Single-Cell RNA-Seq Reveals a Crosstalk between Hyaluronan Receptor LYVE-1-Expressing Macrophages and Vascular Smooth Muscle Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Processing, Cell Staining and Flow Cytometry
2.3. Single-Cell RNA-Sequencing
2.4. scRNAseq Data Analysis
2.5. Human Samples
2.6. The Multiplex Immunoassay
2.7. CCL24 Human ELISA
2.8. Oil Red O Staining for Lipid Content
2.9. Immunohistochemistry
2.10. Immunofluorescent Staining and Quantification
2.11. Tissue Processing, Flow Cytometry, and Cell Sorting
2.12. The Quantification of VSMC Osteoblast-like Cell Transdifferentiation
2.13. Statistical Analyses
3. Results
3.1. The Site-Specific Development of Atherosclerosis Defines the Hypercholesteremia-Associated Transcriptional Signature
3.2. Atherosclerosis-Associated Immune Cell Populations Revealed by scRNAseq
3.3. Atherosclerosis Cluster Gene Expression Signatures
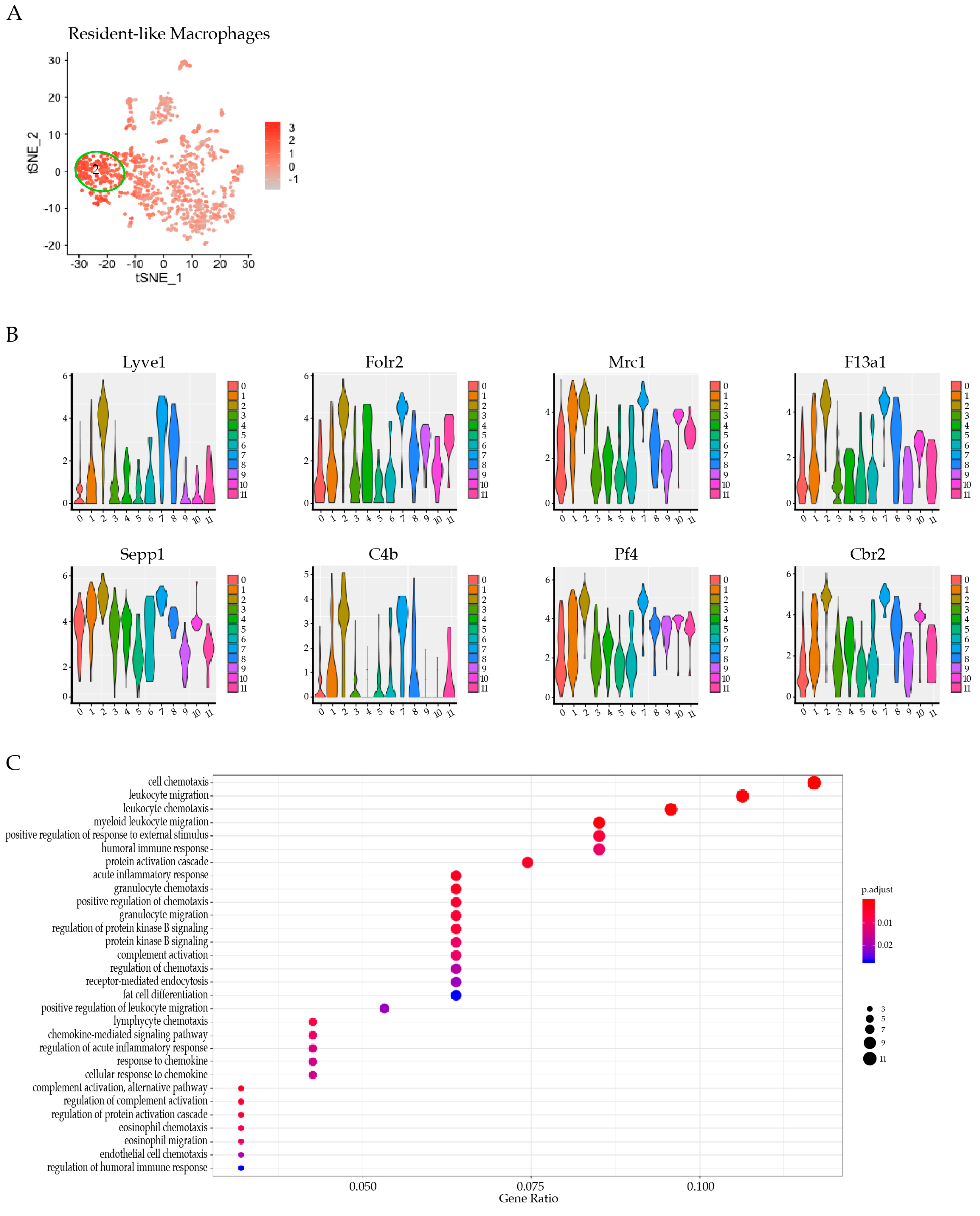
3.4. Res-like Macrophages Populated the Aortic Arch and Roots in Atherosclerosis
3.5. LYVE-1 Res-like Macrophages Expanded in Murine Atherosclerotic Lesions
3.6. LYVE-1 Res-like Macrophages Promoted a VSMC Phenotypic Switch
3.7. LYVE-1 Res-like Macrophage Accumulation was Associated with Plaque Ruptures in Human Carotid Artery Disease
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dickhout, J.G.; Basseri, S.; Austin, R.C. Macrophage Function and Its Impact on Atherosclerotic Lesion Composition, Progression, and Stability. Arter. Thromb. Vasc. Biol. 2008, 28, 1413–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cochain, C.; Vafadarnejad, E.; Arampatzi, P.; Pelisek, J.; Winkels, H.; Ley, K.; Wolf, D.; Saliba, A.-E.; Zernecke, A. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ. Res. 2018, 122, 1661–1674. [Google Scholar] [CrossRef] [PubMed]
- Zernecke, A.; Winkels, H.; Cochain, C.; Williams, J.W.; Wolf, D.; Soehnlein, O.; Robbins, C.S.; Monaco, C.; Park, I.; McNamara, C.A.; et al. Meta-Analysis of Leukocyte Diversity in Atherosclerotic Mouse Aortas. Circ. Res. 2020, 127, 402–426. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B.B.; Siddiqui, A.; et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 2009, 6, 377–382. [Google Scholar] [CrossRef]
- Lim, H.Y.; Lim, S.Y.; Tan, C.K.; Thiam, C.H.; Goh, C.C.; Carbajo, D.; Chew, S.H.S.; See, P.; Chakarov, S.; Wang, X.N.; et al. Hyaluronan Receptor LYVE-1-Expressing Macrophages Maintain Arterial Tone through Hyaluronan-Mediated Regulation of Smooth Muscle Cell Collagen. Immunity 2018, 49, 326–341. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Shim, D.; Lee, J.S.; Zaitsev, K.; Williams, J.; Kim, K.-W.; Jang, M.-Y.; Jang, H.S.; Yun, T.J.; Lee, S.H.; et al. Transcriptome Analysis Reveals Nonfoamy Rather Than Foamy Plaque Macrophages Are Proinflammatory in Atherosclerotic Murine Models. Circ. Res. 2018, 123, 1127–1142. [Google Scholar] [CrossRef]
- Willemsen, L.; De Winther, M.P. Macrophage subsets in atherosclerosis as defined by single-cell technologies. J. Pathol. 2020, 250, 705–714. [Google Scholar] [CrossRef] [Green Version]
- Durham, A.L.; Speer, M.Y.; Scatena, M.; Giachelli, C.M.; Shanahan, C.M. Role of smooth muscle cells in vascular calcification: Implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 2018, 114, 590–600. [Google Scholar] [CrossRef]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef]
- Shankman, L.S.; Gomez, D.; Cherepanova, O.A.; Salmon, M.; Alencar, G.F.; Haskins, R.M.; Swiatlowska, P.; Newman, A.A.C.; Greene, E.S.; Straub, A.C.; et al. Corrigendum: KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat. Med. 2016, 22, 217. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Sun, Z.; Zhang, L.; Yan, J.; Shao, C.; Jing, L.; Li, L.; Wang, Z. Role of Macrophages in the Progression and Regression of Vascular Calcification. Front. Pharmacol. 2020, 11, 661. [Google Scholar] [CrossRef] [PubMed]
- Montecucco, F.; Vuilleumier, N.; Pagano, S.; Lenglet, S.; Bertolotto, M.B.; Braunersreuther, V.; Pelli, G.; Kovari, E.; Pane, B.; Spinella, G.; et al. Anti-Apolipoprotein A-1 auto-antibodies are active mediators of atherosclerotic plaque vulnerability. Eur. Hear. J. 2011, 32, 412–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braunersreuther, V.; Zernecke, A.; Arnaud, C.; Liehn, E.A.; Steffens, S.; Shagdarsuren, E.; Bidzhekov, K.; Burger, F.; Pelli, G.; Luckow, B.; et al. Ccr5 But Not Ccr1 Deficiency Reduces Development of Diet-Induced Atherosclerosis in Mice. Arter. Thromb. Vasc. Biol. 2007, 27, 373–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, D.; Baylis, R.A.; Durgin, B.; Newman, A.A.C.; Alencar, G.F.; Mahan, S.; Hilaire, C.S.; Müller, W.; Waisman, A.; Francis, S.E.; et al. Interleukin-1β has atheroprotective effects in advanced atherosclerotic lesions of mice. Nat. Med. 2018, 24, 1418–1429. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; The Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [Green Version]
- Hennig, C. Cran-Package Fpc. Available online: https://cran.r-project.org/web/packages/fpc/index.html (accessed on 23 December 2021).
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.C.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.-Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef] [Green Version]
- Geer, L.Y.; Marchler-Bauer, A.; Geer, R.C.; Han, L.; He, J.; He, S.; Liu, C.; Shi, W.; Bryant, S.H. The NCBI BioSystems database. Nucleic Acids Res. 2009, 38, D492–D496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Slenter, D.N.; Kutmon, M.; Hanspers, K.; Riutta, A.; Windsor, J.; Nunes, N.; Mélius, J.; Cirillo, E.; Coort, S.L.; Digles, D.; et al. WikiPathways: A multifaceted pathway database bridging metabolomics to other omics research. Nucleic Acids Res. 2018, 46, D661–D667. [Google Scholar] [CrossRef]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [Green Version]
- Donato, M.; Xu, Z.; Tomoiaga, A.; Granneman, J.G.; MacKenzie, R.G.; Bao, R.; Than, N.G.; Westfall, P.H.; Romero, R.; Draghici, S. Analysis and correction of crosstalk effects in pathway analysis. Genome Res. 2013, 23, 1885–1893. [Google Scholar] [CrossRef]
- Montecucco, F.; Lenglet, S.; Gayet-Ageron, A.; Bertolotto, M.; Pelli, G.; Palombo, D.; Pane, B.; Spinella, G.; Steffens, S.; Raffaghello, L.; et al. Systemic and Intraplaque Mediators of Inflammation Are Increased in Patients Symptomatic for Ischemic Stroke. Stroke 2010, 41, 1394–1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montecucco, F.; Matias, I.; Lenglet, S.; Petrosino, S.; Burger, F.; Pelli, G.; Braunersreuther, V.; Mach, F.; Steffens, S.; Di Marzo, V. Regulation and possible role of endocannabinoids and related mediators in hypercholesterolemic mice with atherosclerosis. Atherosclerosis 2009, 205, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Halliday, A.; Mansfield, A.; Marro, J.; Peto, C.; Peto, R.; Potter, J.; Roberto, A.; MRC Asymptomatic Carotid Surgery Trial (ACST) Collaborative Group. Prevention of disabling and fatal strokes by successful carotid endarterectomy in patients without recent neurological symptoms: Randomised controlled trial. Lancet 2004, 363, 1491–1502. [Google Scholar] [CrossRef] [PubMed]
- Randomised Trial of Endarterectomy for Recently Symptomatic Carotid Stenosis: Final Results of the MRC European Carotid Surgery Trial (ECST). Lancet 1998, 351, 1379–1387. Available online: https://www.ncbi.nlm.nih.gov/pubmed/9593407 (accessed on 23 December 2021). [CrossRef]
- Carbone, F.; Crowe, L.A.; Roth, A.; Burger, F.; Lenglet, S.; Braunersreuther, V.; Brandt, K.J.; Quercioli, A.; Mach, F.; Vallée, J.-P.; et al. Treatment with anti-RANKL antibody reduces infarct size and attenuates dysfunction impacting on neutrophil-mediated injury. J. Mol. Cell. Cardiol. 2016, 94, 82–94. [Google Scholar] [CrossRef] [Green Version]
- Montecucco, F.; Bondarenko, A.I.; Lenglet, S.; Burger, F.; Piscitelli, F.; Carbone, F.; Roth, A.; Liberale, L.; Dallegri, F.; Brandt, K.J.; et al. Treatment with the GPR55 antagonist CID16020046 increases neutrophil activation in mouse atherogenesis. Thromb. Haemost. 2016, 116, 987–997. [Google Scholar] [CrossRef] [Green Version]
- Miteva, K.; Baptista, D.; Montecucco, F.; Asrih, M.; Burger, F.; Roth, A.; Fraga-Silva, R.A.; Stergiopulos, N.; Mach, F.; Brandt, K.J. Cardiotrophin-1 Deficiency Abrogates Atherosclerosis Progression. Sci. Rep. 2020, 10, 5791. [Google Scholar] [CrossRef] [Green Version]
- VanderLaan, P.; Reardon, C.A.; Getz, G.S. Site Specificity of Atherosclerosis. Arter. Thromb. Vasc. Biol. 2004, 24, 12–22. [Google Scholar] [CrossRef] [Green Version]
- Finlay, D.; Cantrell, D. The Coordination of T-cell Function by Serine/Threonine Kinases. Cold Spring Harb. Perspect. Biol. 2011, 3, a002261. [Google Scholar] [CrossRef]
- Ripoll, V.M.; Irvine, K.; Ravasi, T.; Sweet, M.; Hume, D. GpnmbIs Induced in Macrophages by IFN-γ and Lipopolysaccharide and Acts as a Feedback Regulator of Proinflammatory Responses. J. Immunol. 2007, 178, 6557–6566. [Google Scholar] [CrossRef] [Green Version]
- Henriksen, P.A.; Hitt, M.; Xing, Z.; Wang, J.; Haslett, C.; Riemersma, R.A.; Webb, D.J.; Kotelevtsev, Y.V.; Sallenave, J.-M. Adenoviral Gene Delivery of Elafin and Secretory Leukocyte Protease Inhibitor Attenuates NF-κB-Dependent Inflammatory Responses of Human Endothelial Cells and Macrophages to Atherogenic Stimuli. J. Immunol. 2004, 172, 4535–4544. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, W.; Zhao, Y.; Wang, F.; Liu, S.; Liu, L.; Zhao, L.; Lu, W.; Li, M.; Xu, Y. Dendritic Cells and T Cells, Partners in Atherogenesis and the Translating Road Ahead. Front. Immunol. 2020, 11, 1456. [Google Scholar] [CrossRef] [PubMed]
- Wolska, A.; Dunbar, R.L.; Freeman, L.A.; Ueda, M.; Amar, M.J.; Sviridov, D.O.; Remaley, A.T. Apolipoprotein C-II: New findings related to genetics, biochemistry, and role in triglyceride metabolism. Atherosclerosis 2017, 267, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Sommers, C.L.; Burshtyn, D.; Stebbins, C.C.; DeJarnette, J.B.; Trible, R.P.; Grinberg, A.; Tsay, H.C.; Jacobs, H.M.; Kessler, C.M.; et al. Essential Role of LAT in T Cell Development. Immunity 1999, 10, 323–332. [Google Scholar] [CrossRef] [Green Version]
- Malarkannan, S. NKG7 makes a better killer. Nat. Immunol. 2020, 21, 1139–1140. [Google Scholar] [CrossRef]
- Huang, C.; Lewis, C.; Borg, N.; Canals, M.; Diep, H.; Drummond, G.R.; Goode, R.; Schittenhelm, R.B.; Vinh, A.; Zhu, M.; et al. Proteomic Identification of Interferon-Induced Proteins with Tetratricopeptide Repeats as Markers of M1 Macrophage Polarization. J. Proteome Res. 2018, 17, 1485–1499. [Google Scholar] [CrossRef]
- Su, C.-T.; Urban, Z. LTBP4 in Health and Disease. Genes 2021, 12, 795. [Google Scholar] [CrossRef]
- Matukumalli, S.R.; Tangirala, R.; Rao, C.M. Clusterin: Full-length protein and one of its chains show opposing effects on cellular lipid accumulation. Sci. Rep. 2017, 7, 41235. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.S.; Kim, B.; Oh, G.T.; Kim, Y.-J. OASL1 inhibits translation of the type I interferon–regulating transcription factor IRF7. Nat. Immunol. 2013, 14, 346–355. [Google Scholar] [CrossRef]
- Zernecke, A. Dendritic Cells in Atherosclerosis. Arter. Thromb. Vasc. Biol. 2015, 35, 763–770. [Google Scholar] [CrossRef] [Green Version]
- Ensan, S.; Li, A.; Besla, R.; Degousee, N.; Cosme, J.; Roufaiel, M.; Shikatani, E.A.; El-Maklizi, M.; Williams, J.W.; Robins, L.; et al. Self-renewing resident arterial macrophages arise from embryonic CX3CR1+ precursors and circulating monocytes immediately after birth. Nat. Immunol. 2016, 17, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Beckers, C.M.L.; Simpson, K.R.; Griffin, K.J.; Brown, J.M.; Cheah, L.T.; Smith, K.A.; Vacher, J.; Cordell, P.A.; Kearney, M.T.; Grant, P.J.; et al. Cre/lox Studies Identify Resident Macrophages as the Major Source of Circulating Coagulation Factor XIII-A. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1494–1502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson, E.; Hultman, K.; Edsfeldt, A.; Persson, A.; Nitulescu, M.; Nilsson, J.; Goncalves, I.; Björkbacka, H. CD163+ macrophages are associated with a vulnerable plaque phenotype in human carotid plaques. Sci. Rep. 2020, 10, 14362. [Google Scholar] [CrossRef]
- Boyle, J.J.; Harrington, H.A.; Piper, E.; Elderfield, K.; Stark, J.; Landis, R.C.; Haskard, D.O. Coronary Intraplaque Hemorrhage Evokes a Novel Atheroprotective Macrophage Phenotype. Am. J. Pathol. 2009, 174, 1097–1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Martínez, E.; Ibarrola, J.F.; Fernández-Celis, A.; Santamaria, E.; Fernández-Irigoyen, J.; Rossignol, P.; Jaisser, F.; López-Andrés, N. Differential Proteomics Identifies Reticulocalbin-3 as a Novel Negative Mediator of Collagen Production in Human Cardiac Fibroblasts. Sci. Rep. 2017, 7, 12192. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; De Bastiani, M.A.; Parisi, M.M.; Guma, F.T.C.R.; Markoski, M.M.; Castro, M.; Kaplan, M.H.; Barbé-Tuana, F.M.; Klamt, F. Integrated Transcriptomics Establish Macrophage Polarization Signatures and have Potential Applications for Clinical Health and Disease. Sci. Rep. 2015, 5, 13351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chinetti-Gbaguidi, G.; Staels, B. Macrophage polarization in metabolic disorders: Functions and regulation. Curr. Opin. Lipidol. 2011, 22, 365–372. [Google Scholar] [CrossRef] [Green Version]
- Plaza-Díaz, J.; Robles-Sánchez, C.; Abadía-Molina, F.; Morón-Calvente, V.; Sáez-Lara, M.J.; Ruiz-Bravo, A.; Jiménez-Valera, M.; Gil, Á.; Gómez-Llorente, C.; Fontana, L. Adamdec1, Ednrb and Ptgs1/Cox1, inflammation genes upregulated in the intestinal mucosa of obese rats, are downregulated by three probiotic strains. Sci. Rep. 2017, 7, 1939. [Google Scholar] [CrossRef] [Green Version]
- Hume, D.A.; MacDonald, K. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 2012, 119, 1810–1820. [Google Scholar] [CrossRef]
- Mor, A.; Afek, A.; Entin-Meer, M.; Keren, G.; George, J. Anti eotaxin-2 antibodies attenuate the initiation and progression of experimental atherosclerosis. World J. Cardiovasc. Dis. 2013, 3, 34775. [Google Scholar] [CrossRef] [Green Version]
- Raghuraman, G.; Hsiung, J.; Zuniga, M.C.; Baughman, B.D.; Hitchner, E.; Guzman, R.J.; Zhou, W. Eotaxin Augments Calcification in Vascular Smooth Muscle Cells. J. Cell. Biochem. 2016, 118, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Gautier, E.L.; Shay, T.; Miller, J.; Greter, M.; Jakubzick, C.; Ivanov, S.; Helft, J.; Chow, A.; Elpek, K.G.; Gordonov, S.; et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012, 13, 1118–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Byon, C.H.; Yuan, K.; Chen, J.; Mao, X.; Heath, J.M.; Javed, A.; Zhang, K.; Anderson, P.G.; Chen, Y. Smooth Muscle Cell–Specific Runx2 Deficiency Inhibits Vascular Calcification. Circ. Res. 2012, 111, 543–552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aikawa, E.; Nahrendorf, M.; Figueiredo, J.-L.; Swirski, F.K.; Shtatland, T.; Kohler, R.H.; Jaffer, F.A.; Aikawa, M.; Weissleder, R. Osteogenesis Associates with Inflammation in Early-Stage Atherosclerosis Evaluated by Molecular Imaging In Vivo. Circulation 2007, 116, 2841–2850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkels, H.; Ehinger, E.; Vassallo, M.; Buscher, K.; Dinh, H.; Kobiyama, K.; Hamers, A.A.; Cochain, C.; Vafadarnejad, E.; Saliba, A.-E.; et al. Atlas of the Immune Cell Repertoire in Mouse Atherosclerosis Defined by Single-Cell RNA-Sequencing and Mass Cytometry. Circ. Res. 2018, 122, 1675–1688. [Google Scholar] [CrossRef] [PubMed]
- Wolf, D.; Ley, K. Immunity and Inflammation in Atherosclerosis. Circ. Res. 2019, 124, 315–327. [Google Scholar] [CrossRef]
- Winkels, H.; Wolf, D. Heterogeneity of T Cells in Atherosclerosis Defined by Single-Cell RNA-Sequencing and Cytometry by Time of Flight. Arter. Thromb. Vasc. Biol. 2021, 41, 549–563. [Google Scholar] [CrossRef]
- Tabas, I.; Bornfeldt, K.E. Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ. Res. 2016, 118, 653–667. [Google Scholar] [CrossRef] [Green Version]
- Koelwyn, G.J.; Corr, E.M.; Erbay, E.; Moore, K.J. Regulation of macrophage immunometabolism in atherosclerosis. Nat. Immunol. 2018, 19, 526–537. [Google Scholar] [CrossRef] [Green Version]
- Kodali, R.B.; Kim, W.J.; Galaria, I.I.; Miller, C.; Schecter, A.D.; Lira, S.A.; Taubman, M.B. CCL11 (Eotaxin) Induces CCR3-Dependent Smooth Muscle Cell Migration. Arter. Thromb. Vasc. Biol. 2004, 24, 1211–1216. [Google Scholar] [CrossRef] [Green Version]
- Mor, A.; Salto, M.S.; Katav, A.; Barashi, N.; Edelshtein, V.; Manetti, M.; Levi, Y.; George, J.; Matucci-Cerinic, M. Blockade of CCL24 with a monoclonal antibody ameliorates experimental dermal and pulmonary fibrosis. Ann. Rheum. Dis. 2019, 78, 1260–1268. [Google Scholar] [CrossRef] [PubMed]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burger, F.; Baptista, D.; Roth, A.; Brandt, K.J.; da Silva, R.F.; Montecucco, F.; Mach, F.; Miteva, K. Single-Cell RNA-Seq Reveals a Crosstalk between Hyaluronan Receptor LYVE-1-Expressing Macrophages and Vascular Smooth Muscle Cells. Cells 2022, 11, 411. https://doi.org/10.3390/cells11030411
Burger F, Baptista D, Roth A, Brandt KJ, da Silva RF, Montecucco F, Mach F, Miteva K. Single-Cell RNA-Seq Reveals a Crosstalk between Hyaluronan Receptor LYVE-1-Expressing Macrophages and Vascular Smooth Muscle Cells. Cells. 2022; 11(3):411. https://doi.org/10.3390/cells11030411
Chicago/Turabian StyleBurger, Fabienne, Daniela Baptista, Aline Roth, Karim J. Brandt, Rafaela Fernandes da Silva, Fabrizio Montecucco, François Mach, and Kapka Miteva. 2022. "Single-Cell RNA-Seq Reveals a Crosstalk between Hyaluronan Receptor LYVE-1-Expressing Macrophages and Vascular Smooth Muscle Cells" Cells 11, no. 3: 411. https://doi.org/10.3390/cells11030411
APA StyleBurger, F., Baptista, D., Roth, A., Brandt, K. J., da Silva, R. F., Montecucco, F., Mach, F., & Miteva, K. (2022). Single-Cell RNA-Seq Reveals a Crosstalk between Hyaluronan Receptor LYVE-1-Expressing Macrophages and Vascular Smooth Muscle Cells. Cells, 11(3), 411. https://doi.org/10.3390/cells11030411






