Sorting and Manipulation of Human PGC-LC Using PDPN and Hanging Drop Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Culture of Naïve-like hiPSCs
2.2. In Vitro Differentiation of Early and Late hPGC-LCs
2.3. Collection of Human Fetal Ovary
2.4. Collection of Mouse Fetal Ovary
2.5. Real-Time Quantitative RT-PCR
2.6. Single Cell RNA Seq
2.7. Immunohistochemistry
2.8. Fluorescence-Activated Cell Sorting
2.9. Aggregate Cultures
2.10. Statistical Analyses
2.11. Data Availability
3. Results
3.1. The In Vitro Specification of hPGC-LCs from Naïve-like hiPSCs
3.2. Single-Cell Transcriptomic Analysis of PGC-LC Reveals Active BMP-Signaling
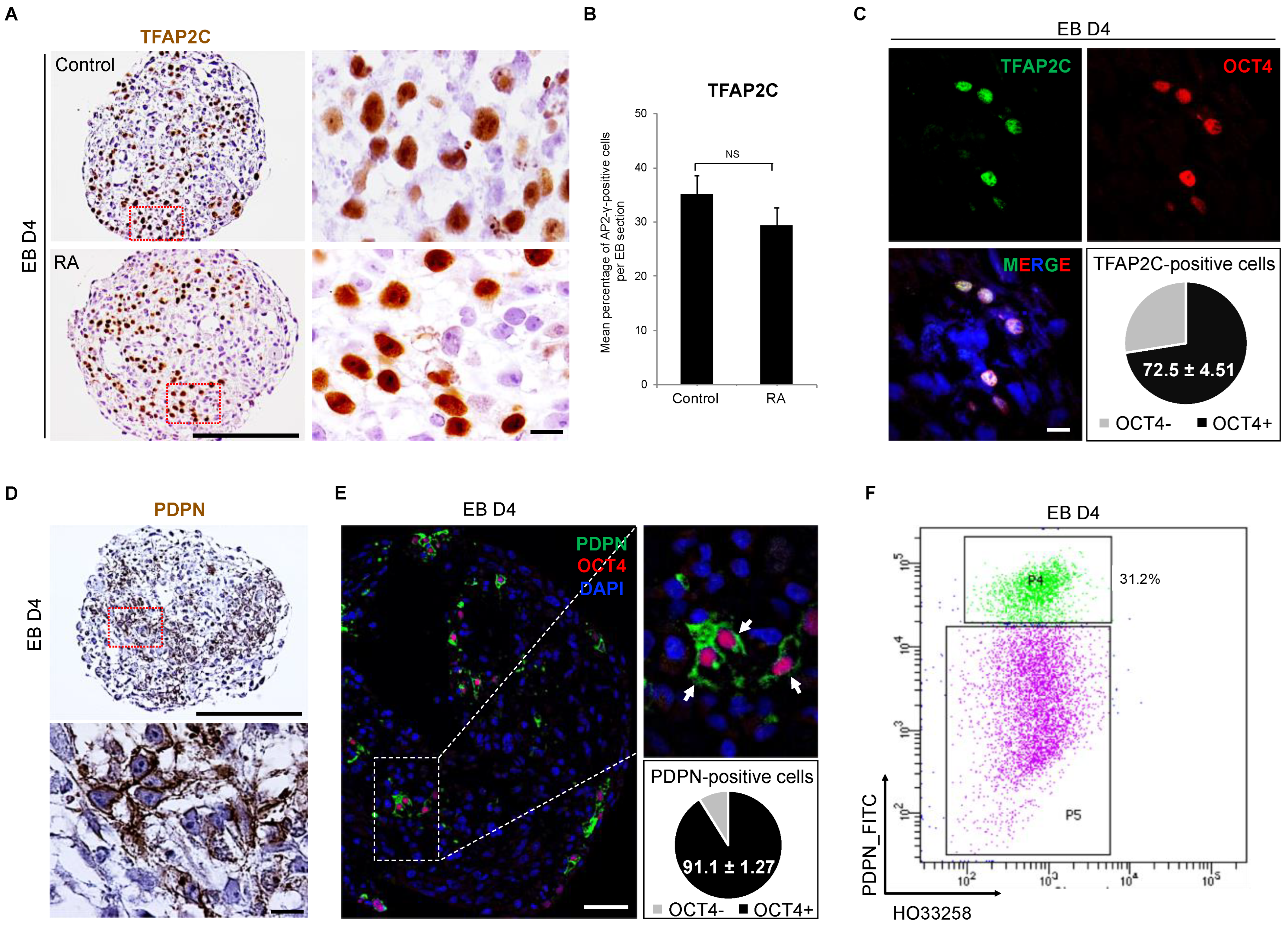
3.3. Detection of Early PGC Protein Markers, TFAP2C and PDPN, in Day 4 Embryoid Bodies
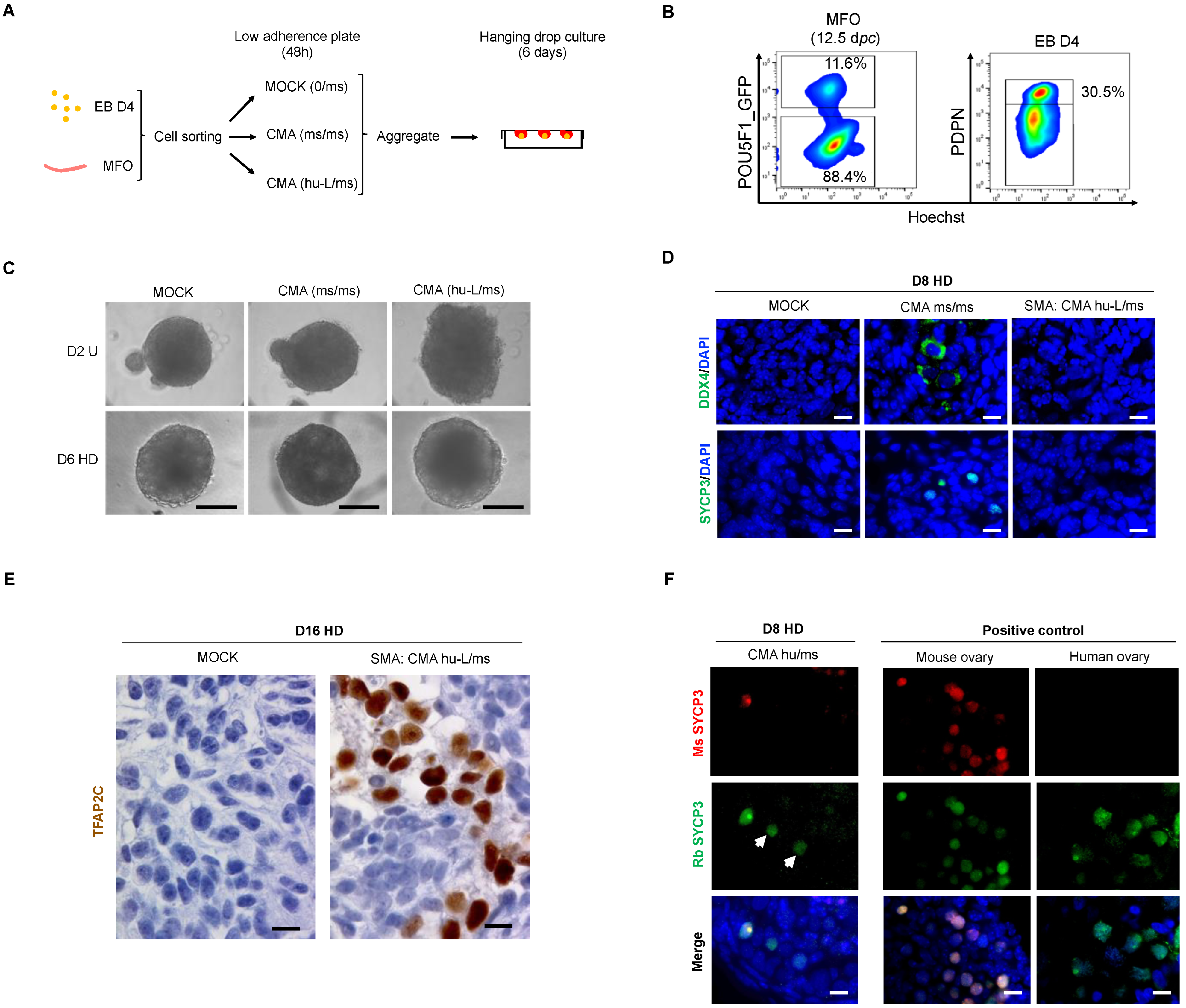
3.4. hPGC-LCs Remain Arrested at Early Stages When Cultured with Human Ovarian Somatic Cells
3.5. The Somatic Environment of Mouse Fetal Ovary Is Unpermissive for hPGC-LC Maturation and Meiotic Onset
3.6. In Vitro Overexpression of Late PGC and Meiotic Gene Markers in Hanging Drop Transferred-EBs
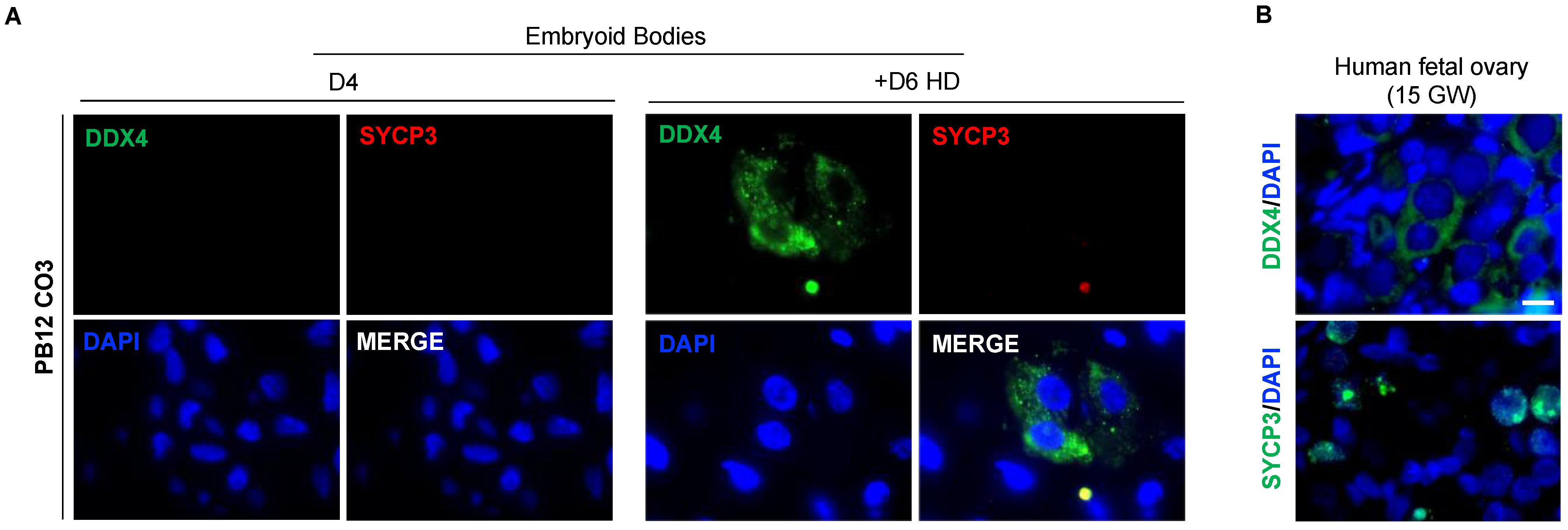
3.7. The In Vitro Induced Germ Cell-like Cells in +D6 HD EBs Express DDX4 but Not SYCP3 Protein
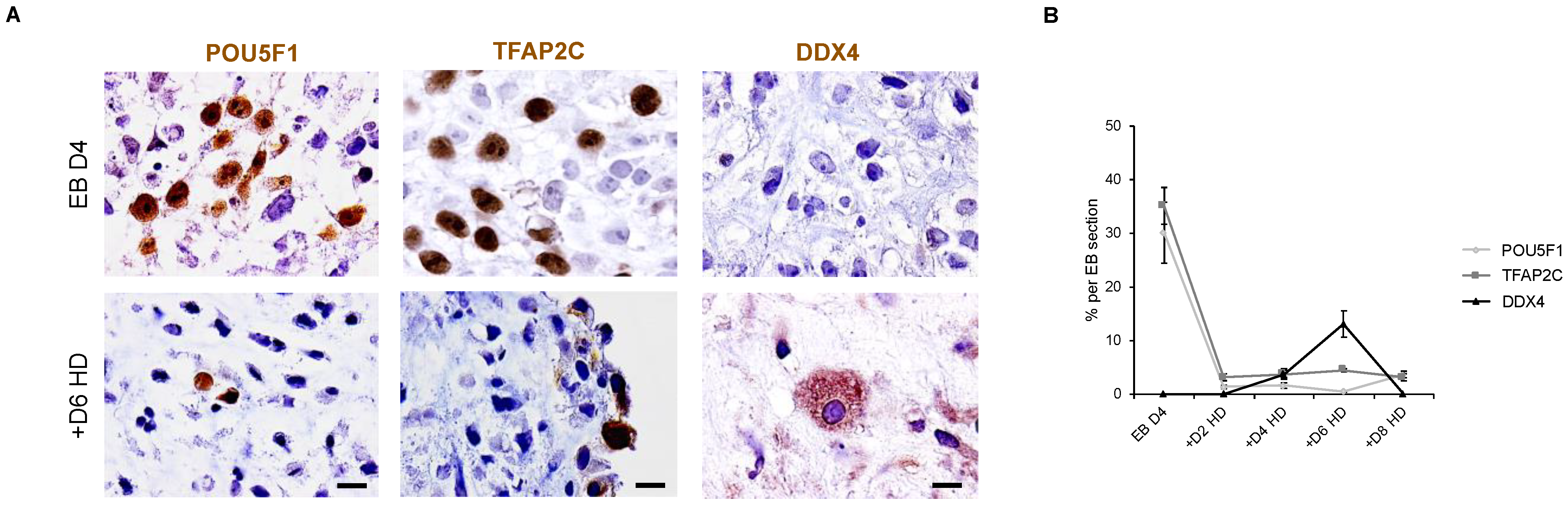
3.8. Down Expression of POU5F1 and TFAP2C Precedes the Apparition of DDX4-Positive Cells in Hanging Drop EBs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Menke, D.-B.; Koubova, J.; Page, D.-C. Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev. Biol. 2003, 262, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Bullejos, M.; Koopman, P. Germ cells enter meiosis in a rostro-caudal wave during development of the mouse ovary. Mol. Reprod. Dev. 2004, 68, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Ungewitter, E.-K.; Yao, H.-H. How to make a gonad: Cellular mechanisms governing formation of the testes and ovaries. Sex. Dev. 2013, 7, 7–20. [Google Scholar] [CrossRef]
- Frydman, N.; Poulain, M.; Arkoun, B.; Duquenne, C.; Tourpin, S.; Messiaen, S.; Habert, R.; Rouiller-Fabre, V.; Benachi, A.; Livera, G. Human foetal ovary shares meiotic preventing factors with the developing testis. Hum. Reprod. 2017, 32, 631–642. [Google Scholar] [CrossRef] [PubMed]
- Haston, K.M.; Tung, J.Y.; Reijo Pera, R.A. Dazl Functions in Maintenance of Pluripotency and Genetic and Epigenetic Programs of Differentiation in Mouse Primordial Germ Cells In Vivo and In Vitro. PLoS ONE 2009, 4, e5654. [Google Scholar] [CrossRef]
- Heeren, A.M.; He, N.; de Souza, A.F.; Goercharn-Ramlal, A.; van Iperen, L.; Roost, M.S.; Gomes Fernandes, M.M.; van der Westerlaken, L.A.; Chuva de Sousa Lopes, S.M. On the development of extragonadal and gonadal human germ cells. Biol. Open. 2016, 5, 185–194. [Google Scholar] [CrossRef]
- Endo, T.; Mikedis, M.M.; Nicholls, P.K.; Page, D.C.; de Rooij, D.G. Retinoic Acid and Germ Cell Development in the Ovary and Testis. Biomolecules 2019, 9, 775. [Google Scholar] [CrossRef]
- Vernet, N.; Condrea, D.; Mayere, C.; Féret, B.; Klopfenstein, M.; Magnant, W.; Alunni, V.; Teletin, M.; Souali-Crespo, S.; Nef, S.; et al. Meiosis occurs normally in the fetal ovary of mice lacking all retinoic acid receptors. Sci. Adv. 2020, 6, eaaz1139. [Google Scholar] [CrossRef]
- Miyauchi, H.; Ohta, H.; Nagaoka, S.; Nakaki, F.; Sasaki, K.; Hayashi, K.; Yabuta, Y.; Nakamura, T.; Yamamoto, T.; Saitou, M. Bone morphogenetic protein and retinoic acid synergistically specify female germ-cell fate in mice. EMBO J. 2017, 36, 3100–3119. [Google Scholar] [CrossRef]
- Imamura, M.; Aoi, T.; Tokumasu, A.; Mise, N.; Abe, K.; Yamanaka, S.; Noce, T. Induction of primordial germ cells from mouse induced pluripotent stem cells derived from adult hepatocytes. Mol. Reprod. Dev. 2010, 77, 802–811. [Google Scholar] [CrossRef]
- Hayashi, K.; Ogushi, S.; Kurimoto, K.; Shimamoto, S.; Ohta, H.; Saitou, M. Offspring from oocytes derived from in vitro primordial germ cell-like cells in mice. Science 2012, 338, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Hikabe, O.; Hamazaki, N.; Nagamatsu, G.; Obata, Y.; Hirao, Y.; Hamada, N.; Shimamito, S.; Imamura, T.; Nakashima, K.; Saitou, M.; et al. Reconstitution in vitro of the entire cycle of the mouse female germ line. Nature 2016, 539, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Gafni, O.; Weinberger, L.; Mansour, A.-A.; Manor, Y.-S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Irie, N.; Weinberger, L.; Tang, W.-W.; Kobayashi, T.; Viukov, S.; Manor, Y.-S.; Dietmann, S.; Hanna, J.-H.; Surani, M.-A. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015, 160, 253–268. [Google Scholar] [CrossRef]
- Abdyyev, V.K.; Sant, D.W.; Kiseleva, E.V.; Spangenberg, V.E.; Kolomiets, O.L.; Andrade, N.S.; Dashinimaev, E.B.; Vorotelyak, E.A.; Vasiliev, A.V. In vitro derived female hPGCLCs are unable to complete meiosis in embryoid bodies. Exp. Cell Res. 2020, 397, 112358. [Google Scholar] [CrossRef]
- Yamashiro, C.; Sasaki, K.; Yabuta, Y.; Kojima, Y.; Nakamura, T.; Okamoto, I.; Yokobayashi, S.; Murase, Y.; Ishikura, Y.; Shirane, K.; et al. Generation of human oogonia from induced pluripotent stem cells in vitro. Science 2018, 362, 356–360. [Google Scholar] [CrossRef]
- Ugorski, M.; Dziegiel, P.; Suchanski, J. Podoplanin—A small glycoprotein with many faces. Am. J. Cancer. Res. 2016, 6, 370–386. [Google Scholar]
- Gomes Fernandes, M.; Bialecka, M.; Salvatori, D.C.F.; Chuva de Sousa Lopes, S.M. Characterization of migratory primordial germ cells in the aorta-gonad-mesonephros of a 4.5-week-old human embryo: A toolbox to evaluate in vitro early gametogenesis. Mol. Hum. Reprod. 2018, 24, 233–243. [Google Scholar] [CrossRef]
- Guerquin, M.J.; Duquenne, C.; Coffigny, H.; Rouiller-Fabre, V.; Lambrot, R.; Bakalska, M.; Frydman, R.; Livera, G. Sex-specific differences in fetal germ cell apoptosis induced by ionizing radiation. Hum. Reprod. 2009, 24, 670–678. [Google Scholar] [CrossRef]
- Evtouchenko, L.; Studer, L.; Spenger, C.; Dreher, E.; Seiler, R.-W. A mathematical model for the estimation of human embryonic and fetal age. Cell. Transplant. 1996, 5, 453–464. [Google Scholar] [CrossRef]
- Yoshimizu, T.; Sugiyama, N.; De Felice, M.; Yeom, Y.-I.; Ohbo, K.; Masuko, K.; Obinata, M.; Abe, K.; Schöler, H.R.; Matsui, Y. Germline-specific expression of the Oct-4/green fluorescent protein (GFP) transgene in mice. Dev. Growth. Differ. 1999, 41, 675–684. [Google Scholar] [CrossRef] [PubMed]
- Guerquin, M.J.; Duquenne, C.; Lahaye, J.-B.; Tourpin, S.; Habert, R.; Livera, G. New testicular mechanisms involved in the prevention of fetal meiotic initiation in mice. Dev. Biol. 2010, 346, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., 3rd; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- Davidson, K.C.; Mason, E.A.; Pera, M.F. The pluripotent state in mouse and human. Development 2015, 142, 3090–3099. [Google Scholar] [CrossRef]
- Anderson, E.L.; Baltus, A.E.; Roepers-Gajadien, H.L.; Hassold, T.J.; de Rooij, D.G.; van Pelt, A.M.; Page, D.-C. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. Proc. Natl. Acad. Sci. USA 2008, 105, 14976–14980. [Google Scholar] [CrossRef]
- Muczynski, V.; Lecureuil, C.; Messiaen, S.; Guerquin, M.J.; N’tumba-Byn, T.; Moison, D.; Hodroj, W.; Benjelloun, H.; Baijer, J.; Livera, G.; et al. Cellular and Molecular Effect of MEHP Involving LXRα in Human Fetal Testis and Ovary. PLoS ONE 2012, 7, e48266. [Google Scholar] [CrossRef]
- Gomes Fernandes, M.; He, N.; Wang, F.; Van Iperen, L.; Eguizabal, C.; Matorras, R.; Roelen, B.A.J.; Chuva De Sousa Lopes, S.M. Human-specific subcellular compartmentalization of P-element induced wimpy testis-like (PIWIL) granules during germ cell development and spermatogenesis. Hum. Reprod. 2018, 33, 258–269. [Google Scholar] [CrossRef]
- Seita, Y.; Cheng, K.; McCarrey, J.R.; Yadu, N.; Cheeseman, I.; Bagwell, A.; Corinna, N.R.; Santana-Toro, I.; Yen, L.H.; Vargas, S.; et al. Efficient generation of marmoset primordial germ cell-like cells using induced pluripotent stem cells. bioRxiv 2022. bioRxiv: 2022.07.25.501382. [Google Scholar] [CrossRef]
- Bendsen, E.; Byskov, A.G.; Andersen, C.Y.; Westergaard, L.G. Number of germ cells and somatic cells in human fetal ovaries during the first weeks after sex differentiation. Hum. Reprod. 2006, 21, 30–35. [Google Scholar] [CrossRef]
- Le Bouffant, R.; Guerquin, M.J.; Duquenne, C.; Frydman, N.; Cofigny, H.; Rouiller-Fabre, V.; Frydman, R.; Habert, R.; Livera, G. Meiosis initiation in the human ovary requires intrinsic retinoic acid synthesis. Hum. Reprod. 2010, 25, 2579–2590. [Google Scholar] [CrossRef]
- Kee, K.; Angeles, V.-T.; Flores, M.; Nguyen, H.N.; Reijo Pera, R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 2009, 462, 222–225. [Google Scholar] [CrossRef] [PubMed]
- Panula, S.; Medrano, J.V.; Kee, K.; Bergström, R.; Nguyen, H.N.; Byers, B.; Wilson, K.D.; Wu, J.C.; Simon, C.; Hovatta, O.; et al. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011, 20, 752–762. [Google Scholar] [CrossRef]
- Medrano, J.V.; Ramathal, C.; Nguyen, H.N.; Simon, C.; Reijo Pera, R.A. Divergent RNA-binding proteins, DAZL and VASA, induce meiotic progression in human germ cells derived in vitro. Stem Cells 2012, 30, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Ruggiu, M.; Speed, R.; Taggart, M.; McKay, S.J.; Kilanowski, F.; Saunders, P.; Dorin, J.; Cooke, H.J. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997, 389, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Schrans-Stassen, B.H.; Saunders, P.T.; Cooke, H.J.; de Rooij, D.G. Nature of the spermatogenic arrest in Dazl−/− mice. Biol. Reprod. 2001, 65, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Yokobayashi, S.; Nakamura, T.; Okamoto, I.; Yabuta, Y.; Kurimoto, K.; Ohta, H.; Moritoki, Y.; Iwatani, C.; Tsuchiya, H.; et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell. 2015, 17, 178–194. [Google Scholar] [CrossRef]
- Saunders, P.T.; Turner, J.M.; Ruggiu, M.; Taggart, M.; Burgoyne, P.S.; Elliott, D.; Cooke, H.-J. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction 2003, 126, 589–597. [Google Scholar] [CrossRef]
- Nagaoka, S.I.; Nakaki, F.; Miyauchi, H.; Nosaka, Y.; Ohta, H.; Yabuta, Y.; Kurimoto, K.; Hayashi, K.; Nakamura, T.; Yamamoto, T.; et al. ZGLP1 is a determinant for the oogenic fate in mice. Science 2020, 367, eaaw4115. [Google Scholar] [CrossRef]
- Hayashi, K.; Ohta, H.; Kurimoto, K.; Aramaki, S.; Saitou, M. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011, 146, 519–532. [Google Scholar] [CrossRef]
- Hackett, J.A.; Huang, Y.; Günesdogan, U.; Gretarsson, K.A.; Kobayashi, T.; Surani, M.A. Tracing the transitions from pluripotency to germ cell fate with CRISPR screening. Nat. Commun. 2018, 9, 4292. [Google Scholar] [CrossRef]
- Leverrier-Penna, S.; Mitchell, R.T.; Becker, E.; Lecante, L.; Ben Maamar, M.; Homer, N.; Lavoué, V.; Kristensen, D.M.; Dejucq-Rainsford, N.; Jégou, B.; et al. Ibuprofen is deleterious for the development of first trimester human fetal ovary ex vivo. Hum. Reprod. 2018, 33, 482–493. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.W.; Bowles, J.; Koopman, P. Control of mammalian germ cell entry into meiosis. Mol. Cell. Endocrinol. 2014, 382, 488–497. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, A.; Nielsen, J.E.; Perlman, S.; Lundvall, L.; Mitchell, R.T.; Juul, A.; Rajpert-De Meyts, E. Ex vivo culture of human fetal gonads: Manipulation of meiosis signalling by retinoic acid treatment disrupts testis development. Hum. Reprod. 2015, 30, 2351–2363. [Google Scholar] [CrossRef] [PubMed]
- Bellutti, L.; Abby, E.; Tourpin, S.; Messiaen, S.; Moison, D.; Trautmann, E.; Guerquin, M.J.; Rouiller-Fabre, V.; Habert, R.; Livera, G. Divergent Roles of CYP26B1 and Endogenous Retinoic Acid in Mouse Fetal Gonads. Biomolecules 2019, 9, 536. [Google Scholar] [CrossRef]
- Chassot, A.A.; Le Rolle, M.; Jolivet, G.; Stevant, I.; Guigonis, J.M.; Da Silva, F.; Nef, S.; Pailhoux, E.; Schedl, A.; Ghyselinck, N.B.; et al. Retinoic acid synthesis by ALDH1A proteins is dispensable for meiosis initiation in the mouse fetal ovary. Sci. Adv. 2020, 6, eaaz1261. [Google Scholar] [CrossRef]
- Yoshino, T.; Suzuki, T.; Nagamatsu, G.; Yabukami, H.; Ikegaya, M.; Kishima, M.; Kita, H.; Imamura, T.; Nakashima, K.; Nishinakamura, R.; et al. Generation of ovarian follicles from mouse pluripotent stem cells. Science 2021, 373, eabe0237. [Google Scholar] [CrossRef]
- Wang, H.; Liu, L.; Liu, C.; Wang, L.; Chen, J.; Wang, H.; Heng, D.; Zeng, M.; Liu, C.; Zhou, Z.; et al. Induction of meiosis by embryonic gonadal somatic cells differentiated from pluripotent stem cells. Stem. Cell. Res. Ther. 2021, 12, 607. [Google Scholar] [CrossRef]
- Gonen, N.; Eozenou, C.; Mitter, R.; Bernardo, A.; Chervova, A.; Frachon, E.; Commère, P.H.; Mazen, I.; McElreavey, S.G.K.; Lovell-Badge, R.; et al. In-vitro cellular reprogramming to model gonad development and its disorders. bioRxiv 2021. bioRxiv: 2021.10.22.465384. [Google Scholar] [CrossRef]
- Hamazaki, N.; Kyogoku, H.; Araki, H.; Miura, F.; Horikawa, C.; Hamada, N.; Shimamoto, S.; Hikabe, O.; Nakashima, K.; Kitajima, T.S.; et al. Reconstitution of the oocyte transcriptional network with transcription factors. Nature 2021, 589, 264–269. [Google Scholar] [CrossRef]
- Mizuta, K.; Katou, Y.; Nakakita, B.; Kishine, A.; Nosaka, Y.; Saito, S.; Iwatani, C.; Tsuchiya, H.; Kawamoto, I.; Nakaya, M.; et al. Ex vivo reconstitution of fetal oocyte development in humans and cynomolgus monkeys. EMBO. J. 2022, 41, e110815. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arkoun, B.; Moison, P.; Guerquin, M.-J.; Messiaen, S.; Moison, D.; Tourpin, S.; Monville, C.; Livera, G. Sorting and Manipulation of Human PGC-LC Using PDPN and Hanging Drop Cultures. Cells 2022, 11, 3832. https://doi.org/10.3390/cells11233832
Arkoun B, Moison P, Guerquin M-J, Messiaen S, Moison D, Tourpin S, Monville C, Livera G. Sorting and Manipulation of Human PGC-LC Using PDPN and Hanging Drop Cultures. Cells. 2022; 11(23):3832. https://doi.org/10.3390/cells11233832
Chicago/Turabian StyleArkoun, Brahim, Pauline Moison, Marie-Justine Guerquin, Sébastien Messiaen, Delphine Moison, Sophie Tourpin, Christelle Monville, and Gabriel Livera. 2022. "Sorting and Manipulation of Human PGC-LC Using PDPN and Hanging Drop Cultures" Cells 11, no. 23: 3832. https://doi.org/10.3390/cells11233832
APA StyleArkoun, B., Moison, P., Guerquin, M.-J., Messiaen, S., Moison, D., Tourpin, S., Monville, C., & Livera, G. (2022). Sorting and Manipulation of Human PGC-LC Using PDPN and Hanging Drop Cultures. Cells, 11(23), 3832. https://doi.org/10.3390/cells11233832





