Zscan4 Contributes to Telomere Maintenance in Telomerase-Deficient Late Generation Mouse ESCs and Human ALT Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Stable Zscan4 RNAi in G4 Terc-/- ES Cells, ALT-U2 OS Cells, and ZSCAN4 KO in U2 OS Cells
2.3. Cell Cycle Analysis
2.4. Gene Expression Analysis by Quantitative Real-Time PCR
2.5. Immunoblot Analysis
2.6. Immunofluorescence Microscopy
2.7. Telomere Quantitative Fluorescence In Situ Hybridization (Q-FISH)
2.8. Telomere Measurement by Quantitative Real-Time PCR (T/S Ratio)
2.9. Telomere Chromatid Orientation-Fluorescence In Situ Hybridization (CO-FISH)
2.10. Global DNA Methylation Analysis
2.11. Bisulfate Sequencing
2.12. ChIP-qPCR Analysis
2.13. Co-Localization of Telomeresand Human Chromosome 19
2.14. Statistical Analysis
3. Results
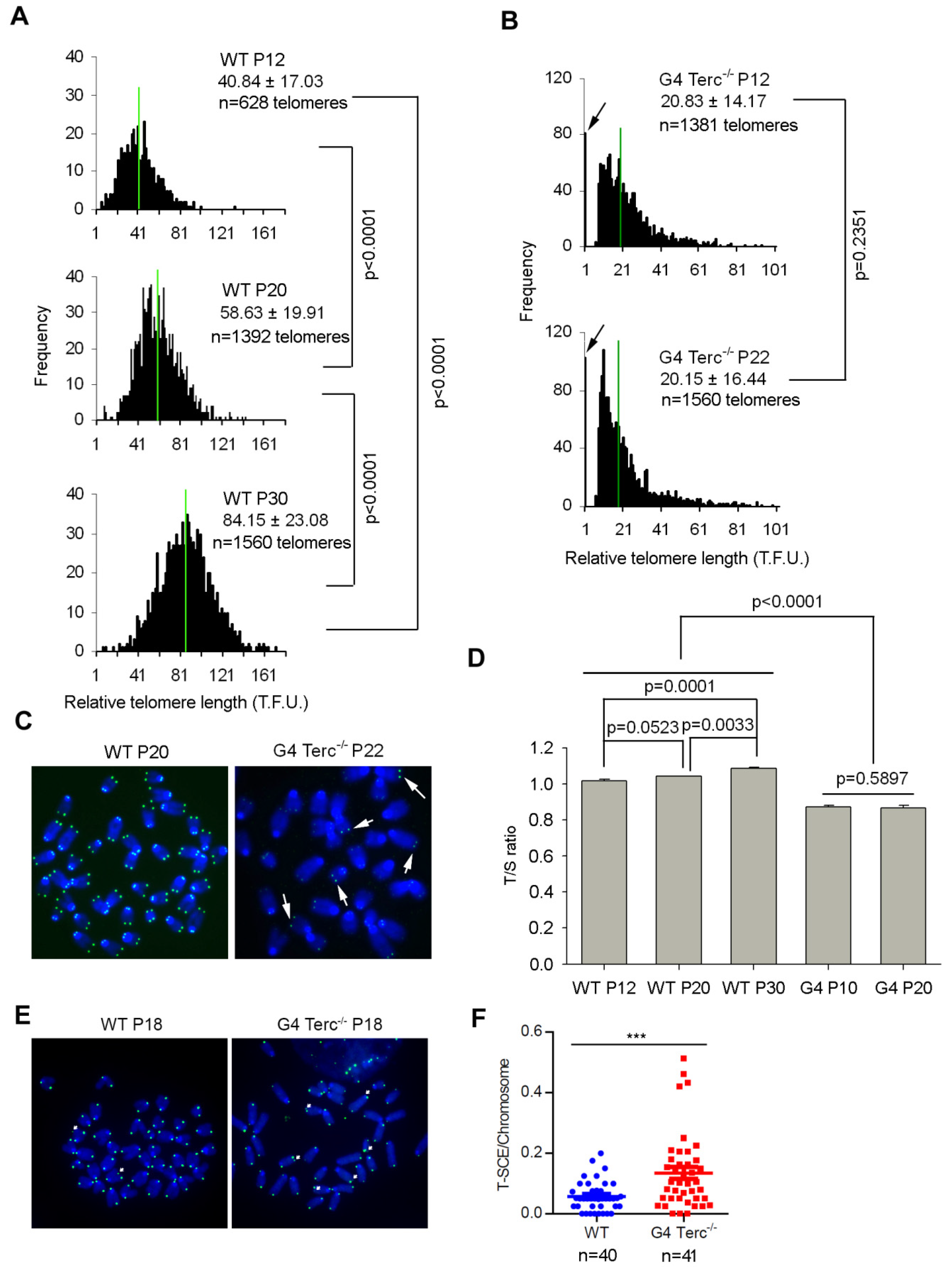
3.1. Late Generation Terc-/- ES Cells Maintains Stably Short Telomere Length during Prolonged Passaging
3.2. Up-Regulation of Zscan4 Is Implicated in Telomere Maintenance of G4 Terc-/- ES Cells
3.3. DNA Methylation and H3K9me3 Regulate Zscan4 of G4 Terc-/- ES Cells
3.4. Inhibition of Repressive DNA and H3K9 Methylation Up-Regulates Zscan4 and Elongates Telomeres
3.5. ZSCAN4 Depletion in Telomerase-Deficient Human ALT U2 OS Cancer Cells Shortens Telomeres
3.6. Human ZSCAN4 Is Also Repressed by Epigenetic Marks at Its Promoter
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blackburn, E.H. Switching and signaling at the telomere. Cell 2001, 106, 661–673. [Google Scholar] [CrossRef] [Green Version]
- Palm, W.; de Lange, T. How shelterin protects mammalian telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [Green Version]
- Hiyama, E.; Hiyama, K. Telomere and telomerase in stem cells. Br. J. Cancer 2007, 96, 1020–1024. [Google Scholar] [CrossRef] [Green Version]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, R.C.; Chang, E.; Kashefi-Aazam, M.; Rogaev, E.I.; Piatyszek, M.A.; Shay, J.W.; Harley, C.B. Telomere shortening is associated with cell division in vitro and in vivo. Exp. Cell Res. 1995, 220, 194–200. [Google Scholar] [CrossRef]
- Blasco, M.A. Telomeres and human disease: Ageing, cancer and beyond. Nat. Rev. Genet. 2005, 6, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Wang, F.; Okuka, M.; Liu, N.; Ji, G.; Ye, X.; Zuo, B.; Li, M.; Liang, P.; Ge, W.W.; et al. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011, 21, 779–792. [Google Scholar] [CrossRef] [Green Version]
- Shay, J.W.; Reddel, R.R.; Wright, W.E. Cancer. Cancer and telomeres—An ALTernative to telomerase. Science 2012, 336, 1388–1390. [Google Scholar] [CrossRef]
- Blasco, M.A. The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 2007, 8, 299–309. [Google Scholar] [CrossRef]
- Ottaviani, A.; Gilson, E.; Magdinier, F. Telomeric position effect: From the yeast paradigm to human pathologies? Biochimie 2008, 90, 93–107. [Google Scholar] [CrossRef] [Green Version]
- Slijepcevic, P. Telomere length regulation—A view from the individual chromosome perspective. Exp. Cell Res. 1998, 244, 268–274. [Google Scholar] [CrossRef]
- Zhimulev, I.F.; Beliaeva, E.S. Heterochromatin, gene position effect and gene silencing. Genetika 2003, 39, 187–201. [Google Scholar]
- Benetti, R.; Garcia-Cao, M.; Blasco, M.A. Telomere length regulates the epigenetic status of mammalian telomeres and subtelomeres. Nat. Genet. 2007, 39, 243–250. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.A.; Svensson, V.; Krueger, C.; Stubbs, T.M.; Giehr, P.; Krueger, F.; Miragaia, R.J.; Kyriakopoulos, C.; Berrens, R.V.; Milagre, I.; et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 2016, 17, 179–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, J.; Rousseau, P.; Hardikar, S.; Veland, N.; Wong, J.; Autexier, C.; Chen, T. Zscan4 Inhibits Maintenance DNA Methylation to Facilitate Telomere Elongation in Mouse Embryonic Stem Cells. Cell Rep. 2017, 20, 1936–1949. [Google Scholar] [CrossRef] [Green Version]
- Zalzman, M.; Falco, G.; Sharova, L.V.; Nishiyama, A.; Thomas, M.; Lee, S.L.; Stagg, C.A.; Hoang, H.G.; Yang, H.T.; Indig, F.E.; et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature 2010, 464, 858–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falco, G.; Lee, S.L.; Stanghellini, I.; Bassey, U.C.; Hamatani, T.; Ko, M.S. Zscan4: A novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev. Biol. 2007, 307, 539–550. [Google Scholar] [CrossRef] [Green Version]
- De Iaco, A.; Planet, E.; Coluccio, A.; Verp, S.; Duc, J.; Trono, D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat. Genet. 2017, 49, 941–945. [Google Scholar] [CrossRef]
- Hendrickson, P.G.; Dorais, J.A.; Grow, E.J.; Whiddon, J.L.; Lim, J.W.; Wike, C.L.; Weaver, B.D.; Pflueger, C.; Emery, B.R.; Wilcox, A.L.; et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat. Genet. 2017, 49, 925–934. [Google Scholar] [CrossRef]
- Whiddon, J.L.; Langford, A.T.; Wong, C.J.; Zhong, J.W.; Tapscott, S.J. Conservation and innovation in the DUX4-family gene network. Nat. Genet. 2017, 49, 935–940. [Google Scholar] [CrossRef] [Green Version]
- Le, R.; Huang, Y.; Zhang, Y.; Wang, H.; Lin, J.; Dong, Y.; Li, Z.; Guo, M.; Kou, X.; Zhao, Y.; et al. Dcaf11 activates Zscan4-mediated alternative telomere lengthening in early embryos and embryonic stem cells. Cell Stem Cell 2021, 28, 732–747. [Google Scholar] [CrossRef] [PubMed]
- Markiewicz-Potoczny, M.; Lobanova, A.; Loeb, A.M.; Kirak, O.; Olbrich, T.; Ruiz, S.; Lazzerini Denchi, E. TRF2-mediated telomere protection is dispensable in pluripotent stem cells. Nature 2021, 589, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Nady, N.; Arora, N.; Hsieh, L.J.; Swigut, T.; Narlikar, G.J.; Wossidlo, M.; Wysocka, J. Zscan4 binds nucleosomal microsatellite DNA and protects mouse two-cell embryos from DNA damage. Sci. Adv. 2020, 6, eaaz9115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dan, J.; Liu, Y.; Liu, N.; Chiourea, M.; Okuka, M.; Wu, T.; Ye, X.; Mou, C.; Wang, L.; Yin, Y.; et al. Rif1 maintains telomere length homeostasis of ESCs by mediating heterochromatin silencing. Dev. Cell 2014, 29, 7–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, S.S.; Martens, U.M.; Ward, R.K.; Lansdorp, P.M. Telomere length measurements using digital fluorescence microscopy. Cytometry 1999, 36, 267–278. [Google Scholar] [CrossRef]
- Callicott, R.J.; Womack, J.E. Real-time PCR assay for measurement of mouse telomeres. Comp. Med. 2006, 56, 17–22. [Google Scholar]
- Liu, L.; Bailey, S.M.; Okuka, M.; Munoz, P.; Li, C.; Zhou, L.; Wu, C.; Czerwiec, E.; Sandler, L.; Seyfang, A.; et al. Telomere lengthening early in development. Nat. Cell Biol. 2007, 9, 1436–1441. [Google Scholar] [CrossRef]
- Dan, J.; Li, M.; Yang, J.; Li, J.; Okuka, M.; Ye, X.; Liu, L. Roles for Tbx3 in regulation of two-cell state and telomere elongation in mouse ES cells. Sci. Rep. 2013, 3, 3492. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Yin, Y.; Ye, X.; Liu, K.; Zhu, H.; Wang, L.; Chiourea, M.; Okuka, M.; Ji, G.; Dan, J.; et al. Molecular insights into the heterogeneity of telomere reprogramming in induced pluripotent stem cells. Cell Res. 2012, 22, 757–768. [Google Scholar] [CrossRef]
- Marion, R.M.; Strati, K.; Li, H.; Tejera, A.; Schoeftner, S.; Ortega, S.; Serrano, M.; Blasco, M.A. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 2009, 4, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Varela, E.; Schneider, R.P.; Ortega, S.; Blasco, M.A. Different telomere-length dynamics at the inner cell mass versus established embryonic stem (ES) cells. Proc. Natl. Acad. Sci. USA 2011, 108, 15207–15212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macfarlan, T.S.; Gifford, W.D.; Driscoll, S.; Lettieri, K.; Rowe, H.M.; Bonanomi, D.; Firth, A.; Singer, O.; Trono, D.; Pfaff, S.L. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 2012, 487, 57–63. [Google Scholar] [CrossRef] [Green Version]
- Nakai-Futatsugi, Y.; Niwa, H. Zscan4 Is Activated after Telomere Shortening in Mouse Embryonic Stem Cells. Stem Cell Rep. 2016, 6, 483–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, L.H. Epigenetic regulation of telomere chromatin integrity in pluripotent embryonic stem cells. Epigenomics 2010, 2, 639–655. [Google Scholar] [CrossRef] [PubMed]
- Pucci, F.; Gardano, L.; Harrington, L. Short telomeres in ESCs lead to unstable differentiation. Cell Stem Cell 2013, 12, 479–486. [Google Scholar] [CrossRef] [Green Version]
- Draskovic, I.; Arnoult, N.; Steiner, V.; Bacchetti, S.; Lomonte, P.; Londono-Vallejo, A. Probing PML body function in ALT cells reveals spatiotemporal requirements for telomere recombination. Proc. Natl. Acad. Sci. USA 2009, 106, 15726–15731. [Google Scholar] [CrossRef] [Green Version]
- Londono-Vallejo, J.A.; Der-Sarkissian, H.; Cazes, L.; Bacchetti, S.; Reddel, R.R. Alternative lengthening of telomeres is characterized by high rates of telomeric exchange. Cancer Res. 2004, 64, 2324–2327. [Google Scholar] [CrossRef] [Green Version]
- Vera, E.; Canela, A.; Fraga, M.F.; Esteller, M.; Blasco, M.A. Epigenetic regulation of telomeres in human cancer. Oncogene 2008, 27, 6817–6833. [Google Scholar] [CrossRef] [Green Version]
- Scheel, C.; Schaefer, K.L.; Jauch, A.; Keller, M.; Wai, D.; Brinkschmidt, C.; van Valen, F.; Boecker, W.; Dockhorn-Dworniczak, B.; Poremba, C. Alternative lengthening of telomeres is associated with chromosomal instability in osteosarcomas. Oncogene 2001, 20, 3835–3844. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Pan, X.; Kalmbach, K.; Seth-Smith, M.L.; Ye, X.; Antumes, D.M.; Yin, Y.; Liu, L.; Keefe, D.L.; Weissman, S.M. Robust measurement of telomere length in single cells. Proc. Natl. Acad. Sci. USA 2013, 110, E1906–E1912. [Google Scholar] [CrossRef] [Green Version]
- Niida, H.; Shinkai, Y.; Hande, M.P.; Matsumoto, T.; Takehara, S.; Tachibana, M.; Oshimura, M.; Lansdorp, P.M.; Furuichi, Y. Telomere maintenance in telomerase-deficient mouse embryonic stem cells: Characterization of an amplified telomeric DNA. Mol. Cell. Biol. 2000, 20, 4115–4127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Erdmann, N.; Giannone, R.J.; Wu, J.; Gomez, M.; Liu, Y. An increase in telomere sister chromatid exchange in murine embryonic stem cells possessing critically shortened telomeres. Proc. Natl. Acad. Sci. USA 2005, 102, 10256–10260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portney, B.A.; Arad, M.; Gupta, A.; Brown, R.A.; Khatri, R.; Lin, P.N.; Hebert, A.M.; Angster, K.H.; Silipino, L.E.; Meltzer, W.A.; et al. ZSCAN4 facilitates chromatin remodeling and promotes the cancer stem cell phenotype. Oncogene 2020, 39, 4970–4982. [Google Scholar] [CrossRef] [PubMed]
- Schuster-Bockler, B.; Lehner, B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 2012, 488, 504–507. [Google Scholar] [CrossRef]
- Silva, B.; Arora, R.; Bione, S.; Azzalin, C.M. TERRA transcription destabilizes telomere integrity to initiate break-induced replication in human ALT cells. Nat. Commun. 2021, 12, 3760. [Google Scholar] [CrossRef]
- Dilley, R.L.; Verma, P.; Cho, N.W.; Winters, H.D.; Wondisford, A.R.; Greenberg, R.A. Break-induced telomere synthesis underlies alternative telomere maintenance. Nature 2016, 539, 54–58. [Google Scholar] [CrossRef] [Green Version]
- Roumelioti, F.M.; Sotiriou, S.K.; Katsini, V.; Chiourea, M.; Halazonetis, T.D.; Gagos, S. Alternative lengthening of human telomeres is a conservative DNA replication process with features of break-induced replication. EMBO Rep. 2016, 17, 1731–1737. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes. Mol. Cell. Biol. 2017, 37, e00226-17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.M.; Yadav, T.; Ouyang, J.; Lan, L.; Zou, L. Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways. Cell Rep. 2019, 26, 955–968.e3. [Google Scholar] [CrossRef] [Green Version]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008, 10, 228–236. [Google Scholar] [CrossRef]
- Arnoult, N.; Van Beneden, A.; Decottignies, A. Telomere length regulates TERRA levels through increased trimethylation of telomeric H3K9 and HP1alpha. Nat. Struct. Mol. Biol. 2012, 19, 948–956. [Google Scholar] [CrossRef]
- Hu, J.; Hwang, S.S.; Liesa, M.; Gan, B.; Sahin, E.; Jaskelioff, M.; Ding, Z.; Ying, H.; Boutin, A.T.; Zhang, H.; et al. Antitelomerase therapy provokes ALT and mitochondrial adaptive mechanisms in cancer. Cell 2012, 148, 651–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechter, O.E.; Zou, Y.; Walker, W.; Wright, W.E.; Shay, J.W. Telomeric recombination in mismatch repair deficient human colon cancer cells after telomerase inhibition. Cancer Res. 2004, 64, 3444–3451. [Google Scholar] [CrossRef] [Green Version]
- Nittis, T.; Guittat, L.; Stewart, S.A. Alternative lengthening of telomeres (ALT) and chromatin: Is there a connection? Biochimie 2008, 90, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Panier, S.; Maric, M.; Hewitt, G.; Mason-Osann, E.; Gali, H.; Dai, A.; Labadorf, A.; Guervilly, J.H.; Ruis, P.; Segura-Bayona, S.; et al. SLX4IP Antagonizes Promiscuous BLM Activity during ALT Maintenance. Mol. Cell 2019, 76, 27–43.e11. [Google Scholar] [CrossRef] [Green Version]
- Barroso-Gonzalez, J.; Garcia-Exposito, L.; Hoang, S.M.; Lynskey, M.L.; Roncaioli, J.L.; Ghosh, A.; Wallace, C.T.; de Vitis, M.; Modesti, M.; Bernstein, K.A.; et al. RAD51AP1 Is an Essential Mediator of Alternative Lengthening of Telomeres. Mol. Cell 2019, 76, 11–26.e17. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dan, J.; Zhou, Z.; Wang, F.; Wang, H.; Guo, R.; Keefe, D.L.; Liu, L. Zscan4 Contributes to Telomere Maintenance in Telomerase-Deficient Late Generation Mouse ESCs and Human ALT Cancer Cells. Cells 2022, 11, 456. https://doi.org/10.3390/cells11030456
Dan J, Zhou Z, Wang F, Wang H, Guo R, Keefe DL, Liu L. Zscan4 Contributes to Telomere Maintenance in Telomerase-Deficient Late Generation Mouse ESCs and Human ALT Cancer Cells. Cells. 2022; 11(3):456. https://doi.org/10.3390/cells11030456
Chicago/Turabian StyleDan, Jiameng, Zhongcheng Zhou, Fang Wang, Hua Wang, Renpeng Guo, David L. Keefe, and Lin Liu. 2022. "Zscan4 Contributes to Telomere Maintenance in Telomerase-Deficient Late Generation Mouse ESCs and Human ALT Cancer Cells" Cells 11, no. 3: 456. https://doi.org/10.3390/cells11030456




