Functional Differences between Proteasome Subtypes
Abstract
:1. Introduction
2. The Proteasome Structure
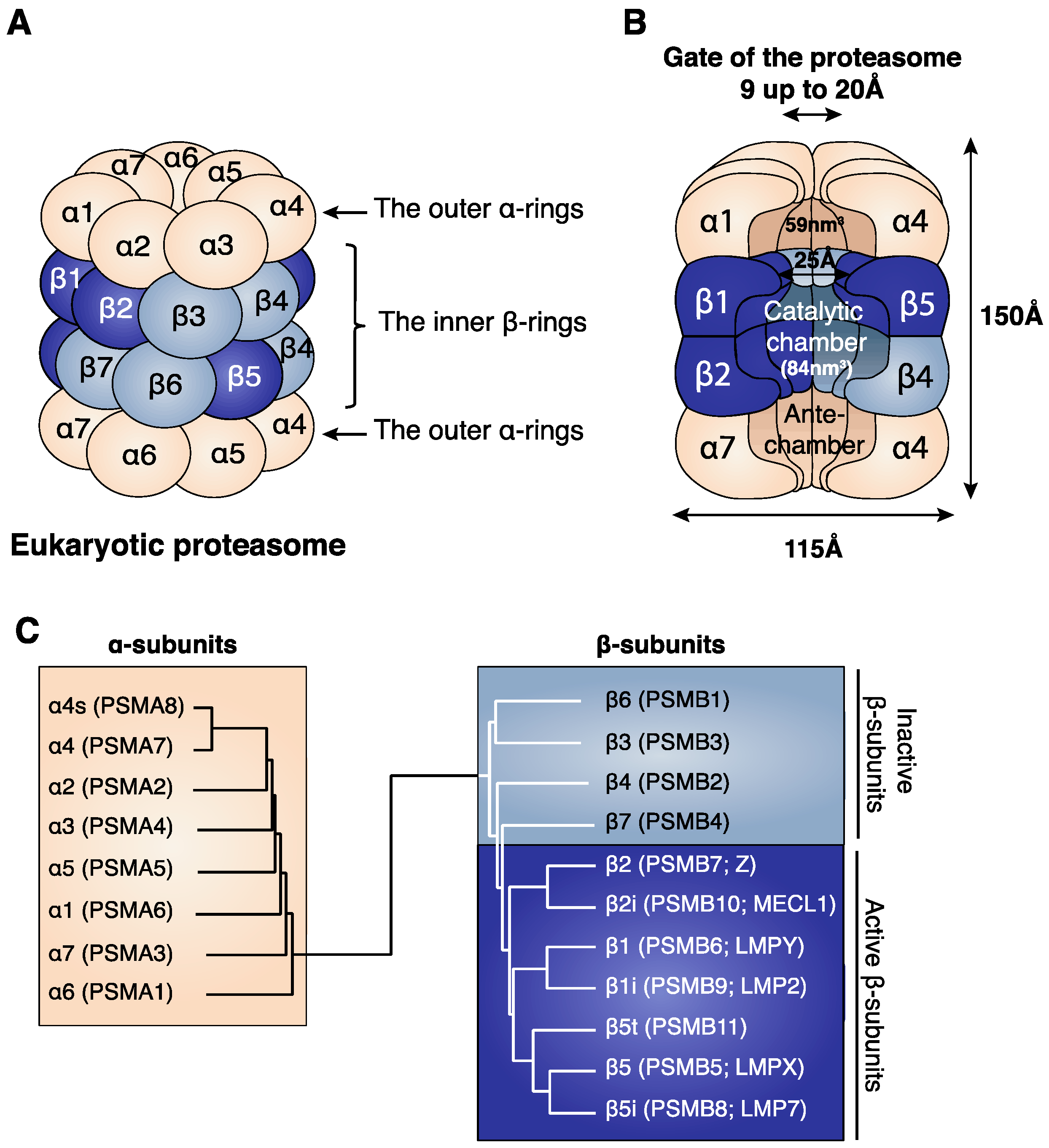
2.1. Structure of the 20S Proteasome
- The two homologous subunits α4 (PSMA7) and α4s (PSMA8).
- The two homologous subunits β1 (PSMB6) and β1i (PSMB9).
- The two homologous subunits β2 (PSMB7) and β2i (PSMB10).
- The three homologous subunits β5 (PSMB5), β5i (PSMB8) and β5t (PSMB11).
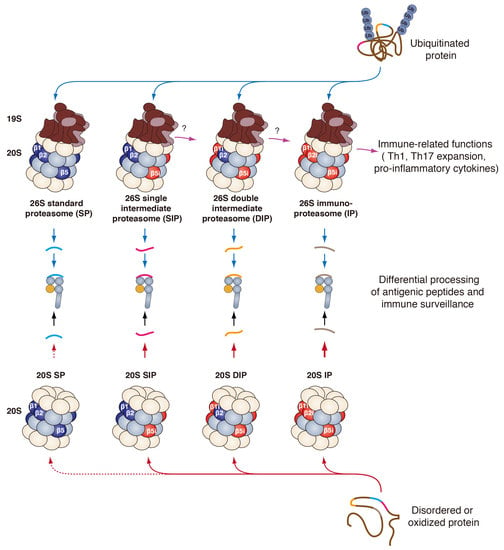
2.2. Proteasome Subtypes
2.2.1. The Standard Proteasome
2.2.2. The Immunoproteasome
2.2.3. The Intermediate Proteasomes
2.2.4. The Thymoproteasome
2.2.5. The Spermatoproteasome
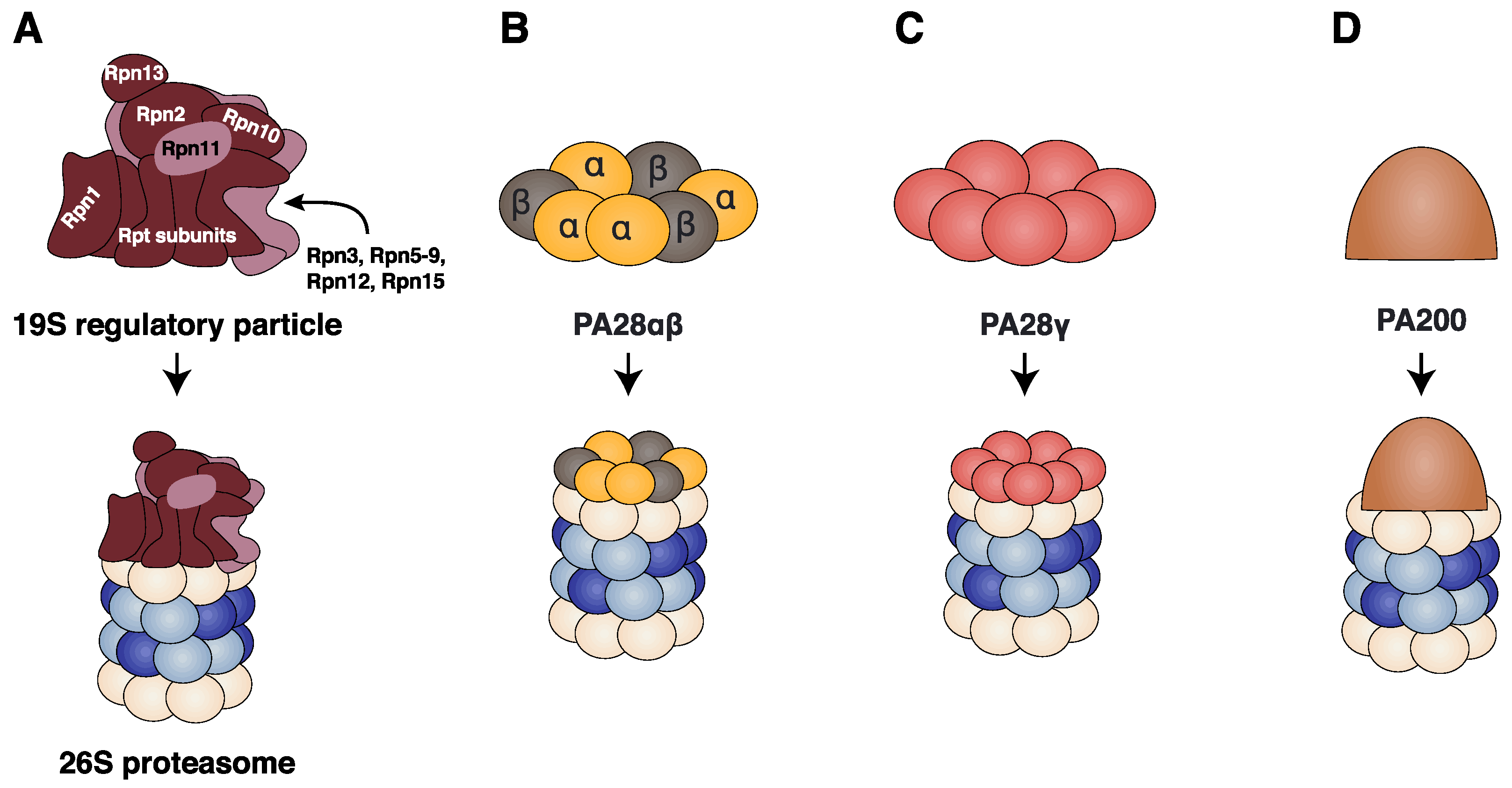
2.3. Regulatory Particles
2.3.1. The 19S Regulatory Particle
2.3.2. The PA28αβ Regulator
2.3.3. The PA28γ Regulator
2.3.4. The PA200 Regulator
3. The Differential Functions of the Proteasome Subtypes
3.1. The Ubiquitin- and ATP-Dependent Proteasomal Degradation
Proteasome Subtypes and the Degradation of Ubiquitinated Proteins
3.2. The Ubiquitin- and ATP-Independent Proteasomal Degradation
3.2.1. Proteasome Subtypes and the ATP- and Ubiquitin-Independent Degradation of Proteins
3.2.2. Role of the Regulators in the ATP- and Ubiquitin-Independent Protein Degradation
3.3. Production of MHC Class I Peptides
3.3.1. Proteasome Subtypes and the Production of Canonical MHC Class I Peptides
3.3.2. Proteasome Subtypes and the Production of Spliced Peptides
3.4. Thymoproteasome and Positive Selection
3.5. Role of the IP in Immune-Related Pathways
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, J.; Zhai, B.; Gygi, S.P.; Goldberg, A.L. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. USA 2015, 112, 15790–15797. [Google Scholar] [CrossRef] [Green Version]
- Kisselev, A.F.; Akopian, T.N.; Goldberg, A.L. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. J. Biol. Chem. 1998, 273, 1982–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigneron, N.; Van den Eynde, B.J. Proteasome subtypes and regulators in the processing of antigenic peptides presented by class I molecules of the major histocompatibility complex. Biomolecules 2014, 4, 994–1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigneron, N.; Abi Habib, J.; Van den Eynde, B.J. Learning from the Proteasome How To Fine-Tune Cancer Immunotherapy. Trends Cancer 2017, 3, 726–741. [Google Scholar] [CrossRef] [PubMed]
- Lowe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Ditzel, L.; Lowe, J.; Stock, D.; Bochtler, M.; Bartunik, H.D.; Huber, R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 1997, 386, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Basler, M.; Schwab, R.; Heinemeyer, W.; Kirk, C.J.; Groettrup, M.; Groll, M. Immuno- and constitutive proteasome crystal structures reveal differences in substrate and inhibitor specificity. Cell 2012, 148, 727–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unno, M.; Mizushima, T.; Morimoto, Y.; Tomisugi, Y.; Tanaka, K.; Yasuoka, N.; Tsukihara, T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure 2002, 10, 609–618. [Google Scholar] [CrossRef] [Green Version]
- Baumeister, W.; Walz, J.; Zuhl, F.; Seemuller, E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell 1998, 92, 367–380. [Google Scholar] [CrossRef] [Green Version]
- Rabl, J.; Smith, D.M.; Yu, Y.; Chang, S.C.; Goldberg, A.L.; Cheng, Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell 2008, 30, 360–368. [Google Scholar] [CrossRef] [Green Version]
- Groll, M.; Bajorek, M.; Kohler, A.; Moroder, L.; Rubin, D.M.; Huber, R.; Glickman, M.H.; Finley, D. A gated channel into the proteasome core particle. Nat. Struct. Biol. 2000, 7, 1062–1067. [Google Scholar] [CrossRef] [PubMed]
- Ruschak, A.M.; Religa, T.L.; Breuer, S.; Witt, S.; Kay, L.E. The proteasome antechamber maintains substrates in an unfolded state. Nature 2010, 467, 868–871. [Google Scholar] [CrossRef] [PubMed]
- Hisamatsu, H.; Shimbara, N.; Saito, Y.; Kristensen, P.; Hendil, K.B.; Fujiwara, T.; Takahashi, E.; Tanahashi, N.; Tamura, T.; Ichihara, A.; et al. Newly identified pair of proteasomal subunits regulated reciprocally by interferon gamma. J. Exp. Med. 1996, 183, 1807–1816. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Sasaki, K.; Kishimoto, T.; Niwa, S.; Hayashi, H.; Takahama, Y.; Tanaka, K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science 2007, 316, 1349–1353. [Google Scholar] [CrossRef]
- Heink, S.; Ludwig, D.; Kloetzel, P.M.; Kruger, E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. USA 2005, 102, 9241–9246. [Google Scholar] [CrossRef] [Green Version]
- Hirano, Y.; Kaneko, T.; Okamoto, K.; Bai, M.; Yashiroda, H.; Furuyama, K.; Kato, K.; Tanaka, K.; Murata, S. Dissecting beta-ring assembly pathway of the mammalian 20S proteasome. EMBO J. 2008, 27, 2204–2213. [Google Scholar] [CrossRef] [Green Version]
- Huber, E.M.; Heinemeyer, W.; Li, X.; Arendt, C.S.; Hochstrasser, M.; Groll, M. A unified mechanism for proteolysis and autocatalytic activation in the 20S proteasome. Nat. Commun. 2016, 7, 10900. [Google Scholar] [CrossRef] [Green Version]
- Groll, M.; Heinemeyer, W.; Jager, S.; Ullrich, T.; Bochtler, M.; Wolf, D.H.; Huber, R. The catalytic sites of 20S proteasomes and their role in subunit maturation: A mutational and crystallographic study. Proc. Natl. Acad. Sci. USA 1999, 96, 10976–10983. [Google Scholar] [CrossRef] [Green Version]
- Qian, M.X.; Pang, Y.; Liu, C.H.; Haratake, K.; Du, B.Y.; Ji, D.Y.; Wang, G.F.; Zhu, Q.Q.; Song, W.; Yu, Y.; et al. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 2013, 153, 1012–1024. [Google Scholar] [CrossRef] [Green Version]
- Kniepert, A.; Groettrup, M. The unique functions of tissue-specific proteasomes. Trends Biochem. Sci. 2014, 39, 17–24. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Fu, J.; Ha, S.W.; Ju, D.; Zheng, J.; Li, L.; Xie, Y. The CCAAT box-binding transcription factor NF-Y regulates basal expression of human proteasome genes. Biochim. Biophys. Acta 2012, 1823, 818–825. [Google Scholar] [CrossRef] [Green Version]
- Vangala, J.R.; Dudem, S.; Jain, N.; Kalivendi, S.V. Regulation of PSMB5 protein and beta subunits of mammalian proteasome by constitutively activated signal transducer and activator of transcription 3 (STAT3): Potential role in bortezomib-mediated anticancer therapy. J. Biol. Chem. 2014, 289, 12612–12622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radhakrishnan, S.K.; Lee, C.S.; Young, P.; Beskow, A.; Chan, J.Y.; Deshaies, R.J. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 2010, 38, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Manning, B.D. mTORC1 signaling activates NRF1 to increase cellular proteasome levels. Cell Cycle 2015, 14, 2011–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Nicholatos, J.; Dreier, J.R.; Ricoult, S.J.; Widenmaier, S.B.; Hotamisligil, G.S.; Kwiatkowski, D.J.; Manning, B.D. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 2014, 513, 440–443. [Google Scholar] [CrossRef] [Green Version]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, M.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J. Biol. Chem. 2003, 278, 21592–21600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickering, A.M.; Linder, R.A.; Zhang, H.; Forman, H.J.; Davies, K.J.A. Nrf2-dependent Induction of Proteasome and Pa28αβ Regulator Are Required for Adaptation to Oxidative Stress. J. Biol. Chem. 2012, 287, 10021–10031. [Google Scholar] [CrossRef] [Green Version]
- Dick, T.P.; Nussbaum, A.K.; Deeg, M.; Heinemeyer, W.; Groll, M.; Schirle, M.; Keilholz, W.; Stevanovic, S.; Wolf, D.H.; Huber, R.; et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J. Biol. Chem. 1998, 273, 25637–25646. [Google Scholar] [CrossRef] [Green Version]
- Heinemeyer, W.; Fischer, M.; Krimmer, T.; Stachon, U.; Wolf, D.H. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 1997, 272, 25200–25209. [Google Scholar] [CrossRef] [Green Version]
- Vigneron, N.; Abi Habib, J.; Van den Eynde, B.J. The capture proteasome assay: A method to measure proteasome activity in vitro. Anal. Biochem. 2015, 482, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Groll, M.; Larionov, O.V.; Huber, R.; de Meijere, A. Inhibitor-binding mode of homobelactosin C to proteasomes: New insights into class I MHC ligand generation. Proc. Natl. Acad. Sci. USA 2006, 103, 4576–4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aki, M.; Shimbara, N.; Takashina, M.; Akiyama, K.; Kagawa, S.; Tamura, T.; Tanahashi, N.; Yoshimura, T.; Tanaka, K.; Ichihara, A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994, 115, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.C.; Seifert, U.; Kato, T.; Rice, C.M.; Feinstone, S.M.; Kloetzel, P.M.; Rehermann, B. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J. Clin. Investig. 2006, 116, 3006–3014. [Google Scholar] [CrossRef] [Green Version]
- Hirano, Y.; Hayashi, H.; Iemura, S.; Hendil, K.B.; Niwa, S.; Kishimoto, T.; Kasahara, M.; Natsume, T.; Tanaka, K.; Murata, S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell. 2006, 24, 977–984. [Google Scholar] [CrossRef]
- Rouette, A.; Trofimov, A.; Haberl, D.; Boucher, G.; Lavallee, V.P.; D’Angelo, G.; Hebert, J.; Sauvageau, G.; Lemieux, S.; Perreault, C. Expression of immunoproteasome genes is regulated by cell-intrinsic and -extrinsic factors in human cancers. Sci. Rep. 2016, 6, 34019. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.G.; Driscoll, J.; Monaco, J.J. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature 1991, 353, 355–357. [Google Scholar] [CrossRef]
- Glynne, R.; Powis, S.H.; Beck, S.; Kelly, A.; Kerr, L.A.; Trowsdale, J. A proteasome-related gene between the two ABC transporter loci in the class II region of the human MHC. Nature 1991, 353, 357–360. [Google Scholar] [CrossRef]
- Groettrup, M.; Kraft, R.; Kostka, S.; Standera, S.; Stohwasser, R.; Kloetzel, P.M. A third interferon-gamma-induced subunit exchange in the 20S proteasome. Eur. J. Immunol. 1996, 26, 863–869. [Google Scholar] [CrossRef]
- Martinez, C.K.; Monaco, J.J. Homology of proteasome subunits to a major histocompatibility complex-linked LMP gene. Nature 1991, 353, 664–667. [Google Scholar] [CrossRef]
- Monaco, J.J.; McDevitt, H.O. H-2-linked low-molecular weight polypeptide antigens assemble into an unusual macromolecular complex. Nature 1984, 309, 797–799. [Google Scholar] [CrossRef] [PubMed]
- Monaco, J.J.; McDevitt, H.O. The LMP antigens: A stable MHC-controlled multisubunit protein complex. Hum. Immunol. 1986, 15, 416–426. [Google Scholar] [CrossRef]
- Nandi, D.; Jiang, H.; Monaco, J.J. Identification of MECL-1 (LMP-10) as the third IFN-gamma-inducible proteasome subunit. J. Immunol. 1996, 156, 2361–2364. [Google Scholar] [PubMed]
- Ortiz-Navarrete, V.; Seelig, A.; Gernold, M.; Frentzel, S.; Kloetzel, P.M.; Hammerling, G.J. Subunit of the ‘20S’ proteasome (multicatalytic proteinase) encoded by the major histocompatibility complex. Nature 1991, 353, 662–664. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee-Kishore, M.; Kishore, R.; Hicklin, D.J.; Marincola, F.M.; Ferrone, S. Different requirements for signal transducer and activator of transcription 1alpha and interferon regulatory factor 1 in the regulation of low molecular mass polypeptide 2 and transporter associated with antigen processing 1 gene expression. J. Biol. Chem. 1998, 273, 16177–16183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foss, G.S.; Prydz, H. Interferon regulatory factor 1 mediates the interferon-gamma induction of the human immunoproteasome subunit multicatalytic endopeptidase complex-like 1. J. Biol. Chem. 1999, 274, 35196–35202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namiki, S.; Nakamura, T.; Oshima, S.; Yamazaki, M.; Sekine, Y.; Tsuchiya, K.; Okamoto, R.; Kanai, T.; Watanabe, M. IRF-1 mediates upregulation of LMP7 by IFN-gamma and concerted expression of immunosubunits of the proteasome. FEBS Lett. 2005, 579, 2781–2787. [Google Scholar] [CrossRef] [Green Version]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar] [CrossRef] [Green Version]
- Kotamraju, S.; Matalon, S.; Matsunaga, T.; Shang, T.; Hickman-Davis, J.M.; Kalyanaraman, B. Upregulation of immunoproteasomes by nitric oxide: Potential antioxidative mechanism in endothelial cells. Free Radic. Biol. Med. 2006, 40, 1034–1044. [Google Scholar] [CrossRef]
- Wright, K.L.; White, L.C.; Kelly, A.; Beck, S.; Trowsdale, J.; Ting, J.P. Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med. 1995, 181, 1459–1471. [Google Scholar] [CrossRef]
- Driscoll, J.; Brown, M.G.; Finley, D.; Monaco, J.J. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature 1993, 365, 262–264. [Google Scholar] [CrossRef] [PubMed]
- Gaczynska, M.; Rock, K.L.; Goldberg, A.L. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature 1993, 365, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, Y.; Xu, C.; Chen, K.; Zhao, Q.; Wang, S.; Yin, Y.; Peng, C.; Ding, Z.; Cong, Y. Cryo-EM of mammalian PA28αβ-iCP immunoproteasome reveals a distinct mechanism of proteasome activation by PA28αβ. Nat. Commun. 2021, 12, 739. [Google Scholar] [CrossRef] [PubMed]
- Griffin, T.A.; Nandi, D.; Cruz, M.; Fehling, H.J.; Kaer, L.V.; Monaco, J.J.; Colbert, R.A. Immunoproteasome assembly: Cooperative incorporation of interferon gamma (IFN-gamma)-inducible subunits. J. Exp. Med. 1998, 187, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Groettrup, M.; Standera, S.; Stohwasser, R.; Kloetzel, P.M. The subunits MECL-1 and LMP2 are mutually required for incorporation into the 20S proteasome. Proc. Natl. Acad. Sci. USA 1997, 94, 8970–8975. [Google Scholar] [CrossRef] [Green Version]
- Hensley, S.E.; Zanker, D.; Dolan, B.P.; David, A.; Hickman, H.D.; Embry, A.C.; Skon, C.N.; Grebe, K.M.; Griffin, T.A.; Chen, W.; et al. Unexpected role for the immunoproteasome subunit LMP2 in antiviral humoral and innate immune responses. J. Immunol. 2010, 184, 4115–4122. [Google Scholar] [CrossRef] [Green Version]
- Hussong, S.A.; Kapphahn, R.J.; Phillips, S.L.; Maldonado, M.; Ferrington, D.A. Immunoproteasome deficiency alters retinal proteasome’s response to stress. J. Neurochem. 2010, 113, 1481–1490. [Google Scholar] [CrossRef] [Green Version]
- Dahlmann, B.; Ruppert, T.; Kuehn, L.; Merforth, S.; Kloetzel, P.M. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J. Mol. Biol. 2000, 303, 643–653. [Google Scholar] [CrossRef]
- Guillaume, B.; Chapiro, J.; Stroobant, V.; Colau, D.; Van Holle, B.; Parvizi, G.; Bousquet-Dubouch, M.P.; Theate, I.; Parmentier, N.; Van den Eynde, B.J. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc. Natl. Acad. Sci. USA 2010, 107, 18599–18604. [Google Scholar] [CrossRef] [Green Version]
- Khilji, M.S.; Verstappen, D.; Dahlby, T.; Burstein Prause, M.C.; Pihl, C.; Bresson, S.E.; Bryde, T.H.; Keller Andersen, P.A.; Klindt, K.; Zivkovic, D.; et al. The intermediate proteasome is constitutively expressed in pancreatic beta cells and upregulated by stimulatory, low concentrations of interleukin 1 β. PLoS ONE 2020, 15, e0222432. [Google Scholar] [CrossRef]
- Vigneron, N.; Abi Habib, J.; Van den Eynde, B.J. The capture proteasome assay (CAPA) to evaluate subtype-specific proteasome inhibitors. Data Brief 2015, 4, 146–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomaru, U.; Ishizu, A.; Murata, S.; Miyatake, Y.; Suzuki, S.; Takahashi, S.; Kazamaki, T.; Ohara, J.; Baba, T.; Iwasaki, S.; et al. Exclusive expression of proteasome subunit {beta}5t in the human thymic cortex. Blood 2009, 113, 5186–5191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ripen, A.M.; Nitta, T.; Murata, S.; Tanaka, K.; Takahama, Y. Ontogeny of thymic cortical epithelial cells expressing the thymoproteasome subunit beta5t. Eur. J. Immunol. 2011, 41, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.M.; Ohigashi, I.; Motosugi, R.; Nakayama, T.; Sakata, M.; Hamazaki, J.; Nishito, Y.; Rode, I.; Tanaka, K.; Takemoto, T.; et al. Foxn1-beta5t transcriptional axis controls CD8(+) T-cell production in the thymus. Nat. Commun. 2017, 8, 14419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuklys, S.; Handel, A.; Zhanybekova, S.; Govani, F.; Keller, M.; Maio, S.; Mayer, C.E.; Teh, H.Y.; Hafen, K.; Gallone, G.; et al. Foxn1 regulates key target genes essential for T cell development in postnatal thymic epithelial cells. Nat. Immunol. 2016, 17, 1206–1215. [Google Scholar] [CrossRef]
- Uechi, H.; Hamazaki, J.; Murata, S. Characterization of the testis-specific proteasome subunit alpha4s in mammals. J. Biol. Chem. 2014, 289, 12365–12374. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.H.; Jiang, T.X.; Chen, L.B.; Zhou, W.; Liu, Y.; Gao, F.; Qiu, X.B. Proteasome subunit α4s is essential for formation of spermatoproteasomes and histone degradation during meiotic DNA repair in spermatocytes. J. Biol. Chem. 2021, 296, 100130. [Google Scholar] [CrossRef]
- Gómez, H.L.; Felipe-Medina, N.; Condezo, Y.B.; Garcia-Valiente, R.; Ramos, I.; Suja, J.A.; Barbero, J.L.; Roig, I.; Sánchez-Martín, M.; de Rooij, D.G.; et al. The PSMA8 subunit of the spermatoproteasome is essential for proper meiotic exit and mouse fertility. PLoS Genet. 2019, 15, e1008316. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Ji, S.Y.; Busayavalasa, K.; Shao, J.; Yu, C. Meiosis I progression in spermatogenesis requires a type of testis-specific 20S core proteasome. Nat. Commun. 2019, 10, 3387. [Google Scholar] [CrossRef] [Green Version]
- Lander, G.C.; Estrin, E.; Matyskiela, M.E.; Bashore, C.; Nogales, E.; Martin, A. Complete subunit architecture of the proteasome regulatory particle. Nature 2012, 482, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Lasker, K.; Forster, F.; Bohn, S.; Walzthoeni, T.; Villa, E.; Unverdorben, P.; Beck, F.; Aebersold, R.; Sali, A.; Baumeister, W. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc. Natl. Acad. Sci. USA 2012, 109, 1380–1387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubin-Tam, M.E.; Olivares, A.O.; Sauer, R.T.; Baker, T.A.; Lang, M.J. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell 2011, 145, 257–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Husnjak, K.; Elsasser, S.; Zhang, N.; Chen, X.; Randles, L.; Shi, Y.; Hofmann, K.; Walters, K.J.; Finley, D.; Dikic, I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 2008, 453, 481–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maillard, R.A.; Chistol, G.; Sen, M.; Righini, M.; Tan, J.; Kaiser, C.M.; Hodges, C.; Martin, A.; Bustamante, C. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell 2011, 145, 459–469. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Chen, X.; Elsasser, S.; Stocks, B.B.; Tian, G.; Lee, B.H.; Shi, Y.; Zhang, N.; de Poot, S.A.; Tuebing, F.; et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 2016, 351. [Google Scholar] [CrossRef] [Green Version]
- van Nocker, S.; Sadis, S.; Rubin, D.M.; Glickman, M.; Fu, H.; Coux, O.; Wefes, I.; Finley, D.; Vierstra, R.D. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol. 1996, 16, 6020–6028. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.M.; Chang, S.C.; Park, S.; Finley, D.; Cheng, Y.; Goldberg, A.L. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol. Cell 2007, 27, 731–744. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.Y.; Tanahashi, N.; Akiyama, K.; Hisamatsu, H.; Noda, C.; Tanaka, K.; Chung, C.H.; Shibmara, N.; Willy, P.J.; Mott, J.D.; et al. Primary structures of two homologous subunits of PA28, a gamma-interferon-inducible protein activator of the 20S proteasome. FEBS Lett. 1995, 366, 37–42. [Google Scholar]
- Realini, C.; Dubiel, W.; Pratt, G.; Ferrell, K.; Rechsteiner, M. Molecular cloning and expression of a gamma-interferon-inducible activator of the multicatalytic protease. J. Biol. Chem. 1994, 269, 20727–20732. [Google Scholar] [CrossRef]
- Dubiel, W.; Pratt, G.; Ferrell, K.; Rechsteiner, M. Purification of an 11 S regulator of the multicatalytic protease. J. Biol. Chem. 1992, 267, 22369–22377. [Google Scholar] [CrossRef]
- Ma, C.P.; Slaughter, C.A.; DeMartino, G.N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J. Biol. Chem. 1992, 267, 10515–10523. [Google Scholar] [CrossRef]
- Preckel, T.; Fung-Leung, W.P.; Cai, Z.; Vitiello, A.; Salter-Cid, L.; Winqvist, O.; Wolfe, T.G.; Von Herrath, M.; Angulo, A.; Ghazal, P.; et al. Impaired immunoproteasome assembly and immune responses in PA28−/− mice. Science 1999, 286, 2162–2165. [Google Scholar] [CrossRef] [PubMed]
- Raule, M.; Cerruti, F.; Benaroudj, N.; Migotti, R.; Kikuchi, J.; Bachi, A.; Navon, A.; Dittmar, G.; Cascio, P. PA28alphabeta reduces size and increases hydrophilicity of 20S immunoproteasome peptide products. Chem. Biol. 2014, 21, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Krutchinsky, A.; Endicott, S.; Realini, C.; Rechsteiner, M.; Standing, K.G. Proteasome activator 11S REG or PA28: Recombinant REG alpha/REG beta hetero-oligomers are heptamers. Biochemistry 1999, 38, 5651–5658. [Google Scholar] [CrossRef] [PubMed]
- Cascio, P.; Call, M.; Petre, B.M.; Walz, T.; Goldberg, A.L. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. EMBO J. 2002, 21, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Huber, E.M.; Groll, M. The Mammalian Proteasome Activator PA28 Forms an Asymmetric alpha4beta3 Complex. Structure 2017, 25, 1473–1480.e1473. [Google Scholar] [CrossRef] [Green Version]
- Knowlton, J.R.; Johnston, S.C.; Whitby, F.G.; Realini, C.; Zhang, Z.; Rechsteiner, M.; Hill, C.P. Structure of the proteasome activator REGalpha (PA28alpha). Nature 1997, 390, 639–643. [Google Scholar] [CrossRef]
- Realini, C.; Jensen, C.C.; Zhang, Z.; Johnston, S.C.; Knowlton, J.R.; Hill, C.P.; Rechsteiner, M. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J. Biol. Chem. 1997, 272, 25483–25492. [Google Scholar] [CrossRef] [Green Version]
- Rechsteiner, M.; Realini, C.; Ustrell, V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem. J. 2000, 345 Pt 1, 1–15. [Google Scholar] [CrossRef]
- Wilk, S.; Chen, W.E.; Magnusson, R.P. Properties of the nuclear proteasome activator PA28gamma (REGgamma). Arch. Biochem. Biophys. 2000, 383, 265–271. [Google Scholar] [CrossRef]
- Ortega, J.; Heymann, J.B.; Kajava, A.V.; Ustrell, V.; Rechsteiner, M.; Steven, A.C. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 2005, 346, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Guan, H.; Wang, Y.; Yu, T.; Huang, Y.; Li, M.; Saeed, A.; Perčulija, V.; Li, D.; Xiao, J.; Wang, D.; et al. Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures. PLoS Biol. 2020, 18, e3000654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, L.; Pratt, G.; Rechsteiner, M. Multiple forms of the 20 S multicatalytic and the 26 S ubiquitin/ATP-dependent proteases from rabbit reticulocyte lysate. J. Biol. Chem. 1992, 267, 22362–22368. [Google Scholar] [CrossRef]
- Ustrell, V.; Hoffman, L.; Pratt, G.; Rechsteiner, M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002, 21, 3516–3525. [Google Scholar] [CrossRef] [Green Version]
- Khor, B.; Bredemeyer, A.L.; Huang, C.Y.; Turnbull, I.R.; Evans, R.; Maggi, L.B., Jr.; White, J.M.; Walker, L.M.; Carnes, K.; Hess, R.A.; et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 2006, 26, 2999–3007. [Google Scholar] [CrossRef] [Green Version]
- Javitt, A.; Shmueli, M.; Kramer, M.; Kolodziejczyk, A.; Cohen, I.; Kamer, I.; Litchfield, K.; Bab-Dinitz, E.; Zadok, O.; Neiens, V.; et al. The proteasome regulator PSME4 drives immune evasion and abrogates anti-tumor immunity in NSCLC. bioRxiv 2021. [Google Scholar] [CrossRef]
- Elsasser, S.; Gali, R.R.; Schwickart, M.; Larsen, C.N.; Leggett, D.S.; Muller, B.; Feng, M.T.; Tubing, F.; Dittmar, G.A.; Finley, D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat. Cell. Biol. 2002, 4, 725–730. [Google Scholar] [CrossRef]
- Funakoshi, M.; Sasaki, T.; Nishimoto, T.; Kobayashi, H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc. Natl. Acad. Sci. USA 2002, 99, 745–750. [Google Scholar] [CrossRef] [Green Version]
- Schauber, C.; Chen, L.; Tongaonkar, P.; Vega, I.; Lambertson, D.; Potts, W.; Madura, K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature 1998, 391, 715–718. [Google Scholar] [CrossRef]
- Inobe, T.; Fishbain, S.; Prakash, S.; Matouschek, A. Defining the geometry of the two-component proteasome degron. Nat. Chem. Biol. 2011, 7, 161–167. [Google Scholar] [CrossRef] [Green Version]
- Kraut, D.A.; Matouschek, A. Proteasomal degradation from internal sites favors partial proteolysis via remote domain stabilization. ACS Chem. Biol. 2011, 6, 1087–1095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prakash, S.; Tian, L.; Ratliff, K.S.; Lehotzky, R.E.; Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol. 2004, 11, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Wu, J.; Lu, Y.; Ma, Y.B.; Lee, B.H.; Yu, Z.; Ouyang, Q.; Finley, D.J.; Kirschner, M.W.; Mao, Y. Structural basis for dynamic regulation of the human 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, 12991–12996. [Google Scholar] [CrossRef] [Green Version]
- de la Pena, A.H.; Goodall, E.A.; Gates, S.N.; Lander, G.C.; Martin, A. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 2018, 362. [Google Scholar] [CrossRef] [Green Version]
- Luan, B.; Huang, X.; Wu, J.; Mei, Z.; Wang, Y.; Xue, X.; Yan, C.; Wang, J.; Finley, D.J.; Shi, Y.; et al. Structure of an endogenous yeast 26S proteasome reveals two major conformational states. Proc. Natl. Acad. Sci. USA 2016, 113, 2642–2647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matyskiela, M.E.; Lander, G.C.; Martin, A. Conformational switching of the 26S proteasome enables substrate degradation. Nat. Struct. Mol. Biol. 2013, 20, 781–788. [Google Scholar] [CrossRef] [Green Version]
- Unverdorben, P.; Beck, F.; Sledz, P.; Schweitzer, A.; Pfeifer, G.; Plitzko, J.M.; Baumeister, W.; Forster, F. Deep classification of a large cryo-EM dataset defines the conformational landscape of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2014, 111, 5544–5549. [Google Scholar] [CrossRef] [Green Version]
- Wehmer, M.; Rudack, T.; Beck, F.; Aufderheide, A.; Pfeifer, G.; Plitzko, J.M.; Forster, F.; Schulten, K.; Baumeister, W.; Sakata, E. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc. Natl. Acad. Sci. USA 2017, 114, 1305–1310. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.M.; Kafri, G.; Cheng, Y.; Ng, D.; Walz, T.; Goldberg, A.L. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol. Cell 2005, 20, 687–698. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Callard, A.; Goldberg, A.L. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J. Biol. Chem. 2006, 281, 8582–8590. [Google Scholar] [CrossRef] [Green Version]
- Raule, M.; Cerruti, F.; Cascio, P. Enhanced rate of degradation of basic proteins by 26S immunoproteasomes. Biochim. Biophys. Acta 2014, 1843, 1942–1947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benaroudj, N.; Zwickl, P.; Seemuller, E.; Baumeister, W.; Goldberg, A.L. ATP hydrolysis by the proteasome regulatory complex PAN serves multiple functions in protein degradation. Mol. Cell 2003, 11, 69–78. [Google Scholar] [CrossRef]
- Henderson, A.; Erales, J.; Hoyt, M.A.; Coffino, P. Dependence of proteasome processing rate on substrate unfolding. J. Biol. Chem. 2011, 286, 17495–17502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bard, J.A.M.; Bashore, C.; Dong, K.C.; Martin, A. The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 2019, 177, 286–298 e215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebstein, F.; Voigt, A.; Lange, N.; Warnatsch, A.; Schroter, F.; Prozorovski, T.; Kuckelkorn, U.; Aktas, O.; Seifert, U.; Kloetzel, P.M.; et al. Immunoproteasomes are important for proteostasis in immune responses. Cell 2013, 152, 935–937. [Google Scholar] [CrossRef] [Green Version]
- Nathan, J.; Spinnenhirn, V.; Schmidtke, G.; Basler, M.; Groettrup, M.; Goldberg, A. Immuno- and Constitutive Proteasomes Do Not Differ in Their Abilities to Degrade Ubiquitinated Proteins. Cell 2013, 152, 1184–1194. [Google Scholar] [CrossRef] [Green Version]
- Seifert, U.; Bialy, L.P.; Ebstein, F.; Bech-Otschir, D.; Voigt, A.; Schroter, F.; Prozorovski, T.; Lange, N.; Steffen, J.; Rieger, M.; et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell 2010, 142, 613–624. [Google Scholar] [CrossRef] [Green Version]
- Abi Habib, J.; De Plaen, E.; Stroobant, V.; Zivkovic, D.; Bousquet, M.P.; Guillaume, B.; Wahni, K.; Messens, J.; Busse, A.; Vigneron, N.; et al. Efficiency of the four proteasome subtypes to degrade ubiquitinated or oxidized proteins. Sci. Rep. 2020, 10, 15765. [Google Scholar] [CrossRef]
- Hewing, B.; Ludwig, A.; Dan, C.; Potzsch, M.; Hannemann, C.; Petry, A.; Lauer, D.; Gorlach, A.; Kaschina, E.; Muller, D.N.; et al. Immunoproteasome subunit ss5i/LMP7-deficiency in atherosclerosis. Sci. Rep. 2017, 7, 13342. [Google Scholar] [CrossRef] [Green Version]
- Kincaid, E.Z.; Che, J.W.; York, I.; Escobar, H.; Reyes-Vargas, E.; Delgado, J.C.; Welsh, R.M.; Karow, M.L.; Murphy, A.J.; Valenzuela, D.M.; et al. Mice completely lacking immunoproteasomes show major changes in antigen presentation. Nat. Immunol. 2012, 13, 129–135. [Google Scholar] [CrossRef] [Green Version]
- Ben-Nissan, G.; Sharon, M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 2014, 4, 862–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar Deshmukh, F.; Yaffe, D.; Olshina, M.A.; Ben-Nissan, G.; Sharon, M. The Contribution of the 20S Proteasome to Proteostasis. Biomolecules 2019, 9, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabre, B.; Lambour, T.; Garrigues, L.; Ducoux-Petit, M.; Amalric, F.; Monsarrat, B.; Burlet-Schiltz, O.; Bousquet-Dubouch, M.P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J. Proteome Res. 2014, 13, 3027–3037. [Google Scholar] [CrossRef] [PubMed]
- Baugh, J.M.; Viktorova, E.G.; Pilipenko, E.V. Proteasomes can degrade a significant proportion of cellular proteins independent of ubiquitination. J. Mol. Biol. 2009, 386, 814–827. [Google Scholar] [CrossRef] [Green Version]
- Asher, G.; Reuven, N.; Shaul, Y. 20S proteasomes and protein degradation "by default". BioEssays 2006, 28, 844–849. [Google Scholar] [CrossRef]
- Liu, C.W.; Corboy, M.J.; DeMartino, G.N.; Thomas, P.J. Endoproteolytic activity of the proteasome. Science 2003, 299, 408–411. [Google Scholar] [CrossRef] [Green Version]
- Myers, N.; Olender, T.; Savidor, A.; Levin, Y.; Reuven, N.; Shaul, Y. The Disordered Landscape of the 20S Proteasome Substrates Reveals Tight Association with Phase Separated Granules. Proteomics 2018, 18, e1800076. [Google Scholar] [CrossRef]
- Tsvetkov, P.; Asher, G.; Paz, A.; Reuven, N.; Sussman, J.L.; Silman, I.; Shaul, Y. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins 2008, 70, 1357–1366. [Google Scholar] [CrossRef]
- Asher, G.; Bercovich, Z.; Tsvetkov, P.; Shaul, Y.; Kahana, C. 20S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 2005, 17, 645–655. [Google Scholar] [CrossRef]
- Asher, G.; Tsvetkov, P.; Kahana, C.; Shaul, Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005, 19, 316–321. [Google Scholar] [CrossRef] [Green Version]
- Moscovitz, O.; Ben-Nissan, G.; Fainer, I.; Pollack, D.; Mizrachi, L.; Sharon, M. The Parkinson’s-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015, 6, 6609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscovitz, O.; Tsvetkov, P.; Hazan, N.; Michaelevski, I.; Keisar, H.; Ben-Nissan, G.; Shaul, Y.; Sharon, M. A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol. Cell 2012, 47, 76–86. [Google Scholar] [CrossRef] [Green Version]
- Olshina, M.A.; Arkind, G.; Kumar Deshmukh, F.; Fainer, I.; Taranavsky, M.; Hayat, D.; Ben-Dor, S.; Ben-Nissan, G.; Sharon, M. Regulation of the 20S Proteasome by a Novel Family of Inhibitory Proteins. Antioxid. Redox Signal. 2020, 32, 636–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiken, C.T.; Kaake, R.M.; Wang, X.; Huang, L. Oxidative stress-mediated regulation of proteasome complexes. Mol. Cell. Proteom. 2011, 10, R110 006924. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.M.; Davies, K.J. Degradation of damaged proteins: The main function of the 20S proteasome. Prog. Mol. Biol. Transl. Sci. 2012, 109, 227–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrington, D.A.; Sun, H.; Murray, K.K.; Costa, J.; Williams, T.D.; Bigelow, D.J.; Squier, T.C. Selective degradation of oxidized calmodulin by the 20 S proteasome. J. Biol. Chem. 2001, 276, 937–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grune, T.; Catalgol, B.; Licht, A.; Ermak, G.; Pickering, A.M.; Ngo, J.K.; Davies, K.J. HSP70 mediates dissociation and reassociation of the 26S proteasome during adaptation to oxidative stress. Free Radic. Biol. Med. 2011, 51, 1355–1364. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chemmama, I.E.; Yu, C.; Huszagh, A.; Xu, Y.; Viner, R.; Block, S.A.; Cimermancic, P.; Rychnovsky, S.D.; Ye, Y.; et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 2017, 292, 16310–16320. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yen, J.; Kaiser, P.; Huang, L. Regulation of the 26S proteasome complex during oxidative stress. Sci. Signal. 2010, 3, ra88. [Google Scholar] [CrossRef] [Green Version]
- Armstrong, J.A.; Cash, N.J.; Ouyang, Y.; Morton, J.C.; Chvanov, M.; Latawiec, D.; Awais, M.; Tepikin, A.V.; Sutton, R.; Criddle, D.N. Oxidative stress alters mitochondrial bioenergetics and modifies pancreatic cell death independently of cyclophilin D, resulting in an apoptosis-to-necrosis shift. J. Biol. Chem. 2018, 293, 8032–8047. [Google Scholar] [CrossRef] [Green Version]
- Green, K.; Brand, M.D.; Murphy, M.P. Prevention of mitochondrial oxidative damage as a therapeutic strategy in diabetes. Diabetes 2004, 53 (Suppl. 1), S110–S118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabharwal, S.S.; Schumacker, P.T. Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 2014, 14, 709–721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Inai, Y.; Nishikimi, M. Increased degradation of oxidized proteins in yeast defective in 26 S proteasome assembly. Arch. Biochem. Biophys. 2002, 404, 279–284. [Google Scholar] [CrossRef]
- Shringarpure, R.; Grune, T.; Mehlhase, J.; Davies, K.J. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J. Biol. Chem. 2003, 278, 311–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, K.J. Degradation of oxidized proteins by the 20S proteasome. Biochimie 2001, 83, 301–310. [Google Scholar] [CrossRef]
- Choi, W.H.; Kim, S.; Park, S.; Lee, M.J. Concept and application of circulating proteasomes. Exp. Mol. Med. 2021, 53, 1539–1546. [Google Scholar] [CrossRef]
- Dianzani, C.; Bellavista, E.; Liepe, J.; Verderio, C.; Martucci, M.; Santoro, A.; Chiocchetti, A.; Gigliotti, C.L.; Boggio, E.; Ferrara, B.; et al. Extracellular proteasome-osteopontin circuit regulates cell migration with implications in multiple sclerosis. Sci. Rep. 2017, 7, 43718. [Google Scholar] [CrossRef]
- Dianzani, C.; Vecchio, D.; Clemente, N.; Chiocchetti, A.; Martinelli Boneschi, F.; Galimberti, D.; Dianzani, U.; Comi, C.; Mishto, M.; Liepe, J. Untangling Extracellular Proteasome-Osteopontin Circuit Dynamics in Multiple Sclerosis. Cells 2019, 8, 262. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Powell, S.R.; Wang, X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. Faseb J. 2011, 25, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.M.; Davies, K.J. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.M.; Koop, A.L.; Teoh, C.Y.; Ermak, G.; Grune, T.; Davies, K.J. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem. J. 2010, 432, 585–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernebring, M.; Fredriksson, A.; Liljevald, M.; Cvijovic, M.; Norrman, K.; Wiseman, J.; Semb, H.; Nystrom, T. Removal of damaged proteins during ES cell fate specification requires the proteasome activator PA28. Sci. Rep. 2013, 3, 1381. [Google Scholar] [CrossRef] [PubMed]
- Lesne, J.; Locard-Paulet, M.; Parra, J.; Zivković, D.; Menneteau, T.; Bousquet, M.P.; Burlet-Schiltz, O.; Marcoux, J. Conformational maps of human 20S proteasomes reveal PA28- and immuno-dependent inter-ring crosstalks. Nat. Commun. 2020, 11, 6140. [Google Scholar] [CrossRef] [PubMed]
- Fabre, B.; Lambour, T.; Garrigues, L.; Amalric, F.; Vigneron, N.; Menneteau, T.; Stella, A.; Monsarrat, B.; Van den Eynde, B.; Burlet-Schiltz, O.; et al. Deciphering preferential interactions within supramolecular protein complexes: The proteasome case. Mol. Syst. Biol. 2015, 11, 771. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Barton, L.F.; Chi, Y.; Clurman, B.E.; Roberts, J.M. Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome. Mol. Cell 2007, 26, 843–852. [Google Scholar] [CrossRef] [Green Version]
- Kanai, K.; Aramata, S.; Katakami, S.; Yasuda, K.; Kataoka, K. Proteasome activator PA28{gamma} stimulates degradation of GSK3-phosphorylated insulin transcription activator MAFA. J. Mol. Endocrinol. 2011, 47, 119–127. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Lonard, D.M.; Jung, S.Y.; Malovannaya, A.; Feng, Q.; Qin, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 2006, 124, 381–392. [Google Scholar] [CrossRef] [Green Version]
- Aladdin, A.; Yao, Y.; Yang, C.; Kahlert, G.; Ghani, M.; Kiraly, N.; Boratko, A.; Uray, K.; Dittmar, G.; Tar, K. The Proteasome Activators Blm10/PA200 Enhance the Proteasomal Degradation of N-Terminal Huntingtin. Biomolecules 2020, 10, 1581. [Google Scholar] [CrossRef]
- Dange, T.; Smith, D.; Noy, T.; Rommel, P.C.; Jurzitza, L.; Cordero, R.J.; Legendre, A.; Finley, D.; Goldberg, A.L.; Schmidt, M. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J. Biol. Chem. 2011, 286, 42830–42839. [Google Scholar] [CrossRef] [Green Version]
- Tar, K.; Dange, T.; Yang, C.; Yao, Y.; Bulteau, A.L.; Salcedo, E.F.; Braigen, S.; Bouillaud, F.; Finley, D.; Schmidt, M. Proteasomes associated with the Blm10 activator protein antagonize mitochondrial fission through degradation of the fission protein Dnm1. J. Biol. Chem. 2014, 289, 12145–12156. [Google Scholar] [CrossRef] [Green Version]
- Schmidtke, G.; Kraft, R.; Kostka, S.; Henklein, P.; Frommel, C.; Lowe, J.; Huber, R.; Kloetzel, P.M.; Schmidt, M. Analysis of mammalian 20S proteasome biogenesis: The maturation of beta-subunits is an ordered two-step mechanism involving autocatalysis. EMBO J. 1996, 15, 6887–6898. [Google Scholar] [CrossRef] [PubMed]
- Seemuller, E.; Lupas, A.; Stock, D.; Lowe, J.; Huber, R.; Baumeister, W. Proteasome from Thermoplasma acidophilum: A threonine protease. Science 1995, 268, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Rock, K.L.; Gramm, C.; Rothstein, L.; Clark, K.; Stein, R.; Dick, L.; Hwang, D.; Goldberg, A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994, 78, 761–771. [Google Scholar] [CrossRef]
- Saric, T.; Graef, C.I.; Goldberg, A.L. Pathway for degradation of peptides generated by proteasomes: A key role for thimet oligopeptidase and other metallopeptidases. J. Biol. Chem. 2004, 279, 46723–46732. [Google Scholar] [CrossRef] [Green Version]
- Ben-Shahar, S.; Cassouto, B.; Novak, L.; Porgador, A.; Reiss, Y. Production of a specific major histocompatibility complex class I-restricted epitope by ubiquitin-dependent degradation of modified ovalbumin in lymphocyte lysate. J. Biol. Chem. 1997, 272, 21060–21066. [Google Scholar] [CrossRef] [Green Version]
- Cerundolo, V.; Benham, A.; Braud, V.; Mukherjee, S.; Gould, K.; Macino, B.; Neefjes, J.; Townsend, A. The proteasome-specific inhibitor lactacystin blocks presentation of cytotoxic T lymphocyte epitopes in human and murine cells. Eur. J. Immunol. 1997, 27, 336–341. [Google Scholar] [CrossRef]
- Harding, C.V.; France, J.; Song, R.; Farah, J.M.; Chatterjee, S.; Iqbal, M.; Siman, R. Novel dipeptide aldehydes are proteasome inhibitors and block the MHC-I antigen-processing pathway. J. Immunol. 1995, 155, 1767–1775. [Google Scholar]
- Michalek, M.T.; Grant, E.P.; Gramm, C.; Goldberg, A.L.; Rock, K.L. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature 1993, 363, 552–554. [Google Scholar] [CrossRef]
- Mo, X.Y.; Cascio, P.; Lemerise, K.; Goldberg, A.L.; Rock, K. Distinct proteolytic processes generate the C and N termini of MHC class I-binding peptides. J. Immunol. 1999, 163, 5851–5859. [Google Scholar]
- Serwold, T.; Gonzalez, F.; Kim, J.; Jacob, R.; Shastri, N. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature 2002, 419, 480–483. [Google Scholar] [CrossRef]
- Guillaume, B.; Stroobant, V.; Bousquet-Dubouch, M.P.; Colau, D.; Chapiro, J.; Parmentier, N.; Dalet, A.; Van den Eynde, B.J. Analysis of the processing of seven human tumor antigens by intermediate proteasomes. J. Immunol. 2012, 189, 3538–3547. [Google Scholar] [CrossRef] [PubMed]
- Morel, S.; Levy, F.; Burlet-Schiltz, O.; Brasseur, F.; Probst-Kepper, M.; Peitrequin, A.L.; Monsarrat, B.; Van Velthoven, R.; Cerottini, J.C.; Boon, T.; et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity 2000, 12, 107–117. [Google Scholar] [CrossRef]
- Sasaki, K.; Takada, K.; Ohte, Y.; Kondo, H.; Sorimachi, H.; Tanaka, K.; Takahama, Y.; Murata, S. Thymoproteasomes produce unique peptide motifs for positive selection of CD8(+) T cells. Nat. Commun. 2015, 6, 7484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olsson, N.; Heberling, M.L.; Zhang, L.; Jhunjhunwala, S.; Phung, Q.T.; Lin, S.; Anania, V.G.; Lill, J.R.; Elias, J.E. An Integrated Genomic, Proteomic, and Immunopeptidomic Approach to Discover Treatment-Induced Neoantigens. Front. Immunol. 2021, 12, 662443. [Google Scholar] [CrossRef]
- Newey, A.; Griffiths, B.; Michaux, J.; Pak, H.S.; Stevenson, B.J.; Woolston, A.; Semiannikova, M.; Spain, G.; Barber, L.J.; Matthews, N.; et al. Immunopeptidomics of colorectal cancer organoids reveals a sparse HLA class I neoantigen landscape and no increase in neoantigens with interferon or MEK-inhibitor treatment. J. Immunother. Cancer 2019, 7, 309. [Google Scholar] [CrossRef]
- Kalaora, S.; Lee, J.S.; Barnea, E.; Levy, R.; Greenberg, P.; Alon, M.; Yagel, G.; Bar Eli, G.; Oren, R.; Peri, A.; et al. Immunoproteasome expression is associated with better prognosis and response to checkpoint therapies in melanoma. Nat. Commun. 2020, 11, 896. [Google Scholar] [CrossRef] [Green Version]
- Chapiro, J.; Claverol, S.; Piette, F.; Ma, W.; Stroobant, V.; Guillaume, B.; Gairin, J.E.; Morel, S.; Burlet-Schiltz, O.; Monsarrat, B.; et al. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J. Immunol. 2006, 176, 1053–1061. [Google Scholar] [CrossRef] [Green Version]
- Zaiss, D.M.; Bekker, C.P.; Grone, A.; Lie, B.A.; Sijts, A.J. Proteasome immunosubunits protect against the development of CD8 T cell-mediated autoimmune diseases. J. Immunol. 2011, 187, 2302–2309. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Lauer, C.; Moebius, J.; Weber, R.; Przybylski, M.; Kisselev, A.F.; Tsu, C.; Groettrup, M. Why the structure but not the activity of the immunoproteasome subunit low molecular mass polypeptide 2 rescues antigen presentation. J. Immunol. 2012, 189, 1868–1877. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Yewdell, J.W.; Yang, J.C. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature 2004, 427, 252–256. [Google Scholar] [CrossRef]
- Vigneron, N.; Stroobant, V.; Chapiro, J.; Ooms, A.; Degiovanni, G.; Morel, S.; van der Bruggen, P.; Boon, T.; Van den Eynde, B.J. An antigenic peptide produced by peptide splicing in the proteasome. Science 2004, 304, 587–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalet, A.; Robbins, P.F.; Stroobant, V.; Vigneron, N.; Li, Y.F.; El-Gamil, M.; Hanada, K.; Yang, J.C.; Rosenberg, S.A.; Van den Eynde, B.J. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. USA 2011, 108, E323–E331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebstein, F.; Textoris-Taube, K.; Keller, C.; Golnik, R.; Vigneron, N.; Van den Eynde, B.J.; Schuler-Thurner, B.; Schadendorf, D.; Lorenz, F.K.; Uckert, W.; et al. Proteasomes generate spliced epitopes by two different mechanisms and as efficiently as non-spliced epitopes. Sci. Rep. 2016, 6, 24032. [Google Scholar] [CrossRef] [PubMed]
- Michaux, A.; Larrieu, P.; Stroobant, V.; Fonteneau, J.F.; Jotereau, F.; Van den Eynde, B.J.; Moreau-Aubry, A.; Vigneron, N. A spliced antigenic peptide comprising a single spliced amino acid is produced in the proteasome by reverse splicing of a longer peptide fragment followed by trimming. J. Immunol. 2014, 192, 1962–1971. [Google Scholar] [CrossRef] [Green Version]
- Warren, E.H.; Vigneron, N.J.; Gavin, M.A.; Coulie, P.G.; Stroobant, V.; Dalet, A.; Tykodi, S.S.; Xuereb, S.M.; Mito, J.K.; Riddell, S.R.; et al. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science 2006, 313, 1444–1447. [Google Scholar] [CrossRef]
- Faridi, P.; Li, C.; Ramarathinam, S.H.; Vivian, J.P.; Illing, P.T.; Mifsud, N.A.; Ayala, R.; Song, J.; Gearing, L.J.; Hertzog, P.J.; et al. A subset of HLA-I peptides are not genomically templated: Evidence for cis- and trans-spliced peptide ligands. Sci. Immunol. 2018, 3, eaar3947. [Google Scholar] [CrossRef] [Green Version]
- Lichti, C.; Vigneron, N.; Clauser, K.; Van den Eynde, B.; Bassani-Sternberg, M. Navigating critical challenges associated with immunopeptidomics-based detection of proteasomal spliced peptide candidates. Cancer Immunol. Res. 2022; in press. [Google Scholar]
- Liepe, J.; Marino, F.; Sidney, J.; Jeko, A.; Bunting, D.E.; Sette, A.; Kloetzel, P.M.; Stumpf, M.P.; Heck, A.J.; Mishto, M. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science 2016, 354, 354–358. [Google Scholar] [CrossRef] [Green Version]
- Liepe, J.; Sidney, J.; Lorenz, F.K.; Sette, A.; Mishto, M. Mapping the MHC class I spliced immunopeptidome of cancer cells. Cancer Immunol. Res. 2018, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Mishto, M.; Mansurkhodzhaev, A.; Ying, G.; Bitra, A.; Cordfunke, R.A.; Henze, S.; Paul, D.; Sidney, J.; Urlaub, H.; Neefjes, J.; et al. An in silico-in vitro Pipeline Identifying an HLA-A(*)02:01(+) KRAS G12V(+) Spliced Epitope Candidate for a Broad Tumor-Immune Response in Cancer Patients. Front. Immunol. 2019, 10, 2572. [Google Scholar] [CrossRef] [Green Version]
- Mylonas, R.; Beer, I.; Iseli, C.; Chong, C.; Pak, H.S.; Gfeller, D.; Coukos, G.; Xenarios, I.; Muller, M.; Bassani-Sternberg, M. Estimating the Contribution of Proteasomal Spliced Peptides to the HLA-I Ligandome. Mol. Cell. Proteom. 2018, 2347–2357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalet, A.; Stroobant, V.; Vigneron, N.; Van den Eynde, B.J. Differences in the production of spliced antigenic peptides by the standard proteasome and the immunoproteasome. Eur. J. Immunol. 2011, 41, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Nitta, T.; Murata, S.; Sasaki, K.; Fujii, H.; Ripen, A.M.; Ishimaru, N.; Koyasu, S.; Tanaka, K.; Takahama, Y. Thymoproteasome shapes immunocompetent repertoire of CD8+ T cells. Immunity 2010, 32, 29–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takada, K.; Van Laethem, F.; Xing, Y.; Akane, K.; Suzuki, H.; Murata, S.; Tanaka, K.; Jameson, S.C.; Singer, A.; Takahama, Y. TCR affinity for thymoproteasome-dependent positively selecting peptides conditions antigen responsiveness in CD8(+) T cells. Nat. Immunol. 2015, 16, 1069–1076. [Google Scholar] [CrossRef] [Green Version]
- Tomaru, U.; Konno, S.; Miyajima, S.; Kimoto, R.; Onodera, M.; Kiuchi, S.; Murata, S.; Ishizu, A.; Kasahara, M. Restricted Expression of the Thymoproteasome Is Required for Thymic Selection and Peripheral Homeostasis of CD8(+) T Cells. Cell Rep. 2019, 26, 639–651.e632. [Google Scholar] [CrossRef] [Green Version]
- Murata, S.; Takahama, Y.; Kasahara, M.; Tanaka, K. The immunoproteasome and thymoproteasome: Functions, evolution and human disease. Nat. Immunol. 2018, 19, 923–931. [Google Scholar] [CrossRef]
- Basler, M.; Dajee, M.; Moll, C.; Groettrup, M.; Kirk, C.J. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J. Immunol. 2010, 185, 634–641. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Li, J.; Groettrup, M. On the role of the immunoproteasome in transplant rejection. Immunogenetics 2018, 71, 263–271. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Maurits, E.; de Bruin, G.; Koerner, J.; Overkleeft, H.S.; Groettrup, M. Amelioration of autoimmunity with an inhibitor selectively targeting all active centres of the immunoproteasome. Br. J. Pharmacol. 2018, 175, 38–52. [Google Scholar] [CrossRef]
- Basler, M.; Mundt, S.; Muchamuel, T.; Moll, C.; Jiang, J.; Groettrup, M.; Kirk, C.J. Inhibition of the immunoproteasome ameliorates experimental autoimmune encephalomyelitis. EMBO Mol. Med. 2014, 6, 226–238. [Google Scholar] [CrossRef]
- Ichikawa, H.T.; Conley, T.; Muchamuel, T.; Jiang, J.; Lee, S.; Owen, T.; Barnard, J.; Nevarez, S.; Goldman, B.I.; Kirk, C.J.; et al. Beneficial effect of novel proteasome inhibitors in murine lupus via dual inhibition of type I interferon and autoantibody-secreting cells. Arthritis Rheum. 2012, 64, 493–503. [Google Scholar] [CrossRef]
- Koerner, J.; Brunner, T.; Groettrup, M. Inhibition and deficiency of the immunoproteasome subunit LMP7 suppress the development and progression of colorectal carcinoma in mice. Oncotarget 2017, 8, 50873–50888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leister, H.; Luu, M.; Staudenraus, D.; Lopez Krol, A.; Mollenkopf, H.J.; Sharma, A.; Schmerer, N.; Schulte, L.N.; Bertrams, W.; Schmeck, B.; et al. Pro- and Antitumorigenic Capacity of Immunoproteasomes in Shaping the Tumor Microenvironment. Cancer Immunol. Res. 2021, 9, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Basler, M.; Alvarez, G.; Brunner, T.; Kirk, C.J.; Groettrup, M. Immunoproteasome inhibition prevents chronic antibody-mediated allograft rejection in renal transplantation. Kidney Int. 2018, 93, 670–680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sula Karreci, E.; Fan, H.; Uehara, M.; Mihali, A.B.; Singh, P.K.; Kurdi, A.T.; Solhjou, Z.; Riella, L.V.; Ghobrial, I.; Laragione, T.; et al. Brief treatment with a highly selective immunoproteasome inhibitor promotes long-term cardiac allograft acceptance in mice. Proc. Natl. Acad. Sci. USA 2016, 113, E8425–E8432. [Google Scholar] [CrossRef] [Green Version]
- Kalim, K.W.; Basler, M.; Kirk, C.J.; Groettrup, M. Immunoproteasome subunit LMP7 deficiency and inhibition suppresses Th1 and Th17 but enhances regulatory T cell differentiation. J. Immunol. 2012, 189, 4182–4193. [Google Scholar] [CrossRef]
- Muchamuel, T.; Basler, M.; Aujay, M.A.; Suzuki, E.; Kalim, K.W.; Lauer, C.; Sylvain, C.; Ring, E.R.; Shields, J.; Jiang, J.; et al. A selective inhibitor of the immunoproteasome subunit LMP7 blocks cytokine production and attenuates progression of experimental arthritis. Nat. Med. 2009, 15, 781–787. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Lin, X.; Tian, J.; Wang, X.; Chen, Q.; Rui, K.; Ma, J.; Wang, S.; Wang, Q.; Wang, X.; et al. Proteasome inhibition suppresses Th17 cell generation and ameliorates autoimmune development in experimental Sjogren’s syndrome. Cell. Mol. Immunol. 2017, 14, 924–934. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Claus, M.; Klawitter, M.; Goebel, H.; Groettrup, M. Immunoproteasome Inhibition Selectively Kills Human CD14(+) Monocytes and as a Result Dampens IL-23 Secretion. J. Immunol. 2019, 203, 1776–1785. [Google Scholar] [CrossRef]
- Moebius, J.; van den Broek, M.; Groettrup, M.; Basler, M. Immunoproteasomes are essential for survival and expansion of T cells in virus-infected mice. Eur. J. Immunol. 2010, 40, 3439–3449. [Google Scholar] [CrossRef] [Green Version]
- Zaiss, D.M.; de Graaf, N.; Sijts, A.J. The proteasome immunosubunit multicatalytic endopeptidase complex-like 1 is a T-cell-intrinsic factor influencing homeostatic expansion. Infect. Immun. 2008, 76, 1207–1213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jenkins, T.W.; Downey-Kopyscinski, S.L.; Fields, J.L.; Rahme, G.J.; Colley, W.C.; Israel, M.A.; Maksimenko, A.V.; Fiering, S.N.; Kisselev, A.F. Activity of immunoproteasome inhibitor ONX-0914 in acute lymphoblastic leukemia expressing MLL-AF4 fusion protein. Sci. Rep. 2021, 11, 10883. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, M.P.; Friese-Hamim, M.; Walter-Bausch, G.; Busch, M.; Gaus, S.; Musil, D.; Rohdich, F.; Zanelli, U.; Downey-Kopyscinski, S.L.; Mitsiades, C.S.; et al. M3258 Is a Selective Inhibitor of the Immunoproteasome Subunit LMP7 (β5i) Delivering Efficacy in Multiple Myeloma Models. Mol. Cancer Ther. 2021, 20, 1378–1387. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abi Habib, J.; Lesenfants, J.; Vigneron, N.; Van den Eynde, B.J. Functional Differences between Proteasome Subtypes. Cells 2022, 11, 421. https://doi.org/10.3390/cells11030421
Abi Habib J, Lesenfants J, Vigneron N, Van den Eynde BJ. Functional Differences between Proteasome Subtypes. Cells. 2022; 11(3):421. https://doi.org/10.3390/cells11030421
Chicago/Turabian StyleAbi Habib, Joanna, Julie Lesenfants, Nathalie Vigneron, and Benoit J. Van den Eynde. 2022. "Functional Differences between Proteasome Subtypes" Cells 11, no. 3: 421. https://doi.org/10.3390/cells11030421
APA StyleAbi Habib, J., Lesenfants, J., Vigneron, N., & Van den Eynde, B. J. (2022). Functional Differences between Proteasome Subtypes. Cells, 11(3), 421. https://doi.org/10.3390/cells11030421