Membrane Contact Sites in Autophagy
Abstract
:1. Introduction
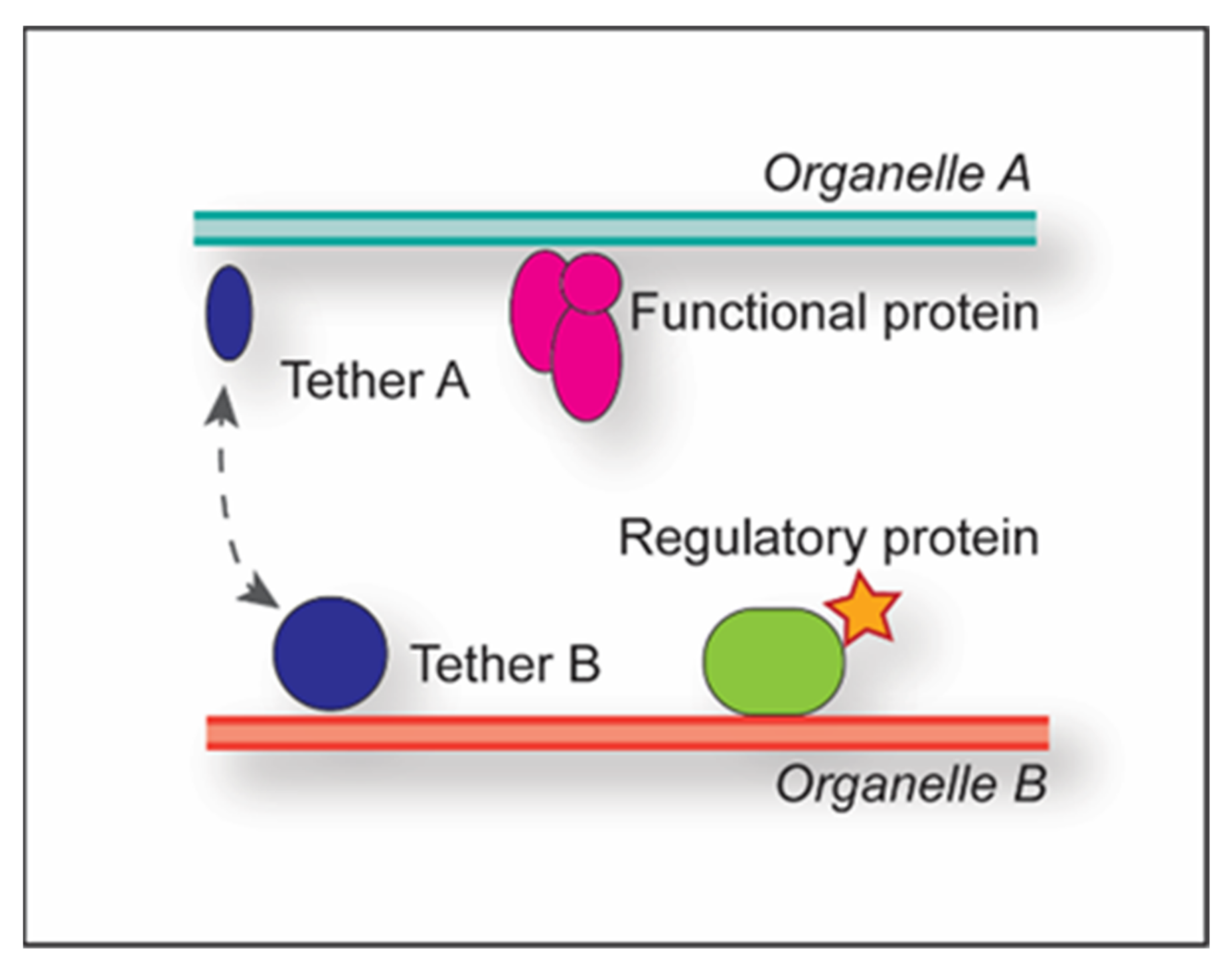
1.1. Membrane Contact Sites
1.2. The Mechanism of Macroautophagy
2. MCSs during Autophagosome Biogenesis
2.1. ER-Phagophore MCSs
2.2. ER-Mitochondria MCSs and Mitochondria-Associated ER Membranes (MAMs)
2.3. ER-PM MCSs
2.4. Phagophore-Vacuole MCSs
2.5. Nucleus-Vacuole Junction MCSs
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scorrano, L.; De Matteis, M.A.; Emr, S.; Giordano, F.; Hajnóczky, G.; Kornmann, B.; Lackner, L.L.; Levine, T.P.; Pellegrini, L.; Reinisch, K.; et al. Coming together to define membrane contact sites. Nat. Commun. 2019, 10, 1287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prinz, W.A.; Toulmay, A.; Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020, 21, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Voeltz, G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol. 2016, 17, 69–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Qian, T.; He, R.; Wan, C.; Liu, Y.; Yu, H. Endoplasmic Reticulum–Plasma Membrane Contact Sites: Regulators, Mechanisms, and Physiological Functions. Front. Cell Dev. Biol. 2021, 9, 627700. [Google Scholar] [CrossRef] [PubMed]
- Lur, G.; Haynes, L.P.; Prior, I.A.; Gerasimenko, O.V.; Feske, S.; Petersen, O.H.; Burgoyne, R.D.; Tepikin, A.V. Ribosome-free Terminals of Rough ER Allow Formation of STIM1 Puncta and Segregation of STIM1 from IP3 Receptors. Curr. Biol. 2009, 19, 1648–1653. [Google Scholar] [CrossRef] [Green Version]
- West, M.; Zurek, N.; Hoenger, A.; Voeltz, G.K. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J. Cell Biol. 2011, 193, 333–346. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Busnadiego, R.; Saheki, Y.; De Camilli, P. Three-dimensional architecture of extended synaptotagmin-mediated endoplasmic reticulum–plasma membrane contact sites. Proc. Natl. Acad. Sci. USA 2015, 112, E2004–E2013. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.M.; Buchanan, J.; Luik, R.M.; Lewis, R.S. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 2006, 174, 803–813. [Google Scholar] [CrossRef]
- Biazik, J.; Ylä-Anttila, P.; Vihinen, H.; Jokitalo, E.; Eskelinen, E.-L. Ultrastructural relationship of the phagophore with surrounding organelles. Autophagy 2015, 11, 439–451. [Google Scholar] [CrossRef] [Green Version]
- Bohnert, M. Tether Me, Tether Me Not—Dynamic Organelle Contact Sites in Metabolic Rewiring. Dev. Cell 2020, 54, 212–225. [Google Scholar] [CrossRef]
- Giordano, F.; Saheki, Y.; Idevall-Hagren, O.; Colombo, S.F.; Pirruccello, M.; Milosevic, I.; Gracheva, E.O.; Bagriantsev, S.N.; Borgese, N.; De Camilli, P. PI(4,5)P2-Dependent and Ca2+-Regulated ER-PM Interactions Mediated by the Extended Synaptotagmins. Cell 2013, 153, 1494–1509. [Google Scholar] [CrossRef] [Green Version]
- Manford, A.G.; Stefan, C.J.; Yuan, H.L.; MacGurn, J.A.; Emr, S.D. ER-to-Plasma Membrane Tethering Proteins Regulate Cell Signaling and ER Morphology. Dev. Cell 2012, 23, 1129–1140. [Google Scholar] [CrossRef] [Green Version]
- Quon, E.; Sere, Y.Y.; Chauhan, N.; Johansen, J.; Sullivan, D.P.; Dittman, J.S.; Rice, W.J.; Chan, R.B.; Di Paolo, G.; Beh, C.T.; et al. Endoplasmic reticulum-plasma membrane contact sites integrate sterol and phospholipid regulation. PLOS Biol. 2018, 16, e2003864. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, L.P.; Wendling, C.; Védie, B.; Kobayashi, T.; Chenard, M.; Tomasetto, C.; Drin, G.; Alpy, F. STARD 3 mediates endoplasmic reticulum-to-endosome cholesterol transport at membrane contact sites. EMBO J. 2017, 36, 1412–1433. [Google Scholar] [CrossRef]
- Stefan, C.J.; Manford, A.G.; Baird, D.; Yamada-Hanff, J.; Mao, Y.; Emr, S.D. Osh Proteins Regulate Phosphoinositide Metabolism at ER-Plasma Membrane Contact Sites. Cell 2011, 144, 389–401. [Google Scholar] [CrossRef] [Green Version]
- Bui, M.; Gilady, S.Y.; Fitzsimmons, R.E.; Benson, M.D.; Lynes, E.M.; Gesson, K.; Alto, N.M.; Strack, S.; Scott, J.D.; Simmen, T. Rab32 Modulates Apoptosis Onset and Mitochondria-associated Membrane (MAM) Properties. J. Biol. Chem. 2010, 285, 31590–31602. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Klionsky, D.J.; Shen, H.-M. The emerging mechanisms and functions of microautophagy. Nat. Rev. Mol. Cell Biol. 2022, in press. [Google Scholar] [CrossRef]
- Bandyopadhyay, U.; Kaushik, S.; Varticovski, L.; Cuervo, A.M. The Chaperone-Mediated Autophagy Receptor Organizes in Dynamic Protein Complexes at the Lysosomal Membrane. Mol. Cell. Biol. 2008, 28, 5747–5763. [Google Scholar] [CrossRef] [Green Version]
- Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef]
- Gómez-Sánchez, R.; Tooze, S.A.; Reggiori, F. Membrane supply and remodeling during autophagosome biogenesis. Curr. Opin. Cell Biol. 2021, 71, 112–119. [Google Scholar] [CrossRef]
- Dikic, I.; Elazar, Z. Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol. 2018, 19, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Lahiri, V.; Hawkins, W.D.; Klionsky, D.J. Watch What You (Self-) Eat: Autophagic Mechanisms that Modulate Metabolism. Cell Metab. 2019, 29, 803–826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licheva, M.; Raman, B.; Kraft, C.; Reggiori, F. Phosphoregulation of the autophagy machinery by kinases and phosphatases. Autophagy 2022, 18, 104–123. [Google Scholar] [CrossRef] [PubMed]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-Mediated Induction of Autophagy via an Apg1 Protein Kinase Complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, L.; Chen, S.; Du, F.; Li, S.; Zhao, L.; Wang, X. Nutrient starvation elicits an acute autophagic response mediated by Ulk1 dephosphorylation and its subsequent dissociation from AMPK. Proc. Natl. Acad. Sci. USA 2011, 108, 4788–4793. [Google Scholar] [CrossRef] [Green Version]
- Mack, H.I.; Zheng, B.; Asara, J.M.; Thomas, S.M. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy 2012, 8, 1197–1214. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef]
- Suzuki, K.; Kubota, Y.; Sekito, T.; Ohsumi, Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007, 12, 209–218. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Huang, W.-P.; Stromhaug, P.E.; Klionsky, D.J. Convergence of Multiple Autophagy and Cytoplasm to Vacuole Targeting Components to a Perivacuolar Membrane Compartment Prior tode Novo Vesicle Formation. J. Biol. Chem. 2002, 277, 763–773. [Google Scholar] [CrossRef]
- Bieber, A.; Capitanio, C.; Erdmann, P.S.; Fiedler, F.; Beck, F.; Lee, C.-W.; Li, D.; Hummer, G.; Schulman, B.A.; Baumeister, W.; et al. In situ structural analysis reveals membrane shape transitions during autophagosome formation. Proc. Natl. Acad. Sci. USA 2022, 119, e2209823119. [Google Scholar] [CrossRef]
- Graef, M.; Friedman, J.; Graham, C.; Babu, M.; Nunnari, J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol. Biol. Cell 2013, 24, 2918–2931. [Google Scholar] [CrossRef]
- Suzuki, K.; Akioka, M.; Kondo-Kakuta, C.; Yamamoto, H.; Ohsumi, Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013, 126, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Reggiori, F. Molecular regulation of autophagosome formation. Biochem. Soc. Trans. 2022, 50, 55–69. [Google Scholar] [CrossRef]
- Sawa-Makarska, J.; Baumann, V.; Coudevylle, N.; von Bülow, S.; Nogellova, V.; Abert, C.; Schuschnig, M.; Graef, M.; Hummer, G.; Martens, S. Reconstitution of autophagosome nucleation defines Atg9 vesicles as seeds for membrane formation. Science 2020, 369, eaaz7714. [Google Scholar] [CrossRef]
- Broadbent, D.; Barnaba, C.; Schmidt, J.C. Systematic Analysis of Autophagy Identifies Atg9 Vesicles as the Origin of the Phagophore. bioRxiv 2022. [Google Scholar]
- Olivas, T.J.; Wu, Y.; Yu, S.; Luan, L.; Choi, P.; Nag, S.; De Camilli, P.; Gupta, K.; Melia, T.J. ATG9 vesicles comprise the seed membrane of mammalian autophagosomes. bioRxiv 2022. [Google Scholar]
- Young, A.R.J.; Chan, E.Y.W.; Hu, X.W.; Köchl, R.; Crawshaw, S.G.; High, S.; Hailey, D.W.; Lippincott-Schwartz, J.; Tooze, S.A. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006, 119, 3888–3900. [Google Scholar] [CrossRef] [Green Version]
- Orsi, A.; Razi, M.; Dooley, H.C.; Robinson, D.; Weston, A.E.; Collinson, L.M.; Tooze, S.A. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol. Biol. Cell 2012, 23, 1860–1873. [Google Scholar] [CrossRef]
- Ge, L.; Melville, D.; Zhang, M.; Schekman, R. The ER–Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. eLife 2013, 2, e00947. [Google Scholar] [CrossRef]
- Reggiori, F.; Ungermann, C. Autophagosome Maturation and Fusion. J. Mol. Biol. 2017, 429, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.A.; Amato, C.; Fortier, T.M.; Velentzas, P.; Wood, W.; Baehrecke, E.H. A conserved myotubularin-related phosphatase regulates autophagy by maintaining autophagic flux. J. Cell Biol. 2020, 219, e201909073. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cheng, S.; Zhao, H.; Zou, W.; Yoshina, S.; Mitani, S.; Zhang, H.; Wang, X. PI 3 P phosphatase activity is required for autophagosome maturation and autolysosome formation. EMBO Rep. 2014, 15, 973–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cebollero, E.; van der Vaart, A.; Zhao, M.; Rieter, E.; Klionsky, D.J.; Helms, J.B.; Reggiori, F. Phosphatidylinositol-3-Phosphate Clearance Plays a Key Role in Autophagosome Completion. Curr. Biol. 2012, 22, 1545–1553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monastyrska, I.; Rieter, E.; Klionsky, D.J.; Reggiori, F. Multiple roles of the cytoskeleton in autophagy. Biol. Rev. 2009, 84, 431–448. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.G.; Zhang, H. Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 2018, 218, 757–770. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Chen, Y.; Miao, G.; Zhao, H.; Qu, W.; Li, D.; Wang, Z.; Liu, N.; Li, L.; Chen, S.; et al. The ER-Localized Transmembrane Protein EPG-3/VMP1 Regulates SERCA Activity to Control ER-Isolation Membrane Contacts for Autophagosome Formation. Mol. Cell 2017, 67, 974–989.e6. [Google Scholar] [CrossRef] [Green Version]
- Alpy, F.; Rousseau, A.; Schwab, Y.; Legueux, F.; Stoll, I.; Wendling, C.; Spiegelhalter, C.; Kessler, P.; Mathelin, C.; Rio, M.-C.; et al. STARD3 or STARD3NL and VAP form a novel molecular tether between late endosomes and the ER. J. Cell Sci. 2013, 126, 5500–5512. [Google Scholar]
- Raiborg, C.; Wenzel, E.M.; Pedersen, N.M.; Olsvik, H.; Schink, K.O.; Schultz, S.W.; Vietri, M.; Nisi, V.; Bucci, C.; Brech, A.; et al. Repeated ER–endosome contacts promote endosome translocation and neurite outgrowth. Nature 2015, 520, 234–238. [Google Scholar] [CrossRef]
- Knoblach, B.; Sun, X.; Coquelle, N.; Fagarasanu, A.; Poirier, R.L.; Rachubinski, A.R. An ER-peroxisome tether exerts peroxisome population control in yeast. EMBO J. 2013, 32, 2439–2453. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Sánchez, R.; Rose, J.; Guimarães, R.; Mari, M.; Papinski, D.; Rieter, E.; Geerts, W.J.; Hardenberg, R.; Kraft, C.; Ungermann, C.; et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J. Cell Biol. 2018, 217, 2743–2763. [Google Scholar] [CrossRef] [Green Version]
- Uemura, T.; Yamamoto, M.; Kametaka, A.; Sou, Y.-S.; Yabashi, A.; Yamada, A.; Annoh, H.; Kametaka, S.; Komatsu, M.; Waguri, S. A Cluster of Thin Tubular Structures Mediates Transformation of the Endoplasmic Reticulum to Autophagic Isolation Membrane. Mol. Cell. Biol. 2014, 34, 1695–1706. [Google Scholar] [CrossRef]
- Ylä-Anttila, P.; Vihinen, H.; Jokitalo, E.; Eskelinen, E.-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009, 5, 1180–1185. [Google Scholar] [CrossRef] [Green Version]
- Hayashi-Nishino, M.; Fujita, N.; Noda, T.; Yamaguchi, A.; Yoshimori, T.; Yamamoto, A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009, 11, 1433–1437. [Google Scholar] [CrossRef]
- Ghanbarpour, A.; Valverde, D.P.; Melia, T.J.; Reinisch, K.M. A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2101562118. [Google Scholar] [CrossRef]
- Kotani, T.; Kirisako, H.; Koizumi, M.; Ohsumi, Y.; Nakatogawa, H. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc. Natl. Acad. Sci. USA 2018, 115, 10363–10368. [Google Scholar] [CrossRef] [Green Version]
- Valverde, D.P.; Yu, S.; Boggavarapu, V.; Kumar, N.; Lees, J.A.; Walz, T.; Reinisch, K.M.; Melia, T.J. ATG2 transports lipids to promote autophagosome biogenesis. J. Cell Biol. 2019, 218, 1787–1798. [Google Scholar] [CrossRef] [Green Version]
- Chowdhury, S.; Otomo, C.; Leitner, A.; Ohashi, K.; Aebersold, R.; Lander, G.C.; Otomo, T. Insights into autophagosome biogenesis from structural and biochemical analyses of the ATG2A-WIPI4 complex. Proc. Natl. Acad. Sci. USA 2018, 115, E9792–E9801. [Google Scholar] [CrossRef] [Green Version]
- Osawa, T.; Ishii, Y.; Noda, N.N. Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes Cells 2020, 25, 65–70. [Google Scholar] [CrossRef]
- Osawa, T.; Kotani, T.; Kawaoka, T.; Hirata, E.; Suzuki, K.; Nakatogawa, H.; Ohsumi, Y.; Noda, N.N. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat. Struct. Mol. Biol. 2019, 26, 281–288. [Google Scholar] [CrossRef]
- Rieter, E.; Vinke, F.; Bakula, D.; Cebollero, E.; Ungermann, C.; Proikas-Cezanne, T.; Reggiori, F. Atg18 function in autophagy is regulated by specific sites within its β-propeller. J. Cell Sci. 2013, 126, 593–604. [Google Scholar] [CrossRef] [Green Version]
- van Vliet, A.R.; Chiduza, G.N.; Maslen, S.L.; Pye, V.E.; Joshi, D.; De Tito, S.; Jefferies, H.B.; Christodoulou, E.; Roustan, C.; Punch, E.; et al. ATG9A and ATG2A form a heteromeric complex essential for autophagosome formation. Mol. Cell 2022, 82, 4324–4339.e8. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Otomo, C.; Otomo, T. The autophagic membrane tether ATG2A transfers lipids between membranes. eLife 2019, 8, e45777. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.T.J.; Herrmann, B.; Antunes, D.; Rapaport, D.; Dimmer, K.S.; Kornmann, B. Vps13-Mcp1 interact at vacuole–mitochondria interfaces and bypass ER–mitochondria contact sites. J. Cell Biol. 2017, 216, 3219–3229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-S.; Thorsness, M.K.; Policastro, R.; McGoldrick, L.L.; Hollingsworth, N.M.; Thorsness, P.E.; Neiman, A.M. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol. Cell 2016, 27, 2435–2449. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [Green Version]
- Matoba, K.; Kotani, T.; Tsutsumi, A.; Tsuji, T.; Mori, T.; Noshiro, D.; Sugita, Y.; Nomura, N.; Iwata, S.; Ohsumi, Y.; et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat. Struct. Mol. Biol. 2020, 27, 1185–1193. [Google Scholar] [CrossRef]
- Maeda, S.; Yamamoto, H.; Kinch, L.N.; Garza, C.M.; Takahashi, S.; Otomo, C.; Grishin, N.V.; Forli, S.; Mizushima, N.; Otomo, T. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat. Struct. Mol. Biol. 2020, 27, 1194–1201. [Google Scholar] [CrossRef]
- Chumpen-Ramirez, S.; Gomez-Sanchez, R.; Verlhac, P.; Margeritis, E.; Cosentino, K.; Reggiori, F.; Ungermann, C. Atg9 interactions via its transmembrane domains are required for phagophore expansion during autophagy. Autophagy 2022, in press. [Google Scholar] [CrossRef]
- Nguyen, A.; Lugarini, F.; David, C.; Hosnani, P.; Knotkova, B.; Patel, A.; Parfentev, I.; Friedrich, A.; Urlaub, H.; Meinecke, M.; et al. Metamorphic proteins at the basis of human autophagy initiation and lipid transfer. bioRxiv 2009. [Google Scholar] [CrossRef]
- Li, Y.E.; Wang, Y.; Du, X.; Zhang, T.; Mak, H.Y.; Hancock, S.E.; McEwen, H.; Pandzic, E.; Whan, R.M.; Aw, Y.C.; et al. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J. Cell Biol. 2021, 220, e202103105. [Google Scholar] [CrossRef]
- Tábara, L.-C.; Escalante, R. VMP1 Establishes ER-Microdomains that Regulate Membrane Contact Sites and Autophagy. PLoS ONE 2016, 11, e0166499. [Google Scholar] [CrossRef]
- Morita, K.; Hama, Y.; Izume, T.; Tamura, N.; Ueno, T.; Yamashita, Y.; Sakamaki, Y.; Mimura, K.; Morishita, H.; Shihoya, W.; et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J. Cell Biol. 2018, 217, 3817–3828. [Google Scholar] [CrossRef] [Green Version]
- Kishi-Itakura, C.; Koyama-Honda, I.; Itakura, E.; Mizushima, N. Ultrastructural analysis of autophagosome organization using mammalian autophagy-deficient cells. J. Cell Sci. 2014, 127, 4089–4102. [Google Scholar] [CrossRef] [Green Version]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef] [Green Version]
- Wong, V.K.; Li, T.; Law, B.Y.; Ma, E.D.; Yip, N.C.; Michelangeli, F.; Law, C.K.; Zhang, M.M.; Lam, K.Y.; Chan, P.L.; et al. Saikosaponin-d, a novel SERCA inhibitor, induces autophagic cell death in apoptosis-defective cells. Cell Death Dis. 2013, 4, e720. [Google Scholar] [CrossRef] [Green Version]
- Fan, M.; Gao, J.; Zhou, L.; Xue, W.; Wang, Y.; Chen, J.; Li, W.; Yu, Y.; Liu, B.; Shen, Y.; et al. Highly expressed SERCA2 triggers tumor cell autophagy and is a druggable vulnerability in triple-negative breast cancer. Acta Pharm. Sin. B. 2022, in press. [Google Scholar] [CrossRef]
- Zhao, Y.G.; Liu, N.; Miao, G.; Chen, Y.; Zhao, H.; Zhang, H. The ER Contact Proteins VAPA/B Interact with Multiple Autophagy Proteins to Modulate Autophagosome Biogenesis. Curr. Biol. 2018, 28, 1234–1245.e4. [Google Scholar] [CrossRef]
- De Vos, K.J.; Mórotz, G.M.; Stoica, R.; Tudor, E.L.; Lau, K.-F.; Ackerley, S.; Warley, A.; Shaw, C.E.; Miller, C.C.J. VAPB interacts with the mitochondrial protein PTPIP51 to regulate calcium homeostasis. Hum. Mol. Genet. 2012, 21, 1299–1311. [Google Scholar] [CrossRef] [Green Version]
- Mao, D.; Lin, G.; Tepe, B.; Zuo, Z.; Tan, K.L.; Senturk, M.; Zhang, S.; Arenkiel, B.R.; Sardiello, M.; Bellen, H.J. VAMP associated proteins are required for autophagic and lysosomal degradation by promoting a PtdIns4P-mediated endosomal pathway. Autophagy 2019, 15, 1214–1233. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhao, H.; Zhao, Y.G.; Hu, J.; Zhang, H. Atlastin 2/3 regulate ER targeting of the ULK1 complex to initiate autophagy. J. Cell Biol. 2021, 220, e202012091. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Chen, Y.; Chen, D.; Zhao, H.; Feng, Y.; Meng, Q.; Zhao, Y.; Zhang, H. Calcium transients on the ER surface trigger liquid-liquid phase separation of FIP200 to specify autophagosome initiation sites. Cell 2022, 185, 4082–4098.e22. [Google Scholar] [CrossRef] [PubMed]
- Csordás, G.; Renken, C.; Várnai, P.; Walter, L.; Weaver, D.; Buttle, K.F.; Balla, T.; Mannella, C.A.; Hajnóczky, G. Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 2006, 174, 915–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzuto, R.; Pinton, P.; Carrington, W.; Fay, F.S.; Fogarty, K.E.; Lifshitz, L.M.; Tuft, R.A.; Pozzan, T. Close Contacts with the Endoplasmic Reticulum as Determinants of Mitochondrial Ca2+ Responses. Science 1998, 280, 1763–1766. [Google Scholar] [CrossRef]
- Manganelli, V.; Matarrese, P.; Antonioli, M.; Gambardella, L.; Vescovo, T.; Gretzmeier, C.; Longo, A.; Capozzi, A.; Recalchi, S.; Riitano, G.; et al. Raft-like lipid microdomains drive autophagy initiation via AMBRA1-ERLIN1 molecular association within MAMs. Autophagy 2021, 17, 2528–2548. [Google Scholar] [CrossRef]
- Hailey, D.W.; Rambold, A.S.; Satpute-Krishnan, P.; Mitra, K.; Sougrat, R.; Kim, P.K.; Lippincott-Schwartz, J. Mitochondria Supply Membranes for Autophagosome Biogenesis during Starvation. Cell 2010, 141, 656–667. [Google Scholar] [CrossRef] [Green Version]
- Reggiori, F.; Shintani, T.; Chong, H.; Nair, U.; Klionsky, D.J. Atg9 Cycles Between Mitochondria and the Pre-Autophagosomal Structure in Yeasts. Autophagy 2005, 1, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER–mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Arasaki, K.; Tagaya, M. Legionella blocks autophagy by cleaving STX17 (syntaxin 17). Autophagy 2017, 13, 2008–2009. [Google Scholar] [CrossRef] [Green Version]
- Vats, S.; Manjithaya, R. A reversible autophagy inhibitor blocks autophagosome-lysosome fusion by preventing Stx17 loading onto autophagosomes. Mol. Biol. Cell 2019, 30, 2283–2295. [Google Scholar] [CrossRef]
- Itakura, E.; Kishi-Itakura, C.; Mizushima, N. The Hairpin-type Tail-Anchored SNARE Syntaxin 17 Targets to Autophagosomes for Fusion with Endosomes/Lysosomes. Cell 2012, 151, 1256–1269. [Google Scholar] [CrossRef] [Green Version]
- Diao, J.; Liu, R.; Rong, Y.; Zhao, M.; Zhang, J.; Lai, Y.; Zhou, Q.; Wilz, L.M.; Li, J.; Vivona, S.; et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 2015, 520, 563–566. [Google Scholar] [CrossRef]
- Vance, E.J. Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 1990, 265, 7248–7256. [Google Scholar] [CrossRef]
- Lahiri, S.; Toulmay, A.; Prinz, W.A. Membrane contact sites, gateways for lipid homeostasis. Curr. Opin. Cell Biol. 2015, 33, 82–87. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Suaga, P.; Paillusson, S.; Stoica, R.; Noble, W.; Hanger, D.P.; Miller, C.C.J. The ER-Mitochondria Tethering Complex VAPB-PTPIP51 Regulates Autophagy. Curr. Biol. 2017, 27, 371–385. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Javed, R.; Mudd, M.; Pallikkuth, S.; Lidke, K.A.; Jain, A.; Tangavelou, K.; Gudmundsson, S.R.; Ye, C.; Rusten, T.E.; et al. Mammalian hybrid pre-autophagosomal structure HyPAS generates autophagosomes. Cell 2021, 184, 5950–5969.e22. [Google Scholar] [CrossRef]
- Yuan, L.; Liu, Q.; Wang, Z.; Hou, J.; Xu, P. EI24 tethers endoplasmic reticulum and mitochondria to regulate autophagy flux. Cell. Mol. Life Sci. 2020, 77, 1591–1606. [Google Scholar] [CrossRef]
- Tian, Y.; Li, Z.; Hu, W.; Ren, H.; Tian, E.; Zhao, Y.; Lu, Q.; Huang, X.; Yang, P.; Li, X.; et al. C. elegans Screen Identifies Autophagy Genes Specific to Multicellular Organisms. Cell 2010, 141, 1042–1055. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Chen, H.; Zhang, L.; Lin, X.; Li, X.; Zhuang, H.; Fan, H.; Meng, T.; He, Z.; Huang, H.; et al. The AMPK-MFN2 axis regulates MAM dynamics and autophagy induced by energy stresses. Autophagy 2021, 17, 1142–1156. [Google Scholar] [CrossRef]
- Chen, H.C.; Detmer, S.A.; Ewald, A.J.; Griffin, E.E.; Fraser, S.E.; Chan, D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003, 160, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, T.; Matarrese, P.; Manganelli, V.; Marconi, M.; Tinari, A.; Gambardella, L.; Faggioni, A.; Misasi, R.; Sorice, M.; Malorni, W. Evidence for the involvement of lipid rafts localized at the ER-mitochondria associated membranes in autophagosome formation. Autophagy 2016, 12, 917–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matarrese, P.; Garofalo, T.; Manganelli, V.; Gambardella, L.; Marconi, M.; Grasso, M.; Tinari, A.; Misasi, R.; Malorni, W.; Sorice, M. Evidence for the involvement of GD3 ganglioside in autophagosome formation and maturation. Autophagy 2014, 10, 750–765. [Google Scholar] [CrossRef] [PubMed]
- Browman, D.T.; Resek, M.E.; Zajchowski, L.D.; Robbins, S. Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 2006, 119, 3149–3160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Bartolomeo, S.; Corazzari, M.; Nazio, F.; Oliverio, S.; Lisi, G.; Antonioli, M.; Pagliarini, V.; Matteoni, S.; Fuoco, C.; Giunta, L.; et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J. Cell Biol. 2010, 191, 155–168. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Wan, D.; Qian, Q.; Yi, B.; He, Z.; Gu, Y.; Wang, L.; He, S. Ambra1 Is an Essential Regulator of Autophagy and Apoptosis in SW620 Cells: Pro-Survival Role of Ambra1. PLoS ONE 2014, 9, e90151. [Google Scholar] [CrossRef] [Green Version]
- Böckler, S.; Westermann, B. Mitochondrial ER Contacts Are Crucial for Mitophagy in Yeast. Dev. Cell 2014, 28, 450–458. [Google Scholar] [CrossRef] [Green Version]
- Gelmetti, V.; De Rosa, P.; Torosantucci, L.; Marini, E.S.; Romagnoli, A.; Di Rienzo, M.; Arena, G.; Vignone, D.; Fimia, G.M.; Valente, E.M. PINK1 and BECN1 relocalize at mitochondria-associated membranes during mitophagy and promote ER-mitochondria tethering and autophagosome formation. Autophagy 2017, 13, 654–669. [Google Scholar] [CrossRef] [Green Version]
- Eiyama, A.; Okamoto, K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr. Opin. Cell Biol. 2015, 33, 95–101. [Google Scholar] [CrossRef]
- Puri, R.; Cheng, X.-T.; Lin, M.-Y.; Huang, N.; Sheng, Z.-H. Mul1 restrains Parkin-mediated mitophagy in mature neurons by maintaining ER-mitochondrial contacts. Nat. Commun. 2019, 10, 3645. [Google Scholar] [CrossRef] [Green Version]
- McLelland, G.-L.; Goiran, T.; Yi, W.; Dorval, G.; Chen, C.X.; Lauinger, N.D.; Krahn, A.I.; Valimehr, S.; Rakovic, A.; Rouiller, I.; et al. Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. eLife 2018, 7, e32866. [Google Scholar] [CrossRef]
- Sarraf, S.A.; Raman, M.; Guarani-Pereira, V.; Sowa, M.E.; Huttlin, E.L.; Gygi, S.P.; Harper, J.W. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 2013, 496, 372–376. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, A.; Cleland, M.M.; Xu, S.; Narendra, D.P.; Suen, D.-F.; Karbowski, M.; Youle, R.J. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 2010, 191, 1367–1380. [Google Scholar] [CrossRef]
- Stefan, C.J.; Manford, A.; Emr, S.D. ER–PM connections: Sites of information transfer and inter-organelle communication. Curr. Opin. Cell Biol. 2013, 25, 434–442. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, B.; Moreau, K.; Jahreiss, L.; Puri, C.; Rubinsztein, D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010, 12, 747–757. [Google Scholar] [CrossRef]
- Nascimbeni, A.C.; Giordano, F.; Dupont, N.; Grasso, D.; Vaccaro, I.M.; Codogno, P.; Morel, E. ER –plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI 3P synthesis. EMBO J. 2017, 36, 2018–2033. [Google Scholar] [CrossRef]
- Molejon, M.I.; Ropolo, A.; Re, A.L.; Boggio, V.; Vaccaro, M.I. The VMP1-Beclin 1 interaction regulates autophagy induction. Sci. Rep. 2013, 3, 1055. [Google Scholar] [CrossRef] [Green Version]
- Molejon, M.I.; Ropolo, A.; Vaccaro, M.I. VMP1 is a new player in the regulation of the autophagy-specific phosphatidylinositol 3-kinase complex activation. Autophagy 2013, 9, 933–935. [Google Scholar] [CrossRef] [Green Version]
- Gatica, D.; Wen, X.; Cheong, H.; Klionsky, D.J. Vac8 determines phagophore assembly site vacuolar localization during nitrogen starvation-induced autophagy. Autophagy 2021, 17, 1636–1648. [Google Scholar] [CrossRef]
- Hollenstein, D.M.; Gómez-Sánchez, R.; Ciftci, A.; Kriegenburg, F.; Mari, M.; Torggler, R.; Licheva, M.; Reggiori, F.; Kraft, C. Vac8 spatially confines autophagosome formation at the vacuole in S. cerevisiae. J. Cell Sci. 2019, 132, jcs235002. [Google Scholar] [CrossRef] [Green Version]
- Stjepanovic, G.; Davies, C.W.; Stanley, R.E.; Ragusa, M.J.; Kim, D.J.; Hurley, J.H. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc. Natl. Acad. Sci. USA 2014, 111, 12793–12798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boutouja, F.; Stiehm, C.M.; Reidick, C.; Mastalski, T.; Brinkmeier, R.; El Magraoui, F.; Platta, H.W. Vac8 Controls Vacuolar Membrane Dynamics during Different Autophagy Pathways in Saccharomyces cerevisiae. Cells 2019, 8, 661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munzel, L.; Neumann, P.; Otto, F.B.; Krick, R.; Metje-Sprink, J.; Kroppen, B.; Karedla, N.; Enderlein, J.; Meinecke, M.; Ficner, R.; et al. Atg21 organizes Atg8 lipidation at the contact of the vacuole with the phagophore. Autophagy 2021, 17, 1458–1478. [Google Scholar] [CrossRef] [PubMed]
- Scott, S.V.; Nice, D.C.; Nau, J.J.; Weisman, L.S.; Kamada, Y.; Keizer-Gunnink, I.; Funakoshi, T.; Veenhuis, M.; Ohsumi, Y.; Klionsky, D.J. Apg13p and Vac8p Are Part of a Complex of Phosphoproteins That Are Required for Cytoplasm to Vacuole Targeting. J. Biol. Chem. 2000, 275, 25840–25849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lei, Y.; Zhang, X.; Xu, Q.; Liu, S.; Li, C.; Jiang, H.; Lin, H.; Kong, E.; Liu, J.; Qi, S.; et al. Autophagic elimination of ribosomes during spermiogenesis provides energy for flagellar motility. Dev. Cell 2021, 56, 2313–2328.e7. [Google Scholar] [CrossRef]
- Lei, Y.; Zhang, X.; Xu, Q.; Liu, S.; Li, C.; Jiang, H.; Lin, H.; Kong, E.; Liu, J.; Qi, S.; et al. A conserved Vac8/ARMC3-PtdIns3K-CI cascade regulates autophagy initiation and functions in spermiogenesis by promoting ribophagy. Autophagy 2021, 17, 4512–4514. [Google Scholar] [CrossRef]
- Hollenstein, D.M.; Licheva, M.; Konradi, N.; Schweida, D.; Mancilla, H.; Mari, M.; Reggiori, F.; Kraft, C. Spatial control of avidity regulates initiation and progression of selective autophagy. Nat. Commun. 2021, 12, 7194. [Google Scholar] [CrossRef]
- Kvam, E.; Goldfarb, D. Nucleus–vacuole junctions in yeast: Anatomy of a membrane contact site. Biochem. Soc. Trans. 2006, 34, 340–342. [Google Scholar] [CrossRef] [Green Version]
- Gatica, D.; Lahiri, V.; Klionsky, D.J. Cargo recognition and degradation by selective autophagy. Nature Cell Biol. 2018, 20, 233–242. [Google Scholar] [CrossRef]
- Otto, F.B.; Thumm, M. Mechanistic dissection of macro- and micronucleophagy. Autophagy 2021, 17, 626–639. [Google Scholar] [CrossRef]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal Microautophagy of Nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Mochida, K.; Oikawa, Y.; Kimura, Y.; Kirisako, H.; Hirano, H.; Ohsumi, Y.; Nakatogawa, H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 2015, 522, 359–362. [Google Scholar] [CrossRef]
- Pan, X.; Roberts, P.; Chen, Y.; Kvam, E.; Shulga, N.; Huang, K.; Lemmon, S.; Goldfarb, D.S. Nucleus–Vacuole Junctions in Saccharomyces cerevisiae Are Formed Through the Direct Interaction of Vac8p with Nvj1p. Mol. Biol. Cell 2000, 11, 2445–2457. [Google Scholar] [CrossRef]
- Kvam, E.; Goldfarb, D.S. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 2004, 117, 4959–4968. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.; Park, J.; Kim, H.-I.; Lee, M.; Ko, Y.-J.; Lee, S.; Jun, Y.; Lee, C. Mechanistic insight into the nucleus–vacuole junction based on the Vac8p–Nvj1p crystal structure. Proc. Natl. Acad. Sci. USA 2017, 114, E4539–E4548. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Kim, H.-I.; Jeong, H.; Lee, M.; Jang, S.H.; Yoon, S.Y.; Park, Z.-Y.; Jun, Y.; Lee, C. Quaternary structures of Vac8 differentially regulate the Cvt and PMN pathways. Autophagy 2020, 16, 991–1006. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zwilling, E.; Reggiori, F. Membrane Contact Sites in Autophagy. Cells 2022, 11, 3813. https://doi.org/10.3390/cells11233813
Zwilling E, Reggiori F. Membrane Contact Sites in Autophagy. Cells. 2022; 11(23):3813. https://doi.org/10.3390/cells11233813
Chicago/Turabian StyleZwilling, Emma, and Fulvio Reggiori. 2022. "Membrane Contact Sites in Autophagy" Cells 11, no. 23: 3813. https://doi.org/10.3390/cells11233813
APA StyleZwilling, E., & Reggiori, F. (2022). Membrane Contact Sites in Autophagy. Cells, 11(23), 3813. https://doi.org/10.3390/cells11233813






