Modeling Movement Disorders via Generation of hiPSC-Derived Motor Neurons
Abstract
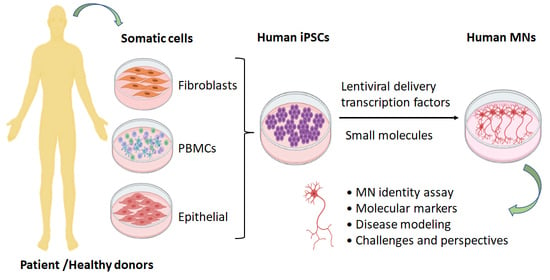
:1. Introduction
2. Generation of hiPSC-Derived MNs
2.1. Generation of NPCs from hiPSCs
2.2. MN Induction via Small Molecules
2.3. MN Induction via Lentiviral Delivery of Transcription Factors
3. Quality Control: Validation of Neuron Identity and Purity
3.1. Markers of Early Induction from hiPSC to NPC
3.2. Markers of MNs at Early Immature Stages
3.3. Markers of MNs at Late Mature Stages
4. Modeling Neurological Diseases Using hiPSC-Derived MNs
5. Future Challenges and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALS | Amyotrophic lateral sclerosis |
| ASCL1 | Achaetescute family bHLH transcription factor 1 |
| ATRA | All-trans retinoic acid |
| bHLH | Basic helix-loop-helix |
| bFGF | Basic fibroblast growth factor |
| BMP | Bone morphogenetic protein |
| BDNF | Brain-derived neurotrophic factor |
| CNS | Central nervous system |
| Wnt | Canonical WNT/β-catenin signaling pathway |
| CNTF | Ciliary neurotrophic factor |
| ChAT | Choline acetyltransferase |
| Cpd E | Compound E |
| CAT7 | Chromatin associated transcript 7 |
| EGF | Epidermal growth factor |
| ESCs | Embryonic stem cells |
| FGF | Fibroblast growth factor |
| FOXP1 | Forkhead box protein 1 |
| FGF2 | Fibroblast growth factor 2 |
| GDNF | Glial cell line-derived neurotrophic factor |
| GSK-3 | Glycogen synthase kinase 3 |
| hiPSCs | Human induced pluripotent stem cells |
| HOXC6 | Homeobox C6 |
| HB9 | Homeodomain |
| HST | Hoechst 33342 |
| ISL1 | Insulin gene enhancer 1 |
| LHX3 | LIM/homeobox 3 |
| LHX1 | Lim homeodomain transcription factors |
| LncRNAs | long non-coding RNAs |
| miRNAs | microRNAs |
| MAP2 | Microtubule Associated Protein 2 |
| MNs | Motor neurons |
| MS1 | Musashi RNA Binding Protein |
| MYT1L | Myelin transcription factor 1 like |
| MNX1 | Motor neuron and pancreas homeobox 1 |
| NPCs | Neural progenitor cells |
| NGN2 | Neurogenin-2 |
| NMJs | Neuromuscular junctions |
| DAPT | N-[N- (3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester |
| NKX2.2 | NK2 homeobox 2 |
| NT3 | Neurotrophin-3 |
| ncRNAs | Non-coding RNAs |
| NEAT1 | Nuclear-enriched abundant transcript 1 |
| NGN1 | Neurogenin 1 |
| NGN2 | Neurogenin 2 |
| NGN3 | Neurogenin 3 |
| NEUROD1 | Neuronal Differentiation 1 |
| NEUROD2 | Neuronal Differentiation 2 |
| Olig2 | Oligodendrocyte transcription factor |
| OC1 | Onecut transcription factors |
| PSA-NCAM | Polysialylated-neural cell adhesion molecule |
| POU5F1 | POU class 5 homeobox 1 |
| PHOX2A | Paired like homeobox 2A |
| POU3F2 | POU class 3 homeobox 2 |
| Pax6 | Paired box protein 6 |
| PUR | Purmorphamine |
| qPCR | Quantitative PCR |
| RA | Retinoic acid signaling pathway |
| ROCK | Rho-associated coiled-coil containing protein kinase |
| Shh | Sonic Hedgehog signaling pathway |
| Sox2 | Sex determining region Y-box 2 |
| SAG | Smoothened agonist |
| SOX1 | SRY-Box Transcription Factor 1 |
| SOX3 | SRY-Box Transcription Factor |
| SOX11 | SRY-box transcription factor 11 |
| Synapsins | Regulation of neurotransmitter release at synapses |
| TGFβ | Transforming growth factor-β |
| TUBB3 | Tubulin Beta 3 Class III |
| TBX20 | T-Box Transcription Factor 20 |
| UTR | 3′Untranslated region |
| VPA | Valproic Acid |
| VAChT | Vesicular acetylcholine transporter |
| WPI | Weeks post viral infection |
| ALS | Amyotrophic lateral sclerosis |
| APCDD1 | Adenomatosis polyposis downregulated |
| 1ASCL1 | Achaetescute family bHLH transcription factor 1 |
| ATRA | All-trans retinoic acidbHLH, Basic helix-loop-helix |
| bFGF | Basic fibroblast growth factor |
| BMP | Bone morphogenetic protein |
| BME | β-mercaptoethanol |
| BDNF | Brain-derived neurotrophic factor |
| CNS | Central nervous system |
| Wnt | Canonical WNT/β-catenin signaling pathway |
| CNTF | Ciliary neurotrophic factor |
| ChAT | Choline acetyltransferase |
| Cpd E | Compound E |
| CAT7 | Chromatin associated transcript 7 |
| Dl1 | Delta-like 1 |
| Dl4 | Delta-like 4 |
| DMH1 | a bone morphogenetic protein (BMP) inhibitor |
| EGF | Epidermal growth factor |
| ESCs | Embryonic stem cells |
| FGF | Fibroblast growth factor |
| FOXP1 | Forkhead box protein 1 |
| FGF2 | Fibroblast growth factor 2 |
| GDNF | Glial cell line-derived neurotrophic factor |
| GSK-3 | Glycogen synthase kinase 3 |
| hiPSCs | Human induced pluripotent stem cells |
| HOXC6 | Homeobox C6 |
| HST | Hoechst 33342 |
| ISL1 | Insulin gene enhancer 1 |
| LHX3 | LIM/homeobox 3 |
| LHX1 | Lim homeodomain transcription factors |
| LncRNAs | Long non-coding RNAs |
| miRNAs | microRNAs |
| MAP2 | Microtubule Associated Protein 2 |
| MEG3 | Maternally expressed gene 3 |
| MNs | Motor neurons |
| MS1 | Musashi RNA Binding Protein |
| MYT1L | Myelin transcription factor 1 like |
| MNX1/HB9 | Motor neuron and pancreas homeobox 1 |
| NPCs | Neural progenitor cells |
| NMJs | Neuromuscular junctions |
| DAPT | N-[N- (3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester |
| NKX2.2 | NK2 homeobox 2 |
| NT3 | Neurotrophin-3 |
| ncRNAs | Non-coding RNAs |
| NEAT1 | Nuclear-enriched abundant transcript 1 |
| NGN1 | Neurogenin 1 |
| NGN2 | Neurogenin 2 |
| NGN3 | Neurogenin 3 |
| NEUROD1 | Neuronal Differentiation 1 |
| NEUROD2 | Neuronal Differentiation 2 |
| Olig2 | Oligodendrocyte transcription factor |
| OC1 | Onecut transcription factors |
| PSA-NCAM | Polysialylated-neural cell adhesion molecule |
| POU5F1 | POU class 5 homeobox 1 |
| PHOX2A | Paired like homeobox 2A |
| POU3F2 | POU class 3 homeobox 2 |
| PAX6 | Paired box protein 6 |
| PUR | Purmorphamine |
| qPCR | Quantitative PCR |
| RA | Retinoic acid signaling pathway |
| ROCK | Rho-associated coiled-coil containing protein kinase |
| Shh | Sonic Hedgehog signaling pathway |
| SOX2 | Sex-determining region Y-box 2 |
| SAG | Smoothened agonist |
| SOX1 | SRY-Box Transcription Factor 1 |
| SOX3 | SRY-Box Transcription Factor |
| SOX11 | SRY-box transcription factor 11 |
| Synapsins | Regulation of neurotransmitter release at synapses |
| SYP | Synaptophysin |
| SMI-32 | Neurofilament |
| TGFβ | Transforming growth factor-β |
| TUBB3 | Tubulin Beta 3 Class III |
| TBX20 | T-Box Transcription Factor 20 |
| UTR | 3′Untranslated region |
| VPA | Valproic Acid |
| VAChT | Vesicular acetylcholine transporter |
| WPI | Weeks post-viral infection |
| Waif1/5T4 | Wnt-activated inhibitory factor 1 |
| ZIC1 | Zic family member 1 |
References
- Schugar, R.; Robbins, P.; Deasy, B. Small molecules in stem cell self-renewal and differentiation. Gene Ther. 2008, 15, 126–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, X.; Vajkoczy, P.; Faust, K. Morphological Abnormalities in the Basal Ganglia of Dystonia Patients. Stereotact. Funct. Neurosurg. 2021, 99, 351–362. [Google Scholar] [CrossRef]
- Balint, B.; Mencacci, N.E.; Valente, E.M.; Pisani, A.; Rothwell, J.; Jankovic, J.; Vidailhet, M.; Bhatia, K.P. Dystonia. Nat. Rev. Dis. Primers 2018, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Augood, S.J.; Keller-McGandy, C.E.; Siriani, A.; Hewett, J.; Ramesh, V.; Sapp, E.; DiFiglia, M.; Breakefield, X.O.; Standaert, D.G. Distribution and ultrastructural localization of torsinA immunoreactivity in the human brain. Brain Res. 2003, 986, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Dickson, D.W. Parkinson’s disease: Experimental models and reality. Acta. Neuropathol. 2018, 135, 13–32. [Google Scholar] [CrossRef]
- Drummond, E.; Wisniewski, T. Alzheimer’s disease: Experimental models and reality. Acta. Neuropathol. 2017, 133, 155–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Genc, B.; Gozutok, O.; Ozdinler, P.H. Complexity of Generating Mouse Models to Study the Upper Motor Neurons: Let Us Shift Focus from Mice to Neurons. Int. J. Mol. Sci. 2019, 20, 3848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, B. Generation of patient-specific motor neurons in modeling movement diseases. Neural. Regen. Res. 2021, 16, 1799. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Shafa, M.; Yang, F.; Fellner, T.; Rao, M.S.; Baghbaderani, B.A. Human-induced pluripotent stem cells manufactured using a current good manufacturing practice-compliant process differentiate into clinically relevant cells from three germ layers. Front. Med. 2018, 5, 69. [Google Scholar] [CrossRef] [Green Version]
- Akter, M.; Cui, H.; Chen, Y.-H.; Ding, B. Generation of two induced pluripotent stem cell lines with heterozygous and homozygous GAG deletion in TOR1A gene from a healthy hiPSC line. Stem Cell Res. 2021, 56, 102536. [Google Scholar] [CrossRef]
- Akter, M.; Cui, H.; Chen, Y.-H.; Ding, B. Generation of gene-corrected isogenic control cell lines from a DYT1 dystonia patient iPSC line carrying a heterozygous GAG mutation in TOR1A gene. Stem Cell Res. 2022, 62, 102807. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Jeong, J.; Choi, D. Small-molecule-mediated reprogramming: A silver lining for regenerative medicine. Exp. Mol. Med. 2020, 52, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Scudellari, M. How iPS cells changed the world. Nature 2016, 534, 310–312. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Inoue, H.; Wu, J.C.; Yamanaka, S. Induced pluripotent stem cell technology: A decade of progress. Nat. Rev. Drug Discov. 2017, 16, 115–130. [Google Scholar] [CrossRef]
- Chang, C.-W.; Lai, Y.-S.; Westin, E.; Khodadadi-Jamayran, A.; Pawlik, K.M.; Lamb, L.S., Jr.; Goldman, F.D.; Townes, T.M. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 2015, 12, 1668–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flynn, R.; Grundmann, A.; Renz, P.; Hänseler, W.; James, W.S.; Cowley, S.A.; Moore, M.D. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp. Hematol. 2015, 43, 838–848.e3. [Google Scholar] [CrossRef] [Green Version]
- Xie, F.; Ye, L.; Chang, J.C.; Beyer, A.I.; Wang, J.; Muench, M.O.; Kan, Y.W. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014, 24, 1526–1533. [Google Scholar] [CrossRef] [Green Version]
- Volpato, V.; Webber, C. Addressing variability in iPSC-derived models of human disease: Guidelines to promote reproducibility. Dis. Model. Mech. 2020, 13, dmm042317. [Google Scholar] [CrossRef]
- Soubannier, V.; Maussion, G.; Chaineau, M.; Sigutova, V.; Rouleau, G.; Durcan, T.M.; Stifani, S. Characterization of human iPSC-derived astrocytes with potential for disease modeling and drug discovery. Neurosci. Lett. 2020, 731, 135028. [Google Scholar] [CrossRef]
- Yamanaka, S. Pluripotent stem cell-based cell therapy-Promise and challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef]
- Sienski, G.; Narayan, P.; Bonner, J.M.; Kory, N.; Boland, S.; Arczewska, A.A.; Ralvenius, W.T.; Akay, L.; Lockshin, E.; He, L. APOE4 disrupts intracellular lipid homeostasis in human iPSC-derived glia. Sci. Transl. Med. 2021, 13, eaaz4564. [Google Scholar] [CrossRef] [PubMed]
- Autar, K.; Guo, X.; Rumsey, J.W.; Long, C.J.; Akanda, N.; Jackson, M.; Narasimhan, N.S.; Caneus, J.; Morgan, D.; Hickman, J.J. A functional hiPSC-cortical neuron differentiation and maturation model and its application to neurological disorders. Stem Cell Rep. 2022, 17, 96–109. [Google Scholar] [CrossRef]
- Gonzalez-Ramos, A.; Waloschková, E.; Mikroulis, A.; Kokaia, Z.; Bengzon, J.; Ledri, M.; Andersson, M.; Kokaia, M. Human stem cell-derived GABAergic neurons functionally integrate into human neuronal networks. Sci. Rep. 2021, 11, 22050. [Google Scholar] [CrossRef]
- Grigor’eva, E.V.; Malankhanova, T.B.; Surumbayeva, A.; Pavlova, S.V.; Minina, J.M.; Kizilova, E.A.; Suldina, L.A.; Morozova, K.N.; Kiseleva, E.; Sorokoumov, E.D. Generation of GABAergic striatal neurons by a novel iPSC differentiation protocol enabling scalability and cryopreservation of progenitor cells. Cytotechnology 2020, 72, 649–663. [Google Scholar] [CrossRef]
- Rakovic, A.; Voß, D.; Vulinovic, F.; Meier, B.; Hellberg, A.-K.; Nau, C.; Klein, C.; Leipold, E. Electrophysiological Properties of Induced Pluripotent Stem Cell-Derived Midbrain Dopaminergic Neurons Correlate With Expression of Tyrosine Hydroxylase. Front. Cell. Neurosci. 2022, 121, 817198. [Google Scholar] [CrossRef]
- Valiulahi, P.; Vidyawan, V.; Puspita, L.; Oh, Y.; Juwono, V.B.; Sittipo, P.; Friedlander, G.; Yahalomi, D.; Sohn, J.-W.; Lee, Y.K. Generation of caudal-type serotonin neurons and hindbrain-fate organoids from hPSCs. Stem Cell Rep. 2021, 16, 1938–1952. [Google Scholar] [CrossRef] [PubMed]
- Akter, M.; Cui, H.; Sepehrimanesh, M.; Hosain, M.A.; Ding, B. Generation of highly pure motor neurons from human induced pluripotent stem cells. STAR Protoc. 2022, 3, 101223. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Akter, M.; Zhang, C.-L. Differential influence of sample sex and neuronal maturation on mRNA and protein transport in induced human neurons. Front. Mol. Neurosci. 2020, 13, 46. [Google Scholar] [CrossRef]
- Ding, B.; Tang, Y.; Ma, S.; Akter, M.; Liu, M.-L.; Zang, T.; Zhang, C.-L. Disease modeling with human neurons reveals LMNB1 dysregulation underlying DYT1 dystonia. J. Neurosci. 2021, 41, 2024–2038. [Google Scholar] [CrossRef]
- Sepehrimanesh, M.; Ding, B. Generation and optimization of highly pure motor neurons from human induced pluripotent stem cells via lentiviral delivery of transcription factors. Am. J. Physiol. Cell Physiol. 2020, 319, C771–C780. [Google Scholar] [CrossRef]
- Voulgaris, D.; Nikolakopoulou, P.; Herland, A. Generation of Human iPSC-Derived Astrocytes with a mature star-shaped phenotype for CNS modeling. Stem Cell Rev. Rep. 2022, 18, 2494–2512. [Google Scholar] [CrossRef]
- Xu, R.; Li, X.; Boreland, A.J.; Posyton, A.; Kwan, K.; Hart, R.P.; Jiang, P. Human iPSC-derived mature microglia retain their identity and functionally integrate in the chimeric mouse brain. Nat. Commun. 2020, 11, 1577. [Google Scholar] [CrossRef] [Green Version]
- Hasselmann, J.; Blurton-Jones, M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 2020, 68, 721–739. [Google Scholar] [CrossRef]
- Ho, R.; Workman, M.J.; Mathkar, P.; Wu, K.; Kim, K.J.; O’Rourke, J.G.; Kellogg, M.; Montel, V.; Banuelos, M.G.; Arogundade, O.A.; et al. Cross-Comparison of Human iPSC Motor Neuron Models of Familial and Sporadic ALS Reveals Early and Convergent Transcriptomic Disease Signatures. Cell Syst. 2021, 12, 159–175.e9. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Zhu, Y.; Hsiao-Nakamoto, J.; Tang, X.; Dugas, J.C.; Moscovitch-Lopatin, M.; Glass, J.D.; Brown, R.H., Jr.; Ladha, S.S.; Lacomis, D. Longitudinal biomarkers in amyotrophic lateral sclerosis. Ann. Clin. Transl. Neurol. 2020, 7, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Ferraiuolo, L.; Maragakis, N.J. Mini-Review: Induced pluripotent stem cells and the search for new cell-specific ALS therapeutic targets. Neurosci. Lett. 2021, 755, 135911. [Google Scholar] [CrossRef] [PubMed]
- Ding, B. Novel insights into the pathogenesis of DYT1 dystonia from induced patient-derived neurons. Neural. Regen. Res. 2022, 17, 561. [Google Scholar] [CrossRef]
- Gilbert, S.F. Developmental Biology; Sinauer Associates, Inc.: Sunderland, MA, USA, 2010. [Google Scholar]
- Leto, K.; Arancillo, M.; Becker, E.B.; Buffo, A.; Chiang, C.; Ding, B.; Dobyns, W.B.; Dusart, I.; Haldipur, P.; Hatten, M.E.; et al. Consensus Paper: Cerebellar Development. Cerebellum 2016, 15, 789–828. [Google Scholar] [CrossRef]
- Briscoe, J.; Ericson, J. Specification of neuronal fates in the ventral neural tube. Curr. Opin. Neurobiol. 2001, 11, 43–49. [Google Scholar] [CrossRef]
- Ericson, J.; Briscoe, J.; Rashbass, P.; Van Heyningen, V.; Jessell, T. Graded sonic hedgehog signaling and the specification of cell fate in the ventral neural tube. In cold Spring Harbor symposia on quantitative biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1997; pp. 451–466. [Google Scholar]
- Muhr, J.; Andersson, E.; Persson, M.; Jessell, T.M.; Ericson, J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell 2001, 104, 861–873. [Google Scholar] [CrossRef]
- Belgacem, Y.H.; Hamilton, A.M.; Shim, S.; Spencer, K.A.; Borodinsky, L.N. The many hats of sonic hedgehog signaling in nervous system development and disease. J. Dev. Biol. 2016, 4, 35. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.-G.; Spassky, N.; Romaguera-Ros, M.; Garcia-Verdugo, J.-M.; Aguilar, A.; Schneider-Maunoury, S.; Alvarez-Buylla, A. Hedgehog signaling and primary cilia are required for the formation of adult neural stem cells. Nat. Neurosci. 2008, 11, 277–284. [Google Scholar] [CrossRef]
- Lai, K.; Kaspar, B.K.; Gage, F.H.; Schaffer, D.V. Sonic hedgehog regulates adult neural progenitor proliferation in vitro and in vivo. Nat. Neurosci. 2003, 6, 21–27. [Google Scholar] [CrossRef]
- Wechsler-Reya, R.J.; Scott, M.P. Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 1999, 22, 103–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chambers, S.M.; Qi, Y.; Mica, Y.; Lee, G.; Zhang, X.-J.; Niu, L.; Bilsland, J.; Cao, L.; Stevens, E.; Whiting, P. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 2012, 30, 715–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.; Wu, T.Y.; Brinker, A.; Peters, E.C.; Hur, W.; Gray, N.S.; Schultz, P.G. Synthetic small molecules that control stem cell fate. Proc. Natl. Acad. Sci. USA 2003, 100, 7632–7637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragdon, B.; Moseychuk, O.; Saldanha, S.; King, D.; Julian, J.; Nohe, A. Bone morphogenetic proteins: A critical review. Cell. Signal. 2011, 23, 609–620. [Google Scholar] [CrossRef]
- Dhara, S.K.; Stice, S.L. Neural differentiation of human embryonic stem cells. J. Cell. Biochem. 2008, 105, 633–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dottori, M.; Pera, M.F. Neural differentiation of human embryonic stem cells. In Neural Stem Cells; Springer: Berlin, Germany, 2008; pp. 19–30. [Google Scholar]
- Gaspard, N.; Vanderhaeghen, P. Mechanisms of neural specification from embryonic stem cells. Curr. Opin. Neurobiol. 2010, 20, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J.; Brivanlou, A.H. Proposal of a model of mammalian neural induction. Dev. Biol. 2007, 308, 247–256. [Google Scholar] [CrossRef] [Green Version]
- Pankratz, M.T.; Li, X.-J.; LaVaute, T.M.; Lyons, E.A.; Chen, X.; Zhang, S.-C. Directed neural differentiation of human embryonic stem cells via an obligated primitive anterior stage. Stem Cells 2007, 25, 1511–1520. [Google Scholar] [CrossRef] [Green Version]
- Kempermann, G.; Song, H.; Gage, F.H. Neurogenesis in the adult hippocampus. Cold Spring Harb. Perspect. Biol. 2015, 7, a018812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koziol, L.F.; Budding, D.; Andreasen, N.; D’Arrigo, S.; Bulgheroni, S.; Imamizu, H.; Ito, M.; Manto, M.; Marvel, C.; Parker, K. Consensus paper: The cerebellum’s role in movement and cognition. Cerebellum 2014, 13, 151–177. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohgushi, M.; Matsumura, M.; Eiraku, M.; Murakami, K.; Aramaki, T.; Nishiyama, A.; Muguruma, K.; Nakano, T.; Suga, H.; Ueno, M. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 2010, 7, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Wataya, T.; Ando, S.; Muguruma, K.; Ikeda, H.; Watanabe, K.; Eiraku, M.; Kawada, M.; Takahashi, J.; Hashimoto, N.; Sasai, Y. Minimization of exogenous signals in ES cell culture induces rostral hypothalamic differentiation. Proc. Natl. Acad. Sci. USA 2008, 105, 11796–11801. [Google Scholar] [CrossRef] [Green Version]
- Fedorova, V.; Vanova, T.; Elrefae, L.; Pospisil, J.; Petrasova, M.; Kolajova, V.; Hudacova, Z.; Baniariova, J.; Barak, M.; Peskova, L. Differentiation of neural rosettes from human pluripotent stem cells in vitro is sequentially regulated on a molecular level and accomplished by the mechanism reminiscent of secondary neurulation. Stem Cell Res. 2019, 40, 101563. [Google Scholar] [CrossRef]
- Patani, R.; Compston, A.; Puddifoot, C.A.; Wyllie, D.J.; Hardingham, G.E.; Allen, N.D.; Chandran, S. Activin/Nodal inhibition alone accelerates highly efficient neural conversion from human embryonic stem cells and imposes a caudal positional identity. PLoS ONE 2009, 4, e7327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amoroso, M.W.; Croft, G.F.; Williams, D.J.; O’Keeffe, S.; Carrasco, M.A.; Davis, A.R.; Roybon, L.; Oakley, D.H.; Maniatis, T.; Henderson, C.E. Accelerated high-yield generation of limb-innervating motor neurons from human stem cells. J. Neurosci. 2013, 33, 574–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.W.; O’Doherty, J.P.; Shimojo, S. Neural computations mediating one-shot learning in the human brain. PLoS Biol. 2015, 13, e1002137. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Mao, X.; Zhou, X.; Su, Y.; Zhou, X.; Shi, K.; Zhao, S. An optimized method for neuronal differentiation of embryonic stem cells in vitro. J. Neurosci. Methods 2020, 330, 108486. [Google Scholar] [CrossRef] [PubMed]
- Maury, Y.; Côme, J.; Piskorowski, R.A.; Salah-Mohellibi, N.; Chevaleyre, V.; Peschanski, M.; Martinat, C.; Nedelec, S. Combinatorial analysis of developmental cues efficiently converts human pluripotent stem cells into multiple neuronal subtypes. Nat. Biotechnol. 2015, 33, 89–96. [Google Scholar] [CrossRef]
- Salimi, A.; Nadri, S.; Ghollasi, M.; Khajeh, K.; Soleimani, M. Comparison of different protocols for neural differentiation of human induced pluripotent stem cells. Mol. Biol. Rep. 2014, 41, 1713–1721. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, P.; Glatza, M.; Hemmer, K.; Tsytsyura, Y.; Thiel, C.S.; Höing, S.; Moritz, S.; Parga, J.A.; Wagner, L.; Bruder, J.M.; et al. Correction: Derivation and Expansion Using Only Small Molecules of Human Neural Progenitors for Neurodegenerative Disease Modeling. PLoS ONE 2013, 8, e59252. [Google Scholar] [CrossRef]
- Smith, K.A.; Chocron, S.; von der Hardt, S.; de Pater, E.; Soufan, A.; Bussmann, J.; Schulte-Merker, S.; Hammerschmidt, M.; Bakkers, J. Rotation and Asymmetric Development of the Zebrafish Heart Requires Directed Migration of Cardiac Progenitor Cells. Dev. Cell 2008, 14, 287–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osumi, N.; Shinohara, H.; Numayama-Tsuruta, K.; Maekawa, M. Concise Review: Pax6 Transcription Factor Contributes to both Embryonic and Adult Neurogenesis as a Multifunctional Regulator. Stem Cells 2008, 26, 1663–1672. [Google Scholar] [CrossRef]
- Zhang, M.; Ngo, J.; Pirozzi, F.; Sun, Y.-P.; Wynshaw-Boris, A. Highly efficient methods to obtain homogeneous dorsal neural progenitor cells from human and mouse embryonic stem cells and induced pluripotent stem cells. Stem Cell Res. Ther. 2018, 9, 67. [Google Scholar] [CrossRef]
- Jiménez-Vaca, A.L.; Benitez-King, G.; Ruiz, V.; Ramírez-Rodríguez, G.B.; Hernández-de la Cruz, B.; Salamanca-Gómez, F.A.; González-Márquez, H.; Ramírez-Sánchez, I.; Ortíz-López, L.; Vélez-del Valle, C. Exfoliated human olfactory neuroepithelium: A source of neural progenitor cells. Mol. Neurobiol. 2018, 55, 2516–2523. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, Z.; Wen, J.; Tang, F.; Liao, B.; Jing, N.; Lai, D.; Jin, Y. SOX1 is required for the specification of rostral hindbrain neural progenitor cells from human embryonic stem cells. Iscience 2020, 23, 101475. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Drakulic, D.; Lazic, A.; Ninkovic, D.S.; Schwirtlich, M.; Mojsin, M. SOX transcription factors as important regulators of neuronal and glial differentiation during nervous system development and adult neurogenesis. Front. Mol. Neurosci. 2021, 14, 654031. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Sepehrimanesh, M. Nucleocytoplasmic transport: Regulatory mechanisms and the implications in neurodegeneration. Int. J. Mol. Sci. 2021, 22, 4165. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Li, C.; Zhang, M.; Wang, H.; Xie, Y.; Tang, Y. A Step-by-Step Refined Strategy for Highly Efficient Generation of Neural Progenitors and Motor Neurons from Human Pluripotent Stem Cells. Cells 2021, 10, 3087. [Google Scholar] [CrossRef]
- Rolletschek, A.; Wobus, A.M. Induced human pluripotent stem cells: Promises and open questions. Biol. Chem. 2009, 390, 845–849. [Google Scholar] [CrossRef]
- Trawczynski, M.; Liu, G.; David, B.T.; Fessler, R.G. Restoring motor neurons in spinal cord injury with induced pluripotent stem cells. Front. Cell. Neurosci. 2019, 13, 369. [Google Scholar] [CrossRef] [Green Version]
- Valizadeh-Arshad, Z.; Shahbazi, E.; Hashemizadeh, S.; Moradmand, A.; Jangkhah, M.; Kiani, S. In vitro differentiation of neural-like cells from human embryonic stem cells by a combination of dorsomorphin, XAV939, and A8301. Cell J. 2018, 19, 545. [Google Scholar]
- Kamishibahara, Y.; Kawaguchi, H.; Shimizu, N. Rho kinase inhibitor Y-27632 promotes neuronal differentiation in mouse embryonic stem cells via phosphatidylinositol 3-kinase. Neurosci. Lett. 2016, 615, 44–49. [Google Scholar] [CrossRef]
- Dworkin, S.; Mantamadiotis, T. Targeting CREB signalling in neurogenesis. Expert Opin. Ther. Targets 2010, 14, 869–879. [Google Scholar] [CrossRef]
- Du, Z.-W.; Chen, H.; Liu, H.; Lu, J.; Qian, K.; Huang, C.-L.; Zhong, X.; Fan, F.; Zhang, S.-C. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 2015, 6, 6626. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, J.; Noakes, P.G.; Bellingham, M.C. The role of altered BDNF/TrkB signaling in amyotrophic lateral sclerosis. Front. Cell. Neurosci. 2019, 13, 368. [Google Scholar] [CrossRef] [PubMed]
- Cintrón-Colón, A.F.; Almeida-Alves, G.; Boynton, A.M.; Spitsbergen, J.M. GDNF synthesis, signaling, and retrograde transport in motor neurons. Cell Tissue Res. 2020, 382, 47–56. [Google Scholar] [CrossRef]
- Zeng, S.; Zhao, X.; Zhang, L.; Pathak, J.L.; Huang, W.; Li, Y.; Guan, H.; Zhao, W.; Ge, L.; Shu, Y. Effect of ciliary neurotrophic factor on neural differentiation of stem cells of human exfoliated deciduous teeth. J. Biol. Eng. 2020, 14, 29. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Ueno, M.; Kamiya, D.; Nishiyama, A.; Matsumura, M.; Wataya, T.; Takahashi, J.B.; Nishikawa, S.; Nishikawa, S.-i.; Muguruma, K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007, 25, 681–686. [Google Scholar] [CrossRef]
- Diez del Corral, R.; Morales, A.V. The multiple roles of FGF signaling in the developing spinal cord. Front. Cell Dev. Biol. 2017, 5, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scalabrino, G. Epidermal growth factor in the CNS: A beguiling journey from integrated cell biology to multiple sclerosis. an extensive translational overview. Cell. Mol. Neurobiol. 2020, 42, 891–916. [Google Scholar] [CrossRef]
- Colombres, M.; Henríquez, J.P.; Reig, G.F.; Scheu, J.; Calderón, R.; Alvarez, A.; Brandan, E.; Inestrosa, N.C. Heparin activates Wnt signaling for neuronal morphogenesis. J. Cell. Physiol. 2008, 216, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Qi, Y.; Sun, Z. The role of sonic hedgehog pathway in the development of the central nervous system and aging-related neurodegenerative diseases. Front. Mol. Biosci. 2021, 8, 711710. [Google Scholar] [CrossRef]
- Tomishima, M. Neural Induction–Dual SMAD Inhibition; Steam Book: Holland Landing, ON, Canada, 2013. [Google Scholar]
- Mitre, M.; Mariga, A.; Chao, M.V. Neurotrophin signalling: Novel insights into mechanisms and pathophysiology. Clin. Sci. 2017, 131, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Westphal, M.; Panza, P.; Kastenhuber, E.; Wehrle, J.; Driever, W. Wnt/β-catenin signaling promotes neurogenesis in the diencephalospinal dopaminergic system of embryonic zebrafish. Sci. Rep. 2022, 12, 1030. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shushan, E.; Feldman, E.; Reubinoff, B.E. Notch signaling regulates motor neuron differentiation of human embryonic stem cells. Stem Cells 2015, 33, 403–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allison, R.L.; Welby, E.; Khayrullina, G.; Burnett, B.G.; Ebert, A.D. Viral mediated knockdown of GATA6 in SMA iPSC-derived astrocytes prevents motor neuron loss and microglial activation. Glia 2022, 70, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Cutarelli, A.; Martínez-Rojas, V.A.; Tata, A.; Battistella, I.; Rossi, D.; Arosio, D.; Musio, C.; Conti, L. A monolayer system for the efficient generation of motor neuron progenitors and functional motor neurons from human pluripotent stem cells. Cells 2021, 10, 1127. [Google Scholar] [CrossRef] [PubMed]
- Solomon, E.; Davis-Anderson, K.; Hovde, B.; Micheva-Viteva, S.; Harris, J.F.; Twary, S.; Iyer, R. Global transcriptome profile of the developmental principles of in vitro iPSC-to-motor neuron differentiation. BMC Mol. Cell Biol. 2021, 22, 13. [Google Scholar] [CrossRef]
- Bianchi, F.; Malboubi, M.; Li, Y.; George, J.H.; Jerusalem, A.; Szele, F.; Thompson, M.S.; Ye, H. Rapid and efficient differentiation of functional motor neurons from human iPSC for neural injury modelling. Stem Cell Res. 2018, 32, 126–134. [Google Scholar] [CrossRef]
- Kiskinis, E.; Kralj, J.M.; Zou, P.; Weinstein, E.N.; Zhang, H.; Tsioras, K.; Wiskow, O.; Ortega, J.A.; Eggan, K.; Cohen, A.E. All-optical electrophysiology for high-throughput functional characterization of a human iPSC-derived motor neuron model of ALS. Stem Cell Rep. 2018, 10, 1991–2004. [Google Scholar] [CrossRef]
- Fujimori, K.; Ishikawa, M.; Otomo, A.; Atsuta, N.; Nakamura, R.; Akiyama, T.; Hadano, S.; Aoki, M.; Saya, H.; Sobue, G. Modeling sporadic ALS in iPSC-derived motor neurons identifies a potential therapeutic agent. Nat. Med. 2018, 24, 1579–1589. [Google Scholar] [CrossRef]
- Goparaju, S.K.; Kohda, K.; Ibata, K.; Soma, A.; Nakatake, Y.; Akiyama, T.; Wakabayashi, S.; Matsushita, M.; Sakota, M.; Kimura, H. Rapid differentiation of human pluripotent stem cells into functional neurons by mRNAs encoding transcription factors. Sci. Rep. 2017, 7, 42367. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Naujock, M.; Fumagalli, L.; Vandoorne, T.; Baatsen, P.; Boon, R.; Ordovás, L.; Patel, A.; Welters, M.; Vanwelden, T. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat. Commun. 2017, 8, 861. [Google Scholar] [CrossRef] [Green Version]
- Ichiyanagi, N.; Fujimori, K.; Yano, M.; Ishihara-Fujisaki, C.; Sone, T.; Akiyama, T.; Okada, Y.; Akamatsu, W.; Matsumoto, T.; Ishikawa, M. Establishment of in vitro FUS-associated familial amyotrophic lateral sclerosis model using human induced pluripotent stem cells. Stem Cell Rep. 2016, 6, 496–510. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Qian, K.; Du, Z.; Cao, J.; Petersen, A.; Liu, H.; Blackbourn IV, L.W.; Huang, C.-L.; Errigo, A.; Yin, Y. Modeling ALS with iPSCs reveals that mutant SOD1 misregulates neurofilament balance in motor neurons. Cell Stem Cell 2014, 14, 796–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, Q.; Li, D.; Louis, K.R.; Li, X.; Yang, H.; Sun, Q.; Crandall, S.R.; Tsang, S.; Zhou, J.; Cox, C.L. High-efficiency motor neuron differentiation from human pluripotent stem cells and the function of Islet-1. Nat. Commun. 2014, 5, 3449. [Google Scholar] [CrossRef] [Green Version]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Briscoe, J.; Pierani, A.; Jessell, T.M.; Ericson, J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell 2000, 101, 435–445. [Google Scholar] [CrossRef] [Green Version]
- Dasen, J.S.; De Camilli, A.; Wang, B.; Tucker, P.W.; Jessell, T.M. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell 2008, 134, 304–316. [Google Scholar] [CrossRef] [Green Version]
- Dasen, J.S.; Liu, J.-P.; Jessell, T.M. Motor neuron columnar fate imposed by sequential phases of Hox-c activity. Nature 2003, 425, 926–933. [Google Scholar] [CrossRef]
- Dasen, J.S.; Tice, B.C.; Brenner-Morton, S.; Jessell, T.M. A Hox regulatory network establishes motor neuron pool identity and target-muscle connectivity. Cell 2005, 123, 477–491. [Google Scholar] [CrossRef]
- Peljto, M.; Dasen, J.S.; Mazzoni, E.O.; Jessell, T.M.; Wichterle, H. Functional diversity of ESC-derived motor neuron subtypes revealed through intraspinal transplantation. Cell Stem Cell 2010, 7, 355–366. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.-H.; Cargnin, F.; Kim, Y.; Lee, B.; Kwon, R.-J.; Nam, H.; Shen, R.; Barnes, A.P.; Lee, J.W.; Lee, S. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 2014, 10, e1004280. [Google Scholar] [CrossRef] [Green Version]
- Palmesino, E.; Rousso, D.L.; Kao, T.-J.; Klar, A.; Laufer, E.; Uemura, O.; Okamoto, H.; Novitch, B.G.; Kania, A. Foxp1 and lhx1 coordinate motor neuron migration with axon trajectory choice by gating Reelin signalling. PLoS Biol. 2010, 8, e1000446. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Liu, H.; Zhang, D.; Xu, G.; Liu, J.; Evans, S.M.; Pan, J.; Cui, S. LIM homeodomain transcription factor Isl1 affects urethral epithelium differentiation and apoptosis via Shh. Cell Death Dis. 2019, 10, 713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novitch, B.G.; Chen, A.I.; Jessell, T.M. Coordinate regulation of motor neuron subtype identity and pan-neuronal properties by the bHLH repressor Olig2. Neuron 2001, 31, 773–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novitch, B.G.; Wichterle, H.; Jessell, T.M.; Sockanathan, S. A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 2003, 40, 81–95. [Google Scholar] [CrossRef] [Green Version]
- Vallstedt, A.; Muhr, J.; Pattyn, A.; Pierani, A.; Mendelsohn, M.; Sander, M.; Jessell, T.M.; Ericson, J. Different levels of repressor activity assign redundant and specific roles to Nkx6 genes in motor neuron and interneuron specification. Neuron 2001, 31, 743–755. [Google Scholar] [CrossRef] [Green Version]
- Lu, Q.R.; Sun, T.; Zhu, Z.; Ma, N.; Garcia, M.; Stiles, C.D.; Rowitch, D.H. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 2002, 109, 75–86. [Google Scholar] [CrossRef] [Green Version]
- Sagner, A.; Gaber, Z.B.; Delile, J.; Kong, J.H.; Rousso, D.L.; Pearson, C.A.; Weicksel, S.E.; Melchionda, M.; Mousavy Gharavy, S.N.; Briscoe, J. Olig2 and Hes regulatory dynamics during motor neuron differentiation revealed by single cell transcriptomics. PLoS Biol. 2018, 16, e2003127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zannino, D.A.; Appel, B. Olig2+ precursors produce abducens motor neurons and oligodendrocytes in the zebrafish hindbrain. J. Neurosci. 2009, 29, 2322–2333. [Google Scholar] [CrossRef] [Green Version]
- Arber, S.; Han, B.; Mendelsohn, M.; Smith, M.; Jessell, T.M.; Sockanathan, S. Requirement for the homeobox gene Hb9 in the consolidation of motor neuron identity. Neuron 1999, 23, 659–674. [Google Scholar] [CrossRef] [Green Version]
- Odden, J.P.; Holbrook, S.; Doe, C.Q. DrosophilaHB9 is expressed in a subset of motoneurons and interneurons, where it regulates gene expression and axon pathfinding. J. Neurosci. 2002, 22, 9143–9149. [Google Scholar] [CrossRef] [Green Version]
- Stifani, N. Motor neurons and the generation of spinal motor neuron diversity. Front. Cell. Neurosci. 2014, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.; Dobner, P.R.; Mullikin-Kilpatrick, D.; Wang, W.; Zhu, H.; Chow, C.W.; Cave, J.W.; Gronostajski, R.M.; Kilpatrick, D.L. BDNF activates an NFI-dependent neurodevelopmental timing program by sequestering NFATc4. Mol. Biol. Cell. 2018, 29, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Frank-Kamenetsky, M.; Zhang, X.M.; Bottega, S.; Guicherit, O.; Wichterle, H.; Dudek, H.; Bumcrot, D.; Wang, F.Y.; Jones, S.; Shulok, J. Small-molecule modulators of Hedgehog signaling: Identification and characterization of Smoothened agonists and antagonists. J. Biol. 2002, 1, 10. [Google Scholar] [CrossRef] [Green Version]
- Wichterle, H.; Lieberam, I.; Porter, J.A.; Jessell, T.M. Directed differentiation of embryonic stem cells into motor neurons. Cell 2002, 110, 385–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wada, T.; Honda, M.; Minami, I.; Tooi, N.; Amagai, Y.; Nakatsuji, N.; Aiba, K. Highly efficient differentiation and enrichment of spinal motor neurons derived from human and monkey embryonic stem cells. PLoS ONE 2009, 4, e6722. [Google Scholar] [CrossRef] [Green Version]
- Bhinge, A.; Namboori, S.C.; Bithell, A.; Soldati, C.; Buckley, N.J.; Stanton, L.W. MiR-375 is essential for human spinal motor neuron development and may be involved in motor neuron degeneration. Stem Cells 2016, 34, 124–134. [Google Scholar] [CrossRef]
- Dajas-Bailador, F.; Bonev, B.; Garcez, P.; Stanley, P.; Guillemot, F.; Papalopulu, N. microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nat. Neurosci. 2012, 15, 697. [Google Scholar] [CrossRef]
- Haramati, S.; Chapnik, E.; Sztainberg, Y.; Eilam, R.; Zwang, R.; Gershoni, N.; McGlinn, E.; Heiser, P.W.; Wills, A.-M.; Wirguin, I. miRNA malfunction causes spinal motor neuron disease. Proc. Natl. Acad. Sci. USA 2010, 107, 13111–13116. [Google Scholar] [CrossRef] [Green Version]
- Luxenhofer, G.; Helmbrecht, M.S.; Langhoff, J.; Giusti, S.A.; Refojo, D.; Huber, A.B. MicroRNA-9 promotes the switch from early-born to late-born motor neuron populations by regulating Onecut transcription factor expression. Dev. Biol. 2014, 386, 358–370. [Google Scholar] [CrossRef] [Green Version]
- Otaegi, G.; Pollock, A.; Hong, J.; Sun, T. MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords. J. Neurosci. 2011, 31, 809–818. [Google Scholar] [CrossRef] [Green Version]
- Otaegi, G.; Pollock, A.; Sun, T. An optimized sponge for microRNA miR-9 affects spinal motor neuron development in vivo. Front. Neurosci. 2012, 5, 146. [Google Scholar] [CrossRef] [PubMed]
- Amin, N.D.; Bai, G.; Klug, J.R.; Bonanomi, D.; Pankratz, M.T.; Gifford, W.D.; Hinckley, C.A.; Sternfeld, M.J.; Driscoll, S.P.; Dominguez, B. Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 2015, 350, 1525–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thiebes, K.P.; Nam, H.; Cambronne, X.A.; Shen, R.; Glasgow, S.M.; Cho, H.-H.; Kwon, J.-S.; Goodman, R.H.; Lee, J.W.; Lee, S. miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1–Lhx3. Nat. Commun. 2015, 6, 7718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, Y.-T.; Lu, Y.-L.; Peng, K.-C.; Yen, Y.-P.; Chang, M.; Li, J.; Jung, H.; Thams, S.; Huang, Y.-P.; Hung, J.-H. Mir-17∼ 92 governs motor neuron subtype survival by mediating nuclear PTEN. Cell Rep. 2015, 11, 1305–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, Y.-T.; Peng, K.-C.; Chen, Y.-C.; Yen, Y.-P.; Chang, M.; Thams, S.; Chen, J.-A. Mir-17∼ 92 confers motor neuron subtype differential resistance to ALS-associated degeneration. Cell Stem Cell 2019, 25, 193–209.e7. [Google Scholar] [CrossRef]
- Li, L.; Luo, Z. Dysregulated miR-27a-3p promotes nasopharyngeal carcinoma cell proliferation and migration by targeting Mapk10. Oncol. Rep. 2017, 37, 2679–2687. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wei, Q.; Chen, X.; Li, C.; Cao, B.; Ou, R.; Hadano, S.; Shang, H.-F. Aberration of miRNAs expression in leukocytes from sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 2016, 9, 69. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Chen, Y.; Chen, X.; Wei, Q.; Ou, R.; Gu, X.; Cao, B.; Shang, H. MicroRNA-183-5p is stress-inducible and protects neurons against cell death in amyotrophic lateral sclerosis. J. Cell. Mol. Med. 2020, 24, 8614–8622. [Google Scholar] [CrossRef]
- Asli, N.S.; Kessel, M. Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8. Dev. Biol. 2010, 344, 857–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Santis, R.; Garone, M.G.; Pagani, F.; de Turris, V.; Di Angelantonio, S.; Rosa, A. Direct conversion of human pluripotent stem cells into cranial motor neurons using a piggyBac vector. Stem Cell Res. 2018, 29, 189–196. [Google Scholar] [CrossRef]
- Rohm, M.; May, C.; Marcus, K.; Steinbach, S.; Theis, V.; Theiß, C.; Matschke, V. The microRNA miR-375-3p and the tumor suppressor NDRG2 are involved in sporadic amyotrophic lateral sclerosis. Cell Physiol. Biochem. 2019, 52, 1412–1426. [Google Scholar] [PubMed]
- Carthew, R.W.; Sontheimer, E.J. Origins and mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- DING, B. Gene expression in maturing neurons: Regulatory mechanisms and related neurodevelopmental disorders. Acta Physiol. Sin. 2015, 67, 113–133. [Google Scholar]
- Williams, A.H.; Valdez, G.; Moresi, V.; Qi, X.; McAnally, J.; Elliott, J.L.; Bassel-Duby, R.; Sanes, J.R.; Olson, E.N. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 2009, 326, 1549–1554. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.-A.; Huang, Y.-P.; Mazzoni, E.O.; Tan, G.C.; Zavadil, J.; Wichterle, H. Mir-17-3p controls spinal neural progenitor patterning by regulating Olig2/Irx3 cross-repressive loop. Neuron 2011, 69, 721–735. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Pfaff, S.L.; Gage, F.H. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007, 21, 531–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, J.W.; Lee, S.-K. The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 2007, 21, 744–749. [Google Scholar] [CrossRef] [Green Version]
- Visvanathan, J.; Lee, S.; Lee, B.; Lee, S.-K. MIR-124 antagonizes the anti-neural rest/scp1 pathway during embryonic development. Dev. Biol. 2008, 2, 574. [Google Scholar] [CrossRef] [Green Version]
- Amin, N.D.; Senturk, G.; Costaguta, G.; Driscoll, S.; O’Leary, B.; Bonanomi, D.; Pfaff, S.L. A hidden threshold in motor neuron gene networks revealed by modulation of miR-218 dose. Neuron 2021, 109, 3252–3267.e6. [Google Scholar] [CrossRef]
- Amin, N.D.; Senturk, G.; Hayashi, M.; Driscoll, S.P.; Pfaff, S.L. Detecting microRNA-mediated gene regulatory effects in murine neuronal subpopulations. STAR Protoc. 2022, 3, 101130. [Google Scholar] [CrossRef] [PubMed]
- Francius, C.; Clotman, F. Generating spinal motor neuron diversity: A long quest for neuronal identity. Cell. Mol. Life Sci. 2014, 71, 813–829. [Google Scholar] [CrossRef] [PubMed]
- Toch, M.; Harris, A.; Schakman, O.; Kondratskaya, E.; Boulland, J.-L.; Dauguet, N.; Debrulle, S.; Baudouin, C.; Hidalgo-Figueroa, M.; Mu, X. Onecut-dependent Nkx6. 2 transcription factor expression is required for proper formation and activity of spinal locomotor circuits. Sci. Rep. 2020, 10, 996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shu, P.; Wu, C.; Liu, W.; Ruan, X.; Liu, C.; Hou, L.; Zeng, Y.; Fu, H.; Wang, M.; Chen, P. The spatiotemporal expression pattern of microRNAs in the developing mouse nervous system. J. Biol. Chem. 2019, 294, 3444–3453. [Google Scholar] [CrossRef] [PubMed]
- Emde, A.; Eitan, C.; Liou, L.L.; Libby, R.T.; Rivkin, N.; Magen, I.; Reichenstein, I.; Oppenheim, H.; Eilam, R.; Silvestroni, A. Dysregulated mi RNA biogenesis downstream of cellular stress and ALS-causing mutations: A new mechanism for ALS. EMBO J. 2015, 34, 2633–2651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoye, M.L.; Koval, E.D.; Wegener, A.J.; Hyman, T.S.; Yang, C.; O’Brien, D.R.; Miller, R.L.; Cole, T.; Schoch, K.M.; Shen, T. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J. Neurosci. 2017, 37, 5574–5586. [Google Scholar] [CrossRef] [Green Version]
- Church, V.A.; Yoo, A.S. MiR-218 steps down to a threshold of motor impairment. Neuron 2021, 109, 3233–3235. [Google Scholar] [CrossRef]
- Emery, B.; Lu, Q.R. Transcriptional and epigenetic regulation of oligodendrocyte development and myelination in the central nervous system. Cold Spring Harb. Perspect. Biol. 2015, 7, a020461. [Google Scholar] [CrossRef] [Green Version]
- Kye, M.J.; Goncalves, I.d.C.G. The role of miRNA in motor neuron disease. Front. Cell. Neurosci. 2014, 8, 15. [Google Scholar] [CrossRef] [Green Version]
- Chalei, V.; Sansom, S.N.; Kong, L.; Lee, S.; Montiel, J.F.; Vance, K.W.; Ponting, C.P. The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 2014, 3, e04530. [Google Scholar] [CrossRef]
- Ng, S.-Y.; Bogu, G.K.; Soh, B.S.; Stanton, L.W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell 2013, 51, 349–359. [Google Scholar] [CrossRef] [Green Version]
- Vance, K.W.; Sansom, S.N.; Lee, S.; Chalei, V.; Kong, L.; Cooper, S.E.; Oliver, P.L.; Ponting, C.P. The long non-coding RNA P aupar regulates the expression of both local and distal genes. EMBO J. 2014, 33, 296–311. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.-G.; Liao, Z.; Xiao, H.; Liu, H.; Hu, Y.-H.; Liao, Q.-D.; Zhong, D. LncRNA KCNQ1OT1 promoted BMP2 expression to regulate osteogenic differentiation by sponging miRNA-214. Exp. Mol. 2019, 107, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Biscarini, S.; Capauto, D.; Peruzzi, G.; Lu, L.; Colantoni, A.; Santini, T.; Shneider, N.A.; Caffarelli, E.; Laneve, P.; Bozzoni, I. Characterization of the lncRNA transcriptome in mESC-derived motor neurons: Implications for FUS-ALS. Stem Cell Res. 2018, 27, 172–179. [Google Scholar] [CrossRef] [PubMed]
- Ray, M.K.; Wiskow, O.; King, M.J.; Ismail, N.; Ergun, A.; Wang, Y.; Plys, A.J.; Davis, C.P.; Kathrein, K.; Sadreyev, R. CAT7 and cat7l long non-coding RNAs tune polycomb repressive complex 1 function during human and zebrafish development. J. Biol. Chem. 2016, 291, 19558–19572. [Google Scholar] [CrossRef] [Green Version]
- Quan, Y.; Wang, J.; Wang, S.; Zhao, J. Association of the plasma long non-coding RNA MEG3 with Parkinson’s disease. Front. Neurol. 2020, 11, 532891. [Google Scholar] [CrossRef]
- Yan, H.; Rao, J.; Yuan, J.; Gao, L.; Huang, W.; Zhao, L.; Ren, J. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate ischemic neuronal death by targeting miR-21/PDCD4 signaling pathway. Cell Death Dis. 2017, 8, 3211. [Google Scholar] [CrossRef]
- Yen, Y.-P.; Hsieh, W.-F.; Tsai, Y.-Y.; Lu, Y.-L.; Liau, E.S.; Hsu, H.-C.; Chen, Y.-C.; Liu, T.-C.; Chang, M.; Li, J. Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife 2018, 7, e38080. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3′-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef] [Green Version]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.-I.; Imai, T.; Murayama, S. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef] [Green Version]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef] [PubMed]
- Shelkovnikova, T.A.; Kukharsky, M.S.; An, H.; Dimasi, P.; Alexeeva, S.; Shabir, O.; Heath, P.R.; Buchman, V.L. Protective paraspeckle hyper-assembly downstream of TDP-43 loss of function in amyotrophic lateral sclerosis. Mol. Neurodegener. 2018, 13, 30. [Google Scholar] [CrossRef]
- Suzuki, H.; Shibagaki, Y.; Hattori, S.; Matsuoka, M. C9-ALS/FTD-linked proline–arginine dipeptide repeat protein associates with paraspeckle components and increases paraspeckle formation. Cell Death Dis. 2019, 10, 746. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Cave, J.W.; Dobner, P.R.; Mullikin-Kilpatrick, D.; Bartzokis, M.; Zhu, H.; Chow, C.W.; Gronostajski, R.M.; Kilpatrick, D.L. Reciprocal autoregulation by NFI occupancy and ETV1 promotes the developmental expression of dendrite-synapse genes in cerebellar granule neurons. Mol. Biol. Cell. 2016, 27, 1488–1499. [Google Scholar] [CrossRef]
- Ding, B.; Wang, W.; Selvakumar, T.; Xi, H.S.; Zhu, H.; Chow, C.W.; Horton, J.D.; Gronostajski, R.M.; Kilpatrick, D.L. Temporal regulation of nuclear factor one occupancy by calcineurin/NFAT governs a voltage-sensitive developmental switch in late maturing neurons. J. Neurosci. 2013, 33, 2860–2872. [Google Scholar] [CrossRef] [Green Version]
- Ding, B.; Kilpatrick, D.L. Lentiviral vector production, titration, and transduction of primary neurons. Methods Mol. Biol. 2013, 1018, 119–131. [Google Scholar] [CrossRef]
- Hulme, A.J.; Maksour, S.; Glover, M.S.-C.; Miellet, S.; Dottori, M. Making neurons, made easy: The use of Neurogenin-2 in neuronal differentiation. Stem Cell Rep. 2021, 17, 13–14. [Google Scholar] [CrossRef]
- Gong, J.; Hu, S.; Huang, Z.; Hu, Y.; Wang, X.; Zhao, J.; Qian, P.; Wang, C.; Sheng, J.; Lu, X. The requirement of Sox2 for the spinal cord motor neuron development of Zebrafish. Front. Mol. Neurosci. 2020, 13, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhati, M.; Llamosas, E.; Jacques, D.A.; Jeffries, C.M.; Dastmalchi, S.; Ripin, N.; Nicholas, H.R.; Matthews, J.M. Interactions between LHX3-and ISL1-family LIM-homeodomain transcription factors are conserved in Caenorhabditis elegans. Sci. Rep. 2017, 7, 4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, Z.; Wang, R.; Li, S.; Shi, Y.; Mo, L.; Yuan, J.; Jing, N.; Cheng, L. Molecular mechanisms underlying ascl1-mediated astrocyte-to-neuron conversion. Stem Cell Rep. 2021, 16, 534–547. [Google Scholar] [CrossRef] [PubMed]
- El Wazan, L.; Urrutia-Cabrera, D.; Wong, R.C.-B. Using transcription factors for direct reprogramming of neurons in vitro. World J. Stem Cells 2019, 11, 431. [Google Scholar] [CrossRef] [PubMed]
- Son, E.Y.; Ichida, J.K.; Wainger, B.J.; Toma, J.S.; Rafuse, V.F.; Woolf, C.J.; Eggan, K. Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 2011, 9, 205–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.-K.; Pfaff, S.L. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron 2003, 38, 731–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazzoni, E.O.; Mahony, S.; Closser, M.; Morrison, C.A.; Nedelec, S.; Williams, D.J.; An, D.; Gifford, D.K.; Wichterle, H. Synergistic binding of transcription factors to cell-specific enhancers programs motor neuron identity. Nat. Neurosci. 2013, 16, 1219–1227. [Google Scholar] [CrossRef] [Green Version]
- Pfaff, S.L.; Mendelsohn, M.; Stewart, C.L.; Edlund, T.; Jessell, T.M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 1996, 84, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Hester, M.E.; Murtha, M.J.; Song, S.; Rao, M.; Miranda, C.J.; Meyer, K.; Tian, J.; Boulting, G.; Schaffer, D.V.; Zhu, M.X. Rapid and efficient generation of functional motor neurons from human pluripotent stem cells using gene delivered transcription factor codes. Mol. Ther. 2011, 19, 1905–1912. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Cuvillier, J.M.; Lee, B.; Shen, R.; Lee, J.W.; Lee, S.-K. Fusion protein Isl1-Lhx3 specifies motor neuron fate by inducing motor neuron genes and concomitantly suppressing the interneuron programs. Proc. Natl. Acad. Sci. USA 2012, 109, 3383–3388. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Shen, R.; Cho, H.-H.; Kwon, R.-J.; Seo, S.Y.; Lee, J.W.; Lee, S.-K. STAT3 promotes motor neuron differentiation by collaborating with motor neuron-specific LIM complex. Proc. Natl. Acad. Sci. USA 2013, 110, 11445–11450. [Google Scholar] [CrossRef] [Green Version]
- Erb, M.; Lee, B.; Seo, S.Y.; Lee, J.W.; Lee, S.; Lee, S.-K. The Isl1-Lhx3 complex promotes motor neuron specification by activating transcriptional pathways that enhance its own expression and formation. eNeuro 2017, 4, ENEURO.0349-16.2017. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.; Park, C.; Jeong, Y.; Song, M.-R. Functional diversification of motor neuron-specific Isl1 enhancers during evolution. PLoS Genet. 2015, 11, e1005560. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lee, B.; Joshi, K.; Pfaff, S.L.; Lee, J.W.; Lee, S.-K. A regulatory network to segregate the identity of neuronal subtypes. Dev. Cell 2008, 14, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.P.; Lee, S.-K.; Jurata, L.W.; Gill, G.N.; Pfaff, S.L. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell 2002, 110, 237–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kania, A. Concocting cholinergy. PLoS Genet. 2014, 10, e1004313. [Google Scholar] [CrossRef] [Green Version]
- Patel, T.; Hammelman, J.; Closser, M.; Gifford, D.K.; Wichterle, H. General and cell-type-specific aspects of the motor neuron maturation transcriptional program. bioRxiv 2021. [Google Scholar] [CrossRef]
- Coppola, E.; Pattyn, A.; Guthrie, S.C.; Goridis, C.; Studer, M. Reciprocal gene replacements reveal unique functions for Phox2 genes during neural differentiation. EMBO J. 2005, 24, 4392–4403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, M.-R.; Glover, J.C.; Dufour, H.D.; Brunet, J.-F.; Goridis, C. Forced expression of Phox2 homeodomain transcription factors induces a branchio-visceromotor axonal phenotype. Dev. Biol. 2007, 303, 687–702. [Google Scholar] [CrossRef] [Green Version]
- Pattyn, A.; Morin, X.; Cremer, H.; Goridis, C.; Brunet, J.-F. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development 1997, 124, 4065–4075. [Google Scholar] [CrossRef]
- Goto, K.; Imamura, K.; Komatsu, K.; Mitani, K.; Aiba, K.; Nakatsuji, N.; Inoue, M.; Kawata, A.; Yamashita, H.; Takahashi, R. Simple derivation of spinal motor neurons from ESCs/iPSCs using sendai virus vectors. Mol. Ther. Methods Clin. Dev. 2017, 4, 115–125. [Google Scholar] [CrossRef] [Green Version]
- Limone, F.; Mitchell, J.M.; San Juan, I.G.; Smith, J.L.; Raghunathan, K.; Couto, A.; Ghosh, S.D.; Meyer, D.; Mello, C.J.; Nemesh, J. Efficient generation of lower induced Motor Neurons by coupling Ngn2 expression with developmental cues. bioRxiv 2022. [Google Scholar] [CrossRef]
- Lee, H.; Lee, H.Y.; Lee, B.E.; Gerovska, D.; Park, S.Y.; Zaehres, H.; Araúzo-Bravo, M.J.; Kim, J.-I.; Ha, Y.; Schöler, H.R. Sequentially induced motor neurons from human fibroblasts facilitate locomotor recovery in a rodent spinal cord injury model. Elife 2020, 9, e52069. [Google Scholar] [CrossRef] [PubMed]
- Garone, M.G.; de Turris, V.; Soloperto, A.; Brighi, C.; De Santis, R.; Pagani, F.; Di Angelantonio, S.; Rosa, A. Conversion of human induced pluripotent stem cells (iPSCs) into functional spinal and cranial motor neurons using PiggyBac vectors. JoVE J. Vis. Exp. 2019, 1, e59321. [Google Scholar] [CrossRef] [PubMed]
- Cantor, E.L.; Shen, F.; Jiang, G.; Tan, Z.; Cunningham, G.M.; Wu, X.; Philips, S.; Schneider, B.P. Passage number affects differentiation of sensory neurons from human induced pluripotent stem cells. Sci. Rep. 2022, 12, 15869. [Google Scholar] [CrossRef] [PubMed]
- Baghbaderani, B.A.; Syama, A.; Sivapatham, R.; Pei, Y.; Mukherjee, O.; Fellner, T.; Zeng, X.; Rao, M.S. Detailed characterization of human induced pluripotent stem cells manufactured for therapeutic applications. Stem Cell Rev. Rep. 2016, 12, 394–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.; Wanczyk, H.; Jensen, T.; Finck, C. Assessment of iPSC teratogenicity throughout directed differentiation toward an alveolar-like phenotype. Differentiation 2019, 105, 45–53. [Google Scholar] [CrossRef]
- Nelakanti, R.V.; Kooreman, N.G.; Wu, J.C. Teratoma formation: A tool for monitoring pluripotency in stem cell research. Curr. Protoc. Stem Cell Biol. 2015, 32, 4A.8.1–4A.8.17. [Google Scholar] [CrossRef] [Green Version]
- Thaler, J.; Harrison, K.; Sharma, K.; Lettieri, K.; Kehrl, J.; Pfaff, S.L. Active suppression of interneuron programs within developing motor neurons revealed by analysis of homeodomain factor HB9. Neuron 1999, 23, 675–687. [Google Scholar] [CrossRef] [Green Version]
- Geula, C.; Schatz, C.; Mesulam, M.-M. Differential localization of NADPH-diaphorase and calbindin-D28k within the cholinergic neurons of the basal forebrain, striatum and brainstem in the rat, monkey, baboon and human. Neuroscience 1993, 54, 461–476. [Google Scholar] [CrossRef]
- Egawa, N.; Kitaoka, S.; Tsukita, K.; Naitoh, M.; Takahashi, K.; Yamamoto, T.; Adachi, F.; Kondo, T.; Okita, K.; Asaka, I. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci. Transl. Med. 2012, 4, ra104–ra145. [Google Scholar] [CrossRef]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar] [CrossRef]
- Kanning, K.C.; Kaplan, A.; Henderson, C.E. Motor neuron diversity in development and disease. Annu. Rev. Neurosci. 2010, 33, 409–440. [Google Scholar] [CrossRef]
- Kiskinis, E.; Sandoe, J.; Williams, L.A.; Boulting, G.L.; Moccia, R.; Wainger, B.J.; Han, S.; Peng, T.; Thams, S.; Mikkilineni, S. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014, 14, 781–795. [Google Scholar] [CrossRef]
- Li, X.-J.; Du, Z.-W.; Zarnowska, E.D.; Pankratz, M.; Hansen, L.O.; Pearce, R.A.; Zhang, S.-C. Specification of motoneurons from human embryonic stem cells. Nat. Biotechnol. 2005, 23, 215–221. [Google Scholar] [CrossRef]
- Nijssen, J.; Comley, L.H.; Hedlund, E. Motor neuron vulnerability and resistance in amyotrophic lateral sclerosis. Acta Neuropathol. 2017, 133, 863–885. [Google Scholar] [CrossRef] [Green Version]
- Faravelli, I.; Bucchia, M.; Rinchetti, P.; Nizzardo, M.; Simone, C.; Frattini, E.; Corti, S. Motor neuron derivation from human embryonic and induced pluripotent stem cells: Experimental approaches and clinical perspectives. Stem Cell Res. Ther. 2014, 5, 87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sances, S.; Bruijn, L.I.; Chandran, S.; Eggan, K.; Ho, R.; Klim, J.R.; Livesey, M.R.; Lowry, E.; Macklis, J.D.; Rushton, D. Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nat. Neurosci. 2016, 19, 542–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lam, D.; Enright, H.A.; Cadena, J.; Peters, S.K.; Sales, A.P.; Osburn, J.J.; Soscia, D.A.; Kulp, K.S.; Wheeler, E.K.; Fischer, N.O. Tissue-specific extracellular matrix accelerates the formation of neural networks and communities in a neuron-glia co-culture on a multi-electrode array. Sci. Rep. 2019, 9, 4159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, J.; Meng, T.; Yang, J.; Hu, N.; Zhao, H.; Tian, T. Three-dimensional in vitro tissue culture models of brain organoids. Exp. Neurol. 2021, 339, 113619. [Google Scholar] [CrossRef]
- Kampmann, M. CRISPR-based functional genomics for neurological disease. Nat. Rev. Neurol. 2020, 16, 465–480. [Google Scholar] [CrossRef]

| Chemicals | Functions | References |
|---|---|---|
| Retinoic acid (RA) | Agonist for RA receptors. Promotes neural differentiation. | [14] |
| Valproic acid (VPA) | Histone deacetylase inhibitor. Facilitates the reprogramming of fibroblasts into iPSCs. Promotes neuronal differentiation. | [79] |
| SB431542 | Inhibitor of TGF-β, Activin and Nodal signaling. Differentiation of human ES and iPSCs into neural progenitors. Increase in reprogramming efficiency in combination with other small molecules. | [49] |
| CHIR99021 | Selective inhibitor of glycogen synthase kinase 3 (GSK-3). Enables reprogramming of fibroblasts into iPSCs. Induces neuronal differentiation. | [49] |
| Purmorphamine (PUR) | Sonic Hedgehog (Shh) activator. Improves the efficiency of MN differentiation. | [80] |
| Dorsomorphin | Inhibitor of both activin/nodal/TGF-β and BMP pathways. Induces rapid and high-efficiency neural conversion in both hESCs and hiPSCs. Induces neuronal differentiation in vitro. | [81] |
| Y-27632 | Highly potent and selective inhibitor of Rho-associated, coiled-coil-containing protein kinase (ROCK). Improves embryoid body (EB) formation efficiency. Enhances survival of hESC during cell passaging. | [82] |
| Forskolin (FSK) | Stimulates adenylate cyclase activity and increases cAMP. Regulates neuronal specification and promotes axonal regeneration. | [83] |
| Compound E | NOTCH signaling inhibitor. Accelerates MN maturation. | [84] |
| Brain-derived neurotrophic factor (BDNF) | Activates TrkB signaling. BDNF enhances the survival and differentiation of neurons in vitro. Critical for neuronal survival, morphogenesis, and plasticity. | [85] |
| Glial cell line-derived neurotrophic factor (GDNF) | Activates tyrosine kinase receptor signaling. Promotes neuronal differentiation in later culture periods. Potential roles in various pathways, mediating growth, differentiation, and migration of neurons. | [86] |
| Ciliary neurotrophic factor (CNTF) | Neurotrophic factor. Promotes the survival of different neurons and the differentiation of neural progenitor cells (NPCs) in vitro. | [87] |
| Neurotrophin-3 (NT3) | Neurotrophic factor-mediated Trk receptor signaling. Neurotrophic factors promote the survival of neurons. Growth factor involved in stem cell differentiation. | [88] |
| Basic fibroblast growth factor (bFGF) | Fibroblast growth factor family. Stimulates hESC to form neural rosettes. Supports the maintenance of undifferentiated human hESCs. | [89] |
| Epidermal growth factor (EGF) | Mitogen. Induces the in vitro and in vivo proliferation of neural stem cells, their migration, and their differentiation towards the neuroglial cell line. | [90] |
| Heparin | Promotes the growth of hESCs. Supports the binding of FGF to its receptor and increases the stability of FGF. Activates Wnt signaling for neuronal morphogenesis. | [91] |
| Cell Signaling | Functions | References |
|---|---|---|
| Sonic Hedgehog (Shh) signaling | Shh signaling is required for the final specification of MNs. Activator: Purmorphamine (PUR); Inhibitors: Cyclopamine and HPI-1. | [92] |
| Dual SMAD inhibition | Block endodermal and mesodermal cell fates and promote neural conversion. Drive the rapid differentiation of hESCs and hiPSCs into a highly enriched population of NPCs. Inhibitors: SB431542, LDN193189, Noggin, LY364947, RepSox, Dorsomorphin, DMH-1 | [93] |
| Neurotrophic factors signaling | Improves MN survival and maturation. Activators: BDNF, GDNF, NGF, NT-3 | [94] |
| Wnt/β-catenin signaling | Contributes to patterning, proliferation, and differentiation throughout vertebrate neural development. Activator: CHIR99021 (CHIR), Kenpaullone, SB216763 Inhibitor: APCDD1, Waif1/5T4 | [95] |
| Notch signaling | Regulates the balance between MN differentiation and the maintenance of the progenitor state. Activator: Dll, Dl4 Inhibitor: DAPT, LY411575 | [96] |
| Chemical Cocktail and Cytokines | Target Signaling Pathways | Cellular Markers | Efficiency | Days | References |
|---|---|---|---|---|---|
| Chir-99021, SB431542, LDN1931899, RA, SAG, DAPT, BME, Ascorbic Acid, and Y-27632 | Wnt, FGF, SHH signaling | OLIG2, ISL1/2, HB9, LHX1/2, FOXP2 | ~77% | 14 | [68,97] |
| RA, PUR, Y-27632, VPA and CHIR99021 | SHH signaling, WNT/b-catenin | HB9, ISLET-1, ChAT | ~85% | 30–40 | [98] |
| SB431542, CHIR99021, RA, PUR, BDNF, GDNF | SHH, WNT/b-catenin, and Notch | TUJ1, MAP2, HB9, ChAT, SYP | >85% | 28 | [99] |
| SB 431542, CHIR99021, dorsomorphin, and Cpd E | Activin/nodal/TGF-β and BMP pathways, SHH signaling | ChAT, HB9, SOX11, PAX6, nestin, OLIG2, TUJ1, MAP2 | ~88% | 21 | [100] |
| SB 431542, dorsomorphin, BDNF, RA, and ISL1/2 | TGF-β, Activin, Nodal, and canonical | FOXP1, OXA5, MAP2, TUJ1 | >40% | 24 | [101] |
| SB 431542, CHIR99021, dorsomorphin, and RA | Activin/nodal/TGF-β, BMP and GSK-3 | ChAT, HB9, SMI-32 | ~80% | 24 | [102] |
| SB 431542, dorsomorphin, B18R, synTFs mRNAs of neurogenin and NeuroD families, FSK, BDNF, GDNF, and NT-3 | Activin/nodal/TGF-β and BMP pathways | ChAT, HB9, and ISL1 | ~86% | 12 | [103] |
| RA, SAG, BDNF, GDNF, and DAPT | Neurotrophic factors, canonical signaling | ChAT, HB9, ISL1, SMI-32, TUJ1 | 70–95% | 32 | [104] |
| Dorsomorphin, SB431542, CHIR99021, RA, PUR, ascorbic acid, dibutyryl cAMP | Activin/nodal/TGF-β and BMP pathways | OLIG2, SOX2, ISLET1, AP2, HB9, SMI32, TUJ1 | ~70 | 45 | [105] |
| SB431542, DMH1, CHIR99021, RA, PUR, Cpd E | BMP, Activin, WNT, SHH and NOTCH | NKX2.2, OLIG2, ISL1, MNX, TUJ1, ChAT, BTX | >90% | 28 | [84] |
| PUR, RA | Shh, Agonist for retinoic acid receptors | HB-9, TUJ1,OLIG2 | >85% | 28 | [106] |
| Compound C, RA, cAMP Neurotrophic factor | BMP and Activin signaling | TUJ1, MAP2 Synapsin I, HB9, ChAT | ~70% | 20 | [107] |
| PUR, RA | Shh signaling agonist | HB9, ISL1/2, ChAT, OLIG2 | ~80% | 15 | [108] |
| Noncoding RNA | Function | References |
|---|---|---|
| miR-9 | miR-9 modifies MN columns by a tuning regulation of transcription factor FoxP1 (Forkhead box protein 1) levels in developing spinal cords. | [133,134,135] |
| miR-218 | Expression of miR-218 is directly upregulated by the Isl1–Lhx3 complex, which drives MN fate. Inhibition of miR-218 suppresses the generation of MNs in both chick neural tube and mouse embryonic stem cells. | [136,137] |
| mir-17~92 | Confers MN subtype survival during development. | [136,138,139] |
| mir-27 | mir-27 as a major regulator coordinates the temporal delay and spatial boundary of Hox protein expression, which contributes to the specification of MN subtype identity. | [140] |
| miR-183-5p | miR-183-5p is a central regulator of MN survival under stress conditions. Increased miR-183-5p is correlated with cell stress in MNs of ALS in pre-symptomatic and early symptomatic stages. | [141,142] |
| miR-196 | The timing and rostro-caudal extent of Hoxb8 activity in the neural tube is tightly regulated by miR-196, a miRNA species encoded within three Hox gene clusters. miR-196 effectively suppresses endogenous Hoxb8. | [143] |
| miR-375 | miR-375 facilitates human spinal MN development and protects MNs from DNA damage-induced degeneration. | [130,144,145] |
| Transcription Factor | Functions | References |
|---|---|---|
| Neurogenin 2 (NEUROG2) | Transcriptional regulator and actively involved in neuronal differentiation. Unique and critical role in determining MN cell-type identity. | [182] |
| Sex determining region Y-box 2 (SOX2) | Critical for early embryogenesis and for maintaining embryonic stem cell pluripotency. | [183] |
| ISL LIM homeobox 1 (ISL1) | ISL1 is a major transcription factor necessary for MN identity. Fusion protein Isl1–Lhx3 specifies MN fate differentiation. | [184] |
| LIM homeobox 3 (LHX3) | Transcriptional activator involved in the development of interneurons and MNs. | [184] |
| POU class 5 homeobox 1 (POU5F1) | Critical for early embryogenesis and for embryonic stem cell pluripotency. Master regulator of initiation, maintenance, and differentiation of pluripotent cells. | [80] |
| Achaete-scute family basic helix-loop-helix transcription factor 1 (ASCL1) | Promotes cell cycle exit and develops neuronal progenitors and differentiation when expressed in neural progenitor cells. | [185] |
| POU Class 3 Homeobox 2 (POU3F2) | Plays potential role in morphological complexity, maturity, and action potentials of the neuronal cells | [186,187] |
| Transcription Factors Delivered | Delivery Vector | Cellular Markers | Efficiency | Days to Reach Maturation | References |
|---|---|---|---|---|---|
| NGN2, ISL1, LHX3 | Lentiviral | MAP2, SMI32, TUBB3, HB9, ChAT | >95% | 35 days | [29,32] |
| NGN2 | Lentiviral | ChAT, HB9, SMI-32, ISL1, FOXP1, MAP2, TUJ1 | ~95% | 30 days | [204] |
| NGN2, SOX1, ISL1, and LHX3 | Lentiviral | HB9, ChAT, TUBB3, MAP2, and synapsin | 45–50% | 35 days | [29,31] |
| POU5F1(OCT4) and LHX3 | Lentiviral | MAP2, TUJ1, HB9, ChAT | 70 ~ 90% | 28 days | [205] |
| NGN2, ISL1, LHX3 and NGN2, ISL1, PHOX2A | Piggy-bac transposable | PHOX2B, TUJ1, ISL1, ChAT | ~90% | 11–12 days | [144,206] |
| NGN2, ISL1, LHX3 | Sendai virus | HB9, MAP2, ChAT, Tuj1, | ~93% | 14 days | [203] |
| NGN2, ISL1, LHX3 | Adenoviral | HB9, CHAT, SMI-31, HOXC6 | 60–70% | 30 days | [191] |
| Ascl1, Brn2 (POU3F2), Myt1l, Hb9 (MNX1), NGN2, ISL1, and LHX3 | Retroviral | MAP2, vChT, HB9, ISL1 | ~60% | 35 days | [187] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akter, M.; Ding, B. Modeling Movement Disorders via Generation of hiPSC-Derived Motor Neurons. Cells 2022, 11, 3796. https://doi.org/10.3390/cells11233796
Akter M, Ding B. Modeling Movement Disorders via Generation of hiPSC-Derived Motor Neurons. Cells. 2022; 11(23):3796. https://doi.org/10.3390/cells11233796
Chicago/Turabian StyleAkter, Masuma, and Baojin Ding. 2022. "Modeling Movement Disorders via Generation of hiPSC-Derived Motor Neurons" Cells 11, no. 23: 3796. https://doi.org/10.3390/cells11233796
APA StyleAkter, M., & Ding, B. (2022). Modeling Movement Disorders via Generation of hiPSC-Derived Motor Neurons. Cells, 11(23), 3796. https://doi.org/10.3390/cells11233796