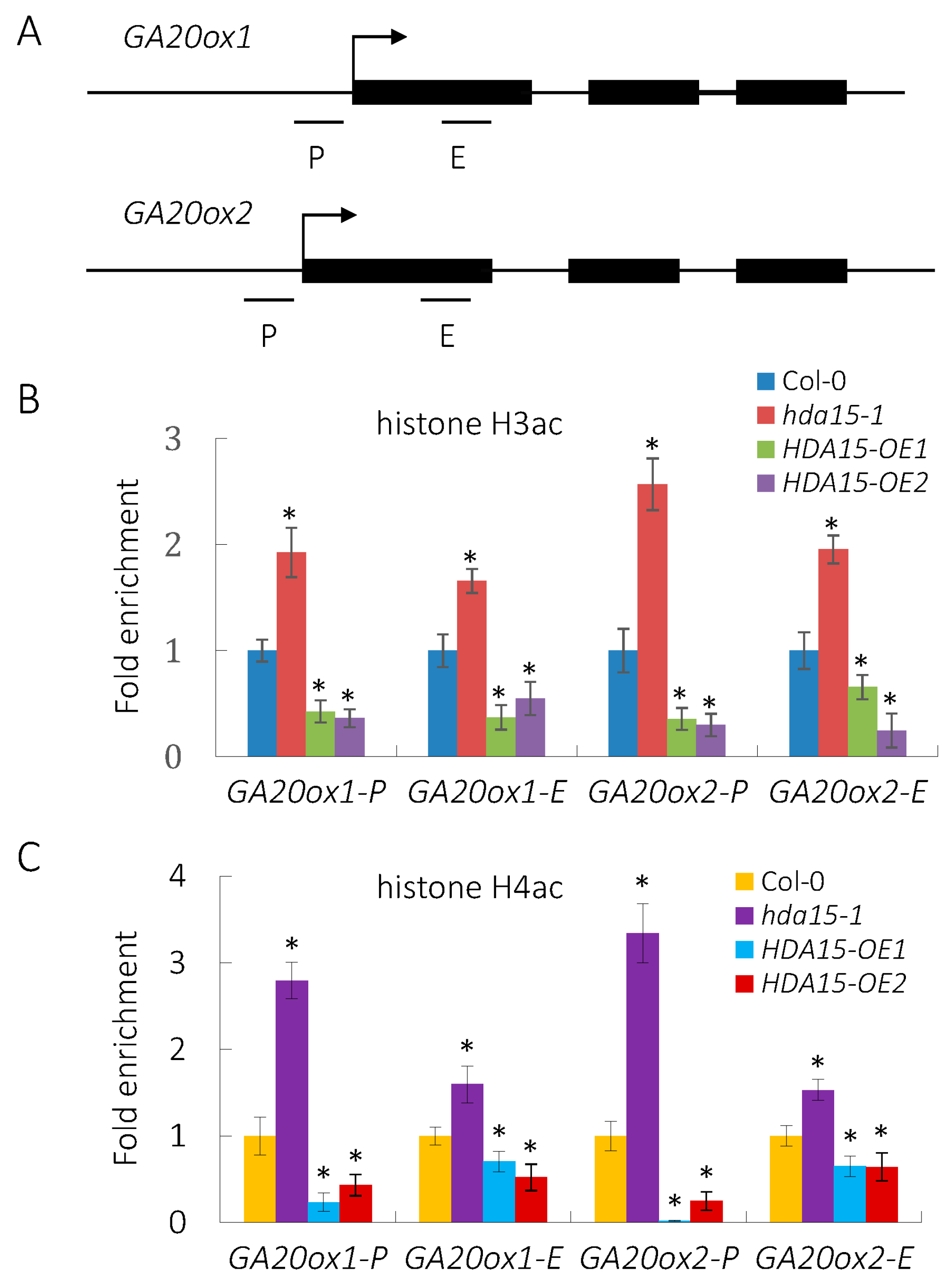
3.4. HDA15 Decreases the Levels of Histone Acetylation of GA20ox1 and GA20ox2
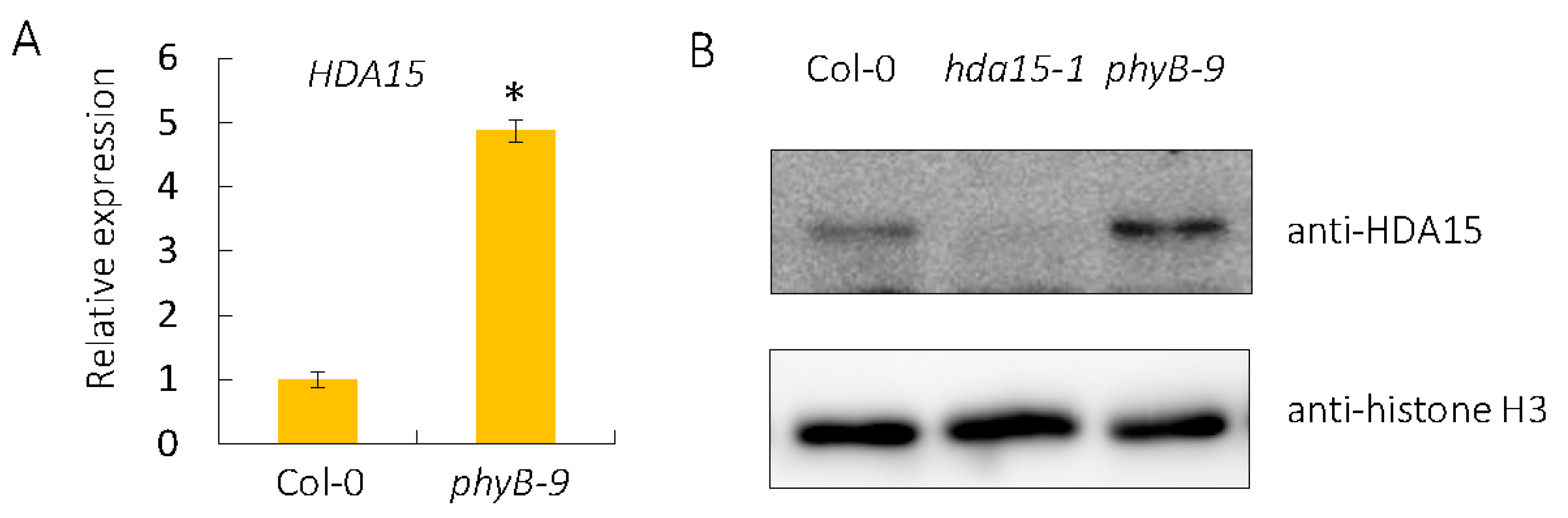
3.5. PHYB Affects the Gene Expression of HDA15 and Protein Level of HDA15
4. Discussion
4.1. HDA15 Plays Dual Roles in PHYB-Dependent Seed Germination
4.2. HDA15 May Interact with Site-Specific GA-Signal Related Transcription Factors to Repress GA20ox1/2 Expression
4.3. PHYB May Promote Seed Germination Partly through Decreasing the Transcription of HDA15
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Quail, P.H. Photosensory perception and signalling in plant cells: New paradigms? Curr. Opin. Cell Biol. 2022, 14, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Rizzini, L.; Favory, J.J.; Cloix, C.; Faggionato, D.; O’Hara, A.; Kaiserli, E.; Baumeister, R.; Schafer, E.; Nagy, F.; Jenkins, G.I.; et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science 2011, 332, 103–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Yamaguchi, S.; Hu, J.; Yusuke, J.; Jung, B.; Paik, I.; Lee, H.S.; Sun, T.P.; Kamiya, Y.; Choi, G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 2007, 19, 1192–1208. [Google Scholar] [CrossRef] [Green Version]
- Daviere, J.M. Seed dormancy and the control of germination Achard, P. Gibberellin signaling in plants. Development 2013, 140, 1147–1151. [Google Scholar]
- Lee, S.; Cheng, H.; King, K.E.; Wang, W.; He, Y.; Hussain, A.; Lo, J.; Harberd, N.P.; Peng, J. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 2020, 16, 646–658. [Google Scholar] [CrossRef] [Green Version]
- Shinomura, T.; Nagatani, A.; Chory, J.; Furuya, M. The Induction of Seed-Germination in Arabidopsis-Thaliana Is Regulated Principally by Phytochrome-B and Secondarily by Phytochrome-a (Vol 104, Pg 363, 1994). Plant Physiol. 1994, 105, 773. [Google Scholar]
- Borthwick, H.A.; Hendricks, S.B.; Parker, M.W.; Toole, E.H.; Toole, V.K. A Reversible Photoreaction Controlling Seed Germination. Proc. Natl. Acad. Sci. USA 1952, 38, 662–666. [Google Scholar] [CrossRef] [Green Version]
- Mathews, S. Phytochrome-mediated development in land plants: Red light sensing evolves to meet the challenges of changing light environments. Mol. Ecol. 2006, 15, 3483–3503. [Google Scholar] [CrossRef]
- Seo, M.; Nambara, E.; Choi, G.; Yamaguchi, S. Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 2009, 69, 463–472. [Google Scholar] [CrossRef]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, E.; Kang, H.; Yamaguchi, S.; Park, J.; Lee, D.; Kamiya, Y.; Choi, G. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 2009, 21, 403–419. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, S.; Zhao, M.; Luo, M.; Yu, C.W.; Chen, C.Y.; Tai, R.; Wu, K. Transcriptional repression by histone deacetylases in plants. Mol. Plant 2014, 7, 764–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Ding, A.B.; Liu, F.; Zhong, X. Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 2020, 71, 5179–5190. [Google Scholar] [CrossRef]
- Chen, X.; Ding, A.B.; Zhong, X. Functions and mechanisms of plant histone deacetylases. Sci. China Life Sci. 2020, 63, 206–216. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Chen, C.Y.; Wang, K.C.; Luo, M.; Tai, R.; Yuan, L.; Zhao, M.; Yang, S.; Tian, G.; Cui, Y.; et al. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 2013, 25, 1258–1273. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.; Liu, X.; Liu, X.; Li, Y.; Wu, K.; Hou, X. Arabidopsis NF-YCs Mediate the Light-Controlled Hypocotyl Elongation via Modulating Histone Acetylation. Mol. Plant 2017, 10, 260–273. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Peng, T.; Chen, C.Y.; Ji, R.; Gu, D.; Li, T.; Zhang, D.; Tu, Y.T.; Wu, K.; Liu, X. HY5 Interacts with the Histone Deacetylase HDA15 to Repress Hypocotyl Cell Elongation in Photomorphogenesis. Plant Physiol. 2018, 180, 1450–1466. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Chen, C.Y.; Zhao, M.; Zhao, L.; Duan, X.; Duan, J.; Wu, K.; Liu, X. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 2017, 45, 7137–7150. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.; Kim, J.; Park, E.; Kim, J.I.; Kang, C.; Choi, G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 2004, 16, 3045–3058. [Google Scholar] [CrossRef] [Green Version]
- Reed, J.W.; Nagpal, P.; Poole, D.S.; Furuya, M.; Chory, J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 1993, 5, 147–157. [Google Scholar]
- Oh, E.; Yamaguchi, S.; Kamiya, Y.; Bae, G.; Chung, W.I.; Choi, G. Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 2006, 47, 124–139. [Google Scholar] [CrossRef] [PubMed]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Teramura, H.; Murakoshi, S.; Nasuno, K.; Nishida, N.; Ito, T.; Yoshida, M.; Kamiya, Y.; Yamaguchi, S.; Takahashi, Y. DELLAs Function as Coactivators of GAI-ASSOCIATED FACTOR1 in Regulation of Gibberellin Homeostasis and Signaling in Arabidopsis. Plant Cell 2014, 26, 2920–2938. [Google Scholar] [CrossRef] [Green Version]
- Fukazawa, J.; Miyamoto, C.; Ando, H.; Mori, K.; Takahashi, Y. DELLA-GAF1 complex is involved in tissue-specific expression and gibberellin feedback regulation of GA20ox1 in Arabidopsis. Plant Mol. Biol. 2021, 107, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Fukazawa, J.; Mori, M.; Watanabe, S.; Miyamoto, C.; Ito, T.; Takahashi, Y. DELLA-GAF1 Complex Is a Main Component in Gibberellin Feedback Regulation of GA20 Oxidase 2. Plant Physiol. 2017, 175, 1395–1406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Kim, J.; Somers, D.E. Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 2013, 110, 761–766. [Google Scholar] [CrossRef] [Green Version]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef] [Green Version]
- Pauwels, L.; Barbero, G.F.; Geerinck, J.; Tilleman, S.; Grunewald, W.; Perez, A.C.; Chico, J.M.; Vanden Bossche, R.; Sewell, J.; Gil, E.; et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 2010, 464, 788-U169. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.J.; Seto, E. The Rpd3/Hda1 family of lysine deacetylases: From bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 2008, 9, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Tu, Y.T.; Chen, C.Y.; Huang, Y.S.; Chang, C.H.; Yen, M.R.; Hsieh, J.A.; Chen, P.Y.; Wu, K. HISTONE DEACETYLASE 15 and MOS4-Associated Complex subunits 3A/3B coregulate intron retention of ABA-responsive genes. Plant Physiol. 2022, 190, 882–897. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, F.; Li, X.; Cao, H.; Ding, M.; Zhang, C.; Zuo, J.; Xu, C.; Xu, J.; Deng, X.; et al. Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1. Nat. Commun. 2016, 7, 13412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Z.M.; Xu, G.; Jing, Y.J.; Tang, W.J.; Lin, R.C. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 2016, 7, 12377. [Google Scholar] [CrossRef]
- Yang, L.W.; Jiang, Z.M.; Jing, Y.J.; Lin, R.C. PIF1 and RVE1 form a transcriptional feedback loop to control light-mediated seed germination in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 1372–1384. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.L.; Sheerin, D.J.; von Roepenack-Lahaye, E.; Stahl, M.; Hiltbrunner, A. The phytochrome interacting proteins ERF55 and ERF58 repress light-induced seed germination in Arabidopsis thaliana. Nat. Commun. 2022, 13, 1656. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, F.; Wang, Y.; Gu, D.; Liu, X. Histone Deacetylase HDA15 Restrains PHYB-Dependent Seed Germination via Directly Repressing GA20ox1/2 Gene Expression. Cells 2022, 11, 3788. https://doi.org/10.3390/cells11233788
Zheng F, Wang Y, Gu D, Liu X. Histone Deacetylase HDA15 Restrains PHYB-Dependent Seed Germination via Directly Repressing GA20ox1/2 Gene Expression. Cells. 2022; 11(23):3788. https://doi.org/10.3390/cells11233788
Chicago/Turabian StyleZheng, Feng, Yahan Wang, Dachuan Gu, and Xuncheng Liu. 2022. "Histone Deacetylase HDA15 Restrains PHYB-Dependent Seed Germination via Directly Repressing GA20ox1/2 Gene Expression" Cells 11, no. 23: 3788. https://doi.org/10.3390/cells11233788
APA StyleZheng, F., Wang, Y., Gu, D., & Liu, X. (2022). Histone Deacetylase HDA15 Restrains PHYB-Dependent Seed Germination via Directly Repressing GA20ox1/2 Gene Expression. Cells, 11(23), 3788. https://doi.org/10.3390/cells11233788






