Are Cell Junctions Implicated in the Regulation of Vitellogenin Uptake? Insights from an RNAseq-Based Study in Eel, Anguilla australis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Histological Analysis
2.3. Total RNA Extraction, cDNA Library Preparation, and High-Throughput Sequencing
2.4. Bioinformatics: De Novo Assembly, Annotation, and Differential Expression Analysis
2.5. Target Genes
2.6. Statistics
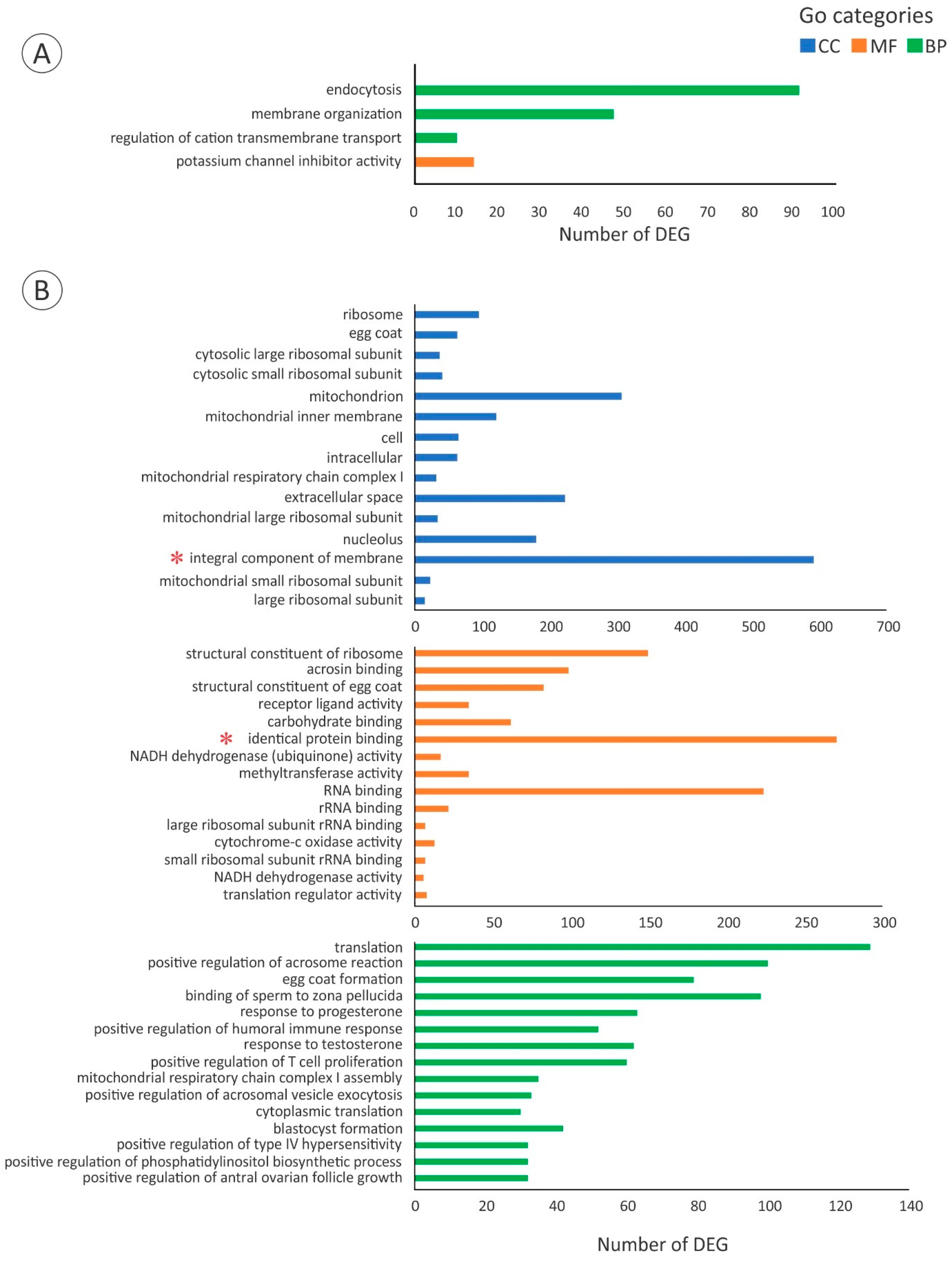
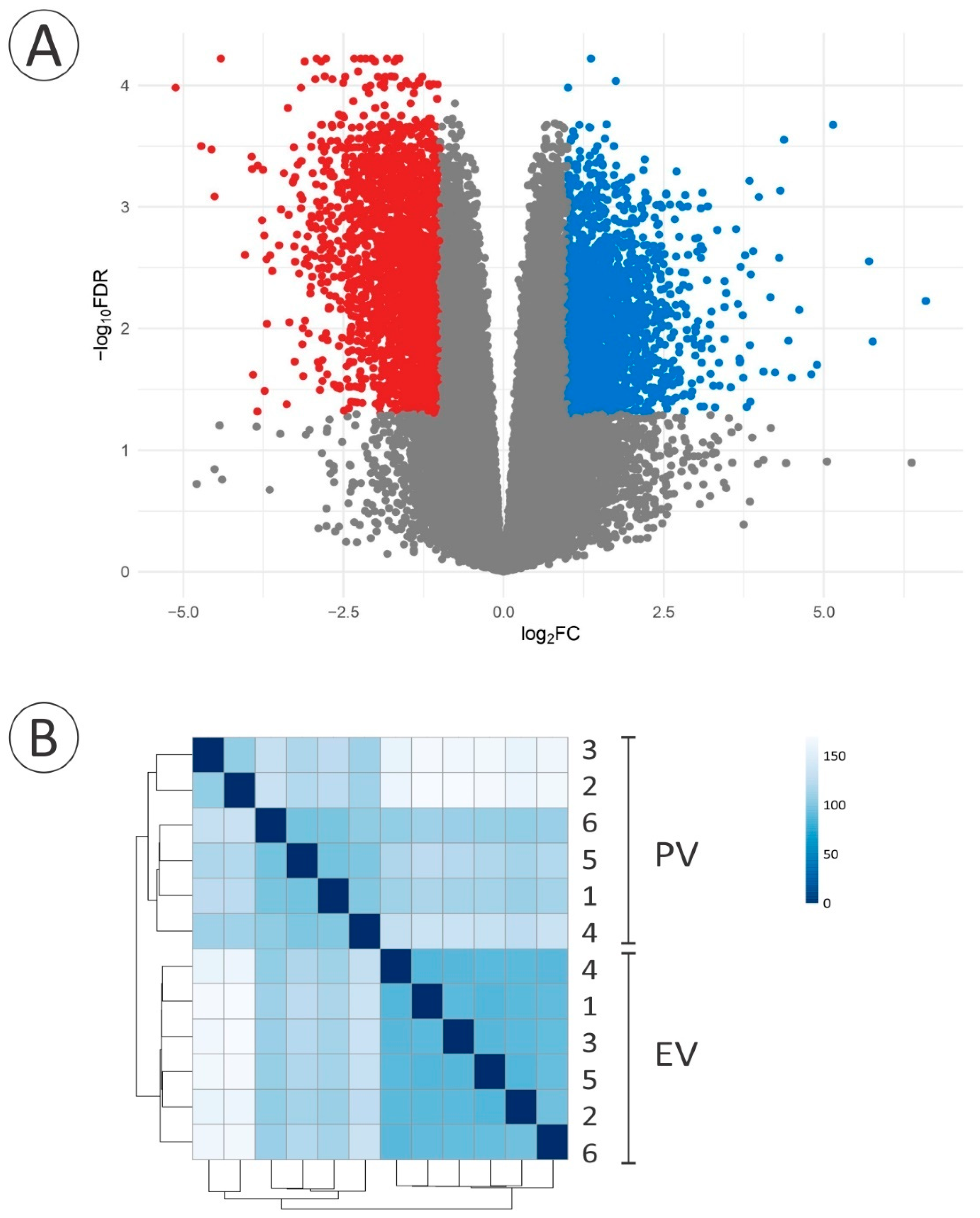
3. Results
3.1. Characterisation of Ovarian Developmental Stage
3.2. De Novo Transcriptome and Downstream Analysis
3.3. Gene Expression of Cell Junctions during the PV-EV Transition
3.4. Expression of Genes Related to Receptor-Mediated Endocytosis Machinery and Vtg Processing during the PV-EV Transition
4. Discussion
4.1. Mechanical Barrier Hypothesis and Expression of TJ-Constituting Proteins during the PV-EV Transition
4.2. Chemical Barrier Hypothesis and Expression of GJ Proteins during the PV-EV Transition
4.3. Expression of Genes Related to Receptor-Mediated Endocytosis Machinery and Vtg Processing during the PV-EV Transition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lancaster, P.M.; Tyler, C.R. Developmental expression and modulation of the vitellogenin receptor in ovarian follicles of the rainbow trout, Oncorhynchus mykiss. J. Exp. Zool. 1994, 269, 458–466. [Google Scholar] [CrossRef]
- Mizuta, H.; Luo, W.; Ito, Y.; Mushirobira, Y.; Todo, T.; Hara, A.; Reading, B.J.; Sullivan, C.V.; Hiramatsu, N. Ovarian expression and localization of a vitellogenin receptor with eight ligand binding repeats in the cutthroat trout (Oncorhynchus clarki). Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2013, 166, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Reading, B.J.; Hiramatsu, N.; Schilling, J.; Molloy, K.T.; Glassbrook, N.; Mizuta, H.; Luo, W.; Baltzegar, D.A.; Williams, V.N.; Todo, T.; et al. Lrp13 is a novel vertebrate lipoprotein receptor that binds vitellogenins in teleost fishes [S]. J. Lipid Res. 2014, 55, 2287–2295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, Ø.; Xu, C.; Timmerhaus, G.; Kirste, K.H.; Naeve, I.; Mommens, M.; Tveiten, H. Resolving the complexity of vitellogenins and their receptors in the tetraploid Atlantic salmon (Salmo salar): Ancient origin of the phosvitinless VtgC in chondrichthyean fishes. Mol. Reprod. Dev. 2017, 84, 1191–1202. [Google Scholar] [CrossRef] [PubMed]
- Morini, M.; Lafont, A.G.; Maugars, G.; Baloche, S.; Dufour, S.; Asturiano, J.F.; Pérez, L. Identification and stable expression of vitellogenin receptor through vitellogenesis in the European eel. Animal 2020, 14, 1213–1222. [Google Scholar] [CrossRef]
- Hiramatsu, N.; Todo, T.; Sullivan, C.V.; Schilling, J.; Reading, B.J.; Matsubara, T.; Ryu, Y.W.; Mizuta, H.; Luo, W.; Nishimiya, O.; et al. Ovarian yolk formation in fishes: Molecular mechanisms underlying formation of lipid droplets and vitellogenin-derived yolk proteins. Gen. Comp. Endocrinol. 2015, 221, 9–15. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, C.; Yilmaz, O. Vitellogenesis and Yolk Proteins, Fish. In Encyclopedia of Reproduction, 2nd ed.; Skinner, M.K., Ed.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 6, pp. 266–277. [Google Scholar] [CrossRef]
- Wallace, R.A. Vitellogenesis and Oocyte Growth in Nonmammalian Vertebrates. In Oogenesis. Developmental Biology (A Comprehensive Synthesis); Browder, L.W., Ed.; Springer: Boston, MA, USA, 1985; Volume 1, pp. 127–177. [Google Scholar] [CrossRef]
- Ito, Y.; Kihara, M.; Nakamura, E.; Yonezawa, S.; Yoshizaki, N. Vitellogenin transport and yolk formation in the quail ovary. Zool. Sci. 2003, 20, 717–726. [Google Scholar] [CrossRef]
- Selman, K.; Wallace, R.A. Oogenesis in Fundulus heteroclitus. III. vitellogenesis. J. Exp. Zool. 1983, 226, 441–457. [Google Scholar] [CrossRef]
- Dumont, J.N. Oogenesis in Xenopus laevis (Daudin) VI. The route of injected tracer transport in the follicle and developing oocyte. J. Exp. Zool. 1978, 204, 193–217. [Google Scholar] [CrossRef]
- Yoshizaki, N. In vivo study of vitellogenin-gold transport in the ovarian follicle and oocyte of Xenopus laevis. Dev. Growth Differ. 1992, 34, 517–527. [Google Scholar] [CrossRef]
- Günzel, D.; Yu, A.S.L. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolosov, D.; Bui, P.; Chasiotis, H.; Kelly, S.P. Claudins in teleost fishes. Tissue Barriers 2013, 1, e25391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chasiotis, H.; Kolosov, D.; Bui, P.; Kelly, S.P. Tight junctions, tight junction proteins and paracellular permeability across the gill epithelium of fishes: A review. Respir. Physiol. Neurobiol. 2012, 184, 269–281. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.Y.; Kwon, H.B.; Ahn, J.C.; Kang, D.; Kwon, S.H.; Park, J.A.; Kim, K.W. Functional and developmental analysis of the blood–brain barrier in zebrafish. Brain Res. Bull. 2008, 75, 619–628. [Google Scholar] [CrossRef]
- Meneguelli De Souza, L.C.; Retamal, C.A.; Rocha, G.M.; Lopez, M.L. Morphological evidence for a permeability barrier in the testis and spermatic duct of Gymnotus carapo (Teleostei: Gymnotidae). Mol. Reprod. Dev. 2015, 82, 663–678. [Google Scholar] [CrossRef]
- Clelland, E.S.; Kelly, S.P. Tight junction proteins in zebrafish ovarian follicles: Stage specific mRNA abundance and response to 17β-estradiol, human chorionic gonadotropin, and maturation inducing hormone. Gen. Comp. Endocrinol. 2010, 168, 388–400. [Google Scholar] [CrossRef]
- Le Menn, F.L.; Cerdà, J.; Babin, P.J. Ultrastructural Aspects of the Ontogeny and Differentiation of Ray-Finned Fish Ovarian Follicles. In The Fish Oocyte: From Basic Studies to Biotechnological Applications; Babin, P.J., Cerdà, J., Lubzens, E., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 1–37. [Google Scholar] [CrossRef]
- Monaco, M.E.; Villecco, E.I.; Sanchez, S. Implication of gap junction coupling in amphibian vitellogenin uptake. Zygote 2007, 15, 149–157. [Google Scholar] [CrossRef]
- Luque, M.; Serrano, M.; Monaco, M.E.; Villecco, E.; Sanchez, S. Involvement of cAMP and calmodulin in endocytic yolk uptake during Xenopus laevis oogenesis. Zygote 2011, 21, 1–9. [Google Scholar] [CrossRef]
- Richardson, J. On some new or little known fishes from the Australian seas. Proc. Zool. Soc. Lond. 1841, 21–22. [Google Scholar]
- Lokman, P.M.; Rohr, D.H.; Davie, P.S.; Young, G. The physiology of silvering in anguillid eels: Androgens and control of metamorphosis from the yellow to silver stage. In Eel Biology; Aida, K., Tsukamoto, K., Yamauchi, K., Eds.; Springer: Tokyo, Japan, 2003; pp. 331–349. [Google Scholar] [CrossRef]
- Tanaka, H.; Kagawa, H.; Ohta, H.; Unuma, T.; Nomura, K. The first production of glass eel in captivity: Fish reproductive physiology facilitates great progress in aquaculture. Fish Physiol. Biochem. 2003, 28, 493–497. [Google Scholar] [CrossRef]
- Di Biase, A.; Lokman, P.M.; Govoni, N.; Casalini, A.; Emmanuele, P.; Parmeggiani, A.; Mordenti, O. Co-treatment with androgens during artificial induction of maturation in female eel, Anguilla anguilla: Effects on egg production and early development. Aquaculture 2017, 479, 508–515. [Google Scholar] [CrossRef]
- Lokman, P.M.; Vermeulen, G.J.; Lambert, J.G.D.; Young, G. Gonad histology and plasma steroid profiles in wild New Zealand freshwater eels (Anguilla dieffenbachii and A. australis) before and at the onset of the natural spawning migration. I. Females*. Fish Physiol. Biochem. 1998, 19, 325–338. [Google Scholar] [CrossRef]
- Lokman, P.M.; George, K.A.N.; Divers, S.L.; Algie, M.; Young, G. 11-Ketotestosterone and IGF-I increase the size of previtellogenic oocytes from shortfinned eel, Anguilla australis, in vitro. Reproduction 2007, 133, 955–967. [Google Scholar] [CrossRef] [PubMed]
- FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 26 August 2021).
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinform. 2010, 32, 11–17. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 genome project data processing subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Bryant, D.M.; Johnson, K.; Di Tommaso, T.; Tickle, T.; Couger, M.B.; Payzin-Dogru, D.; Lee, T.J.; Leigh, N.D.; Kuo, T.H.; Davis, F.G. A tissue-mapped axolotl de novo transcriptome enables identification of limb regeneration factors. Cell Rep. 2017, 18, 762–776. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Madden, T.; Coulouris, G.; Ma, N.; Tao, T.; Agarwala, R. BLAST® Command Line Applications User Manual. National Center for Biotechnology Information: Bethesda, MD, USA, 2008. [Google Scholar]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package Version 1.0.12. 2019. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 5 January 2019).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing, Version 3.5. 1. R Foundation for Statistical Computing, Vienna, Austria. 2018. Available online: https://www.R-project.org/ (accessed on 25 April 2018).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smith, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smith, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [Green Version]
- Phipson, B.; Lee, S.; Majewski, I.J.; Alexander, W.S.; Smyth, G.K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 2016, 10, 946–963. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Coordinators, N.R.; Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018, 46, D8–D13. [Google Scholar] [CrossRef] [Green Version]
- Marchler-Bauer, A.; Bryant, S.H. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Shapiro, S.S.; Wilk, M.B. An analysis of variance test for normality (complete samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Levene, H. Robust Tests for Equality of Variances. In Contributions to Probability and Statistics; Olkins, I., Ed.; Stanford University Press: Palo Alto, CA, USA, 1960; pp. 278–292. [Google Scholar]
- Di Rienzo, J.A.; Casanoves, F.; Balzarini, M.G.; Gonzalez, L.; Tablada, M.; Robledo, C.W. InfoStat Version 2018. InfoStat Group, Córdoba, Argentina. Available online: http://www.infostat.com.ar (accessed on 1 May 2019).
- Babio, L.; Damsteegt, E.L.; Lokman, P.M. Lipoprotein Receptors in Anguilla Australis Ovary. Molecular Characterisation of Putative Vitellogenin Receptors; Department of Zoology, University of Otago: Dunedin, New Zealand, 2022; (manuscript in preparation). [Google Scholar]
- Wegnez, M.; Denis, H.; Mazabraud, A.; Clérot, J.C. Biochemical research on oogenesis. RNA accumulation during oogenesis of the dogfish Scyliorhinus caniculus. Dev. Biol. 1978, 62, 99–111. [Google Scholar] [CrossRef]
- Kroupova, H.; Trubiroha, A.; Wuertz, S.; Kloas, W. Stage-dependent differences in RNA composition and content affect the outcome of expression profiling in roach (Rutilus rutilus) ovary. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2011, 159, 141–149. [Google Scholar] [CrossRef]
- Rojo-Bartolomé, I.; Diaz de Cerio, O.; Diez, G.; Cancio, I. Identification of sex and female’s reproductive stage in commercial fish species through the quantification of ribosomal transcripts in gonads. PLoS ONE 2016, 11, e0149711. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.G.; Yao, H.; Guo, L.; Li, X.X.; Wang, H.P. Ribosome RNA profiling to quantify ovarian development and identify sex in fish. Sci. Rep. 2017, 7, 4196. [Google Scholar] [CrossRef] [Green Version]
- Zihni, C.; Mills, C.; Matter, K. Tight junctions: From simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol. 2016, 17, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Schuetz, A. Structural features of tight-junction proteins. Int. J. Mol. Sci. 2019, 20, 6020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleppe, L.; Edvardsen, R.B.; Furmanek, T.; Taranger, G.L.; Wargelius, A. Global transcriptome analysis identifies regulated transcripts and pathways activated during oogenesis and early embryogenesis in atlantic cod. Mol. Reprod. Dev. 2014, 81, 619–635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster, M.K.; Schmierer, B.; Shkumatava, A.; Kuchler, K. Activin A and follicle-stimulating hormone control tight junctions in avian granulosa cells by regulating occludin expression. Biol. Reprod. 2004, 70, 1493–1499. [Google Scholar] [CrossRef] [Green Version]
- Ikenouchi, J.; Furuse, M.; Furuse, K.; Sasaki, H.; Tsukita, S.; Tsukita, S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J. Cell Biol. 2005, 171, 939–945. [Google Scholar] [CrossRef]
- Staehelin, L.A. Further observations on the fine structure of freeze-cleaved tight junctions. J. Cell Sci. 1973, 13, 763–786. [Google Scholar] [CrossRef]
- Higashi, T.; Tokuda, S.; Kitajiri, S.I.; Masuda, S.; Nakamura, H.; Oda, Y.; Furuse, M. Analysis of the ‘angulin’ proteins LSR, ILDR1 and ILDR2–tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J. Cell Sci. 2013, 126, 966–977. [Google Scholar] [CrossRef] [Green Version]
- Masuda, S.; Oda, Y.; Sasaki, H.; Ikenouchi, J.; Higashi, T.; Akashi, M.; Nishi, E.; Furuse, M. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J. Cell Sci. 2011, 124, 548–555. [Google Scholar] [CrossRef] [Green Version]
- Isasti-Sanchez, J.; Münz-Zeise, F.; Lancino, M.; Luschnig, S. Transient opening of tricellular vertices controls paracellular transport through the follicle epithelium during Drosophila oogenesis. Dev. Cell 2021, 56, 1083–1099. [Google Scholar] [CrossRef]
- Krug, S.M.; Amasheh, S.; Richter, J.F.; Milatz, S.; Günzel, D.; Westphal, J.K.; Huber, O.; Schulzke, J.D.; Fromm, M. Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol. Biol. Cell 2009, 20, 3713–3724. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Luckenbach, J.A.; Middleton, M.A.; Swanson, P. The spatiotemporal expression of multiple coho salmon ovarian connexin genes and their hormonal regulation in vitro during oogenesis. Reprod. Biol. Endocrinol. 2011, 9, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russel, P.J.; Hertz, P.E.; McMillan, B. Biology: The Dynamic Science, 3rd ed.; Brooks/Cole: Belmont, CA, USA, 2014; p. 142. [Google Scholar]
- Patiño, R.; Sullivan, C.V. Ovarian follicle growth, maturation, and ovulation in teleost fish. Fish Physiol. Biochem. 2002, 26, 57–70. [Google Scholar] [CrossRef]
- Serrano, M.; Luque, M.; Sanchez, S. Xepac protein and IP3/Ca2+ pathway implication during Xenopus laevis vitellogenesis. Zygote 2013, 23, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Opresko, L.K.; Wiley, H.S. Receptor-mediated endocytosis in Xenopus oocytes. I. Characterization of the vitellogenin receptor system. J. Biol. Chem. 1987, 262, 4109–4115. [Google Scholar] [CrossRef]
- Wallace, R.A.; Ho, T.; Salter, D.W.; Jared, D.W. Protein incorporation by isolated amphibian oocytes: IV. The role of follicle cells and calcium during protein uptake. Exp. Cell Res. 1973, 82, 287–295. [Google Scholar] [CrossRef]
- Fujita, H.; Sugimoto, K.; Inatomi, S.; Maeda, T.; Osanai, M.; Uchiyama, Y.; Yamamoto, Y.; Wada, T.; Kojima, T.; Yokozaki, H.; et al. Tight junction proteins claudin-2 and -12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Mol. Biol. Cell 2008, 19, 1912–1921. [Google Scholar] [CrossRef] [Green Version]
- Plain, A.; Pan, W.; O’Neill, D.; Ure, M.; Beggs, M.R.; Farhan, M.; Dimke, H.; Cordat, E.; Alexander, R.T. Claudin-12 knockout mice demonstrate reduced proximal tubule calcium permeability. Int. J. Mol. Sci. 2020, 21, 2074. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Na, T.; Peng, J.B. WNK3 positively regulates epithelial calcium channels TRPV5 and TRPV6 via a kinase-dependent pathway. Am. J. Physiol. Renal Physiol. 2008, 295, 1472–1484. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.C.; Davis, T.P. Calcium modulation of adherens and tight junction function: A potential mechanism for blood-brain barrier disruption after stroke. Stroke 2002, 33, 1706–1711. [Google Scholar] [CrossRef]
- Davail, B.; Pakdel, F.; Bujo, H.; Perazzolo, L.M.; Waclawek, M.; Schneider, W.J.; Le Menn, F. Evolution of oogenesis: The receptor for vitellogenin from the rainbow trout. J. Lipid Res. 1998, 39, 1929–1937. [Google Scholar] [CrossRef]
- Mushirobira, Y.; Mizuta, H.; Luo, W.; Todo, T.; Hara, A.; Reading, B.J.; Sullivan, C.V.; Hiramatsu, N. Molecular cloning and partial characterization of a low-density lipoprotein receptor-related protein 13 (Lrp13) involved in vitellogenin uptake in the cutthroat trout (Oncorhynchus clarki). Mol. Reprod. Dev. 2015, 82, 986–1000. [Google Scholar] [CrossRef]
- Damsteegt, E.L.; Falahatimarvast, A.; McCormick, S.P.; Lokman, P.M. Triacylglyceride physiology in the short-finned eel, Anguilla australis—Changes throughout early oogenesis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R935–R944. [Google Scholar] [CrossRef] [PubMed]
- Jéhannet, P.; Kruijt, L.; Damsteegt, E.L.; Swinkels, W.; Heinsbroek, L.T.N.; Lokman, P.M.; Palstra, A.P. A mechanistic model for studying the initiation of anguillid vitellogenesis by comparing the European eel (Anguilla anguilla) and the shortfinned eel (A. australis). Gen. Comp. Endocrinol. 2019, 279, 129–138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Opresko, L.K.; Wiley, H.S. Receptor-mediated endocytosis in Xenopus oocytes. II. Evidence for two novel mechanisms of hormonal regulation. J. Biol. Chem. 1987, 262, 4116–4123. [Google Scholar] [CrossRef]
- Michaely, P.; Li, W.P.; Anderson, R.G.; Cohen, J.C.; Hobbs, H.H. The modular adaptor protein ARH is required for low density lipoprotein (LDL) binding and internalization but not for LDL receptor clustering in coated pits. J. Biol. Chem. 2004, 279, 34023–34031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Zhang, J.; King, M.L. Xenopus autosomal recessive hypercholesterolemia protein couples lipoprotein receptors with the AP-2 complex in oocytes and embryos and is required for vitellogenesis. J. Biol. Chem. 2003, 278, 44584–44592. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Zhang, J.; King, M.L. Polarized distribution of mRNAs encoding a putative LDL receptor adaptor protein, xARH (autosomal recessive hypercholesterolemia) in Xenopus oocytes. Mech. Dev. 2004, 121, 1249–1258. [Google Scholar] [CrossRef]
- Hirayama, S.; Bajari, T.M.; Nimpf, J.; Schneider, W.J. Receptor-mediated chicken oocyte growth: Differential expression of endophilin isoforms in developing follicles. Biol. Reprod. 2003, 68, 1850–1860. [Google Scholar] [CrossRef]
- Gioacchini, G.; Marisaldi, L.; Basili, D.; Candelma, M.; Pignalosa, P.; Cigliano, R.A.; Sanseverino, W.; Hardiman, G.; Carnevali, O. A de novo transcriptome assembly approach elucidates the dynamics of ovarian maturation in the swordfish (Xiphias gladius). Sci. Rep. 2019, 9, 7375. [Google Scholar] [CrossRef] [Green Version]
- Borrelli, L.; De Stasio, R.; Filosa, S.; Parisi, E.; Riggio, M.; Scudiero, R.; Trinchella, F. Evolutionary fate of duplicate genes encoding aspartic proteinases. Nothepsin case study. Gene 2006, 368, 101–109. [Google Scholar] [CrossRef]
- Brooks, S.E.; Tyler, C.R.; Sumpter, J.P. Egg quality in fish: What makes a good egg? Rev. Fish Biol. Fish. 1997, 7, 387–416. [Google Scholar] [CrossRef]
- Carnevali, O.; Cionna, C.; Tosti, L.; Lubzens, E.; Maradonna, F. Role of cathepsins in ovarian follicle growth and maturation. Gen. Comp. Endocrinol. 2006, 146, 195–203. [Google Scholar] [CrossRef]
- Luckenbach, J.A.; Iliev, D.B.; Goetz, F.W.; Swanson, P. Identification of differentially expressed ovarian genes during primary and early secondary oocyte growth in coho salmon, Oncorhynchus kisutch. Reprod. Biol. Endocrinol. 2008, 6, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeong, K.J.; Prat, F.; Randall, C.; Tyler, C.R. Molecular characterization of putative yolk processing enzymes and their expression during oogenesis and embryogenesis in rainbow trout (Oncorhynchus mykiss). Biol. Reprod. 2001, 65, 1701–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Down-Regulated Genes during the PV-EV Transition | ||||||
|---|---|---|---|---|---|---|
| Gene ID TRINITY_ | Annotation | A. anguilla BLASTp Hit/Gene Associated | Log2FC | q Value | PV Reads ± SEM | EV Reads ± SEM |
| DN13517_c0_g1 | claudin-14 [H. sapiens] | claudin-like protein ZF-A9 [XP_035248997.1]/cldng | 2.24 | 0.02 | 45.5 ± 12.1 | 14.7 ± 3.8 |
| DN36018_c0_g1 | claudin-3 [R. norvegicus] | claudin-3-like [XP_035290832.1]/LOC118236501 | 1.70 | 0.02 | 20.8 ± 4.7 | 9.8 ± 2.1 |
| DN7995_c0_g1 | MARVELD3 [H. sapiens] | MarvelD3 [XP_035251135.1]/marveld3 | 1.22 | <0.01 | 391.3 ± 20.6 | 257.5 ± 36.6 |
| DN55532_c0_g1 | claudin-like protein ZF-A89 [D. rerio] | claudin-like protein ZF-A89 [XP_035288223.1]/LOC118235193 | 1.13 | <0.01 | 542.7 ± 65.3 | 356.2 ± 39.2 |
| DN1688_c0_g3 | Jam2a [D. rerio] | Jam2a [XP_035249352.1]/jam2a | 1.12 | <0.01 | 252.2 ± 13.5 | 174.2 ± 8.2 |
| Up-regulated genes during the PV-EV transition | ||||||
| DN10265_c0_g1 | claudin-12 [P. abelii] | claudin-12 [XP_035258512.1]/cldn12 | −1.03 | <0.01 | 74.5 ± 14.4 | 220.3 ± 10.4 |
| DN4344_c0_g2 | MARVELD2 [X. tropicalis] | MarvelD2-like [XP_035245633.1]/LOC118212123 | −1.24 | <0.01 | 92.2 ± 12.4 | 323.5 ± 29.5 |
| Not differentially expressed between PV and EV stages | ||||||
| DN19581_c0_g1 | claudin-7a [D. rerio] | claudin-7b-like [XP_035288086.1]/LOC118235138 | 0.87 | 0.04 | 309.3 ± 20.6 | 266.8 ± 42.6 |
| DN2234_c0_g1 | claudin-7a [D. rerio] | claudin-7-a [XP_035265529.1]/cldn7a | 0.13 | 0.81 | 2309.4 ± 395.7 | 2580 ± 490 |
| DN11172_c0_g1 | claudin-like protein ZF-A89 [D. rerio] | claudin-like protein ZF-A89 [XP_035287488.1]/ LOC118234813 | 0.37 | <0.01 | 52,673.3 ± 3151.4 | 30,285.6 ± 1014.1 |
| DN23602_c0_g2 | claudin-8 [M. musculus] | claudin-8-like [XP_035242392.1]/LOC118210375 | −0.43 | 0.36 | 10.2 ± 1.9 | 22.2 ± 3.2 |
| DN19953_c0_g1 | claudin-11 [B. taurus] | claudin-11a [XP_035278421.1]/cldn11a | −0.70 | 0.08 | 14.8 ± 2.4 | 35.8 ± 3.6 |
| DN11142_c0_g1 | Jam3b [D. rerio] | Jam3b [XP_035242387.1]/jam3a | −1.13 | 0.14 | 16.8 ± 6.3 | 55.7 ± 15.9 |
| DN21932_c0_g1 | Jam2a [D. rerio] | Jamb-like [XP_035266823.1]/LOC118223892 | 0.59 | 0.34 | 10.7 ± 1.9 | 11.7 ± 2.3 |
| DN6434_c0_g1 | occludin [R. norvegicus] | occludin-like [XP_035245592.1]/LOC118212103 | 0.24 | 0.23 | 877.5 ± 99.6 | 958.8 ± 43.3 |
| DN5516_c0_g1 | LSR (Angulin-1) [M. musculus] | Lsr [XP_035263346.1]/lsr | 0.28 | 0.31 | 623.3 ± 72.6 | 698.7 ± 74.5 |
| DN4811_c0_g1 | ILDR1 (Angulin-2) [X. laevis] | Ildr1a [XP_035250686.1]/ildr1a | −0.29 | 0.07 | 3746.9 ± 339.2 | 5119.4 ± 317.7 |
| DN9449_c0_g1 | ILDR2 (Angulin-3) [M. musculus] | Ildr2 [XP_035250586.1]/LOC118214609 | −0.51 | 0.15 | 11.2 ± 0.9 | 24.6 ± 1.5 |
| DN198_c0_g1 | tight junction ZO-1 [C. familiaris] | tight junction ZO-1-like [XP_035273256.1]/LOC118227177 | −0.48 | 0.02 | 1066.7 ± 132.1 | 1836.1 ± 99.2 |
| DN5322_c0_g1 | tight junction ZO-1 [C. familiaris] | tight junction ZO1-like [XP_035251267.1]/LOC118215021 | 0.04 | 0.89 | 84.3 ± 5.8 | 126.5 ± 10.9 |
| DN7765_c0_g1 | tight junction ZO-2 [M. musculus] | tight junction ZO-2-like [XP_035292052.1]/LOC118237445 | 0.87 | 0.01 | 460.5 ± 51.0 | 363.5 ± 48.4 |
| DN5748_c0_g1 | tight junction ZO-2 [H. sapiens] | tight junction ZO-2a [XP_035247383.1]/tjp2a | <−0.01 | 0.99 | 1619.3 ± 200.9 | 1968.6 ± 124.7 |
| DN2057_c0_g1 | tight junction ZO-3 [H. sapiens] | tight junction ZO-3 [XP_035271907.1]/tjp3 | −0.40 | 0.03 | 2773 ± 270.3 | 4170.8 ± 221.7 |
| DN2926_c0_g1 | MARVELD3 [H. sapiens] | MarvelD3 [XP_035270459.1]/si:ch211-191a24.4 | 0.33 | 0.42 | 525.5 ± 82.8 | 584.505 ± 94.0 |
| DN4344_c0_g1 | MARVELD2 [X. tropicalis] | MarvelD2b [XP_035235988.1]/marveld2b | −0.1 | 0.50 | 1209.8 ± 100.3 | 1624.2 ± 93.8 |
| DN7029_c0_g4 | gj gamma 1 protein [D. rerio] | gj gamma 1 protein-like [XP_035250527.1]/LOC118214562 | −0.31 | 0.03 | 787 ± 81 | 1270.8 ± 47.1 |
| DN998_c2_g1 | gj beta-3 protein [R. norvegicus] | gj beta-3 protein-like [XP_035290867.1]/LC118236520 | −0.15 | 0.23 | 5111.3 ± 429.3 | 6064.8 ± 236.4 |
| DN379_c0_g1 | gj beta-4 protein [R. norvegicus] | gj beta-4 protein-like [XP_035242057.1]/LOC118210192 | 0.70 | <0.01 | 180.6 ± 18.3 | 167.3 ± 13.1 |
| DN15444_c0_g1 | gj beta-7 protein [H. sapiens] | connexin 28.8 [XP_035277334.1]/cx28.8 | 1.27 | 0.09 | 36.8 ± 8.1 | 24.8 ± 6.3 |
| DN12469_c0_g1 | gj 32.7 protein [M. undulatus] | connexin 34.5 [XP_035280040.1]/cx34.5 | −0.88 | 0.02 | 41.7 ± 6.1 | 114.8 ± 15.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babio, L.; Lokman, P.M.; Damsteegt, E.L.; Dutoit, L. Are Cell Junctions Implicated in the Regulation of Vitellogenin Uptake? Insights from an RNAseq-Based Study in Eel, Anguilla australis. Cells 2022, 11, 550. https://doi.org/10.3390/cells11030550
Babio L, Lokman PM, Damsteegt EL, Dutoit L. Are Cell Junctions Implicated in the Regulation of Vitellogenin Uptake? Insights from an RNAseq-Based Study in Eel, Anguilla australis. Cells. 2022; 11(3):550. https://doi.org/10.3390/cells11030550
Chicago/Turabian StyleBabio, Lucila, P. Mark Lokman, Erin L. Damsteegt, and Ludovic Dutoit. 2022. "Are Cell Junctions Implicated in the Regulation of Vitellogenin Uptake? Insights from an RNAseq-Based Study in Eel, Anguilla australis" Cells 11, no. 3: 550. https://doi.org/10.3390/cells11030550
APA StyleBabio, L., Lokman, P. M., Damsteegt, E. L., & Dutoit, L. (2022). Are Cell Junctions Implicated in the Regulation of Vitellogenin Uptake? Insights from an RNAseq-Based Study in Eel, Anguilla australis. Cells, 11(3), 550. https://doi.org/10.3390/cells11030550







