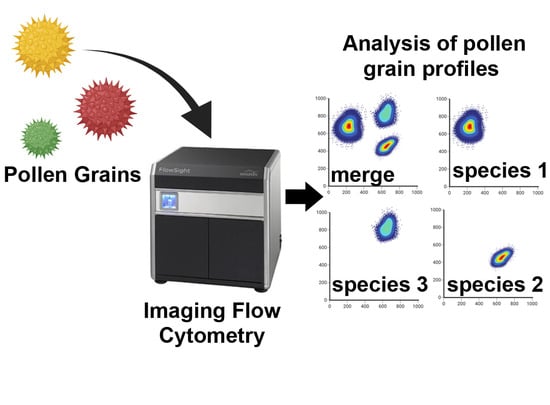
Imaging Flow Cytometry as a Quick and Effective Identification Technique of Pollen Grains from Betulaceae, Oleaceae, Urticaceae and Asteraceae
Abstract
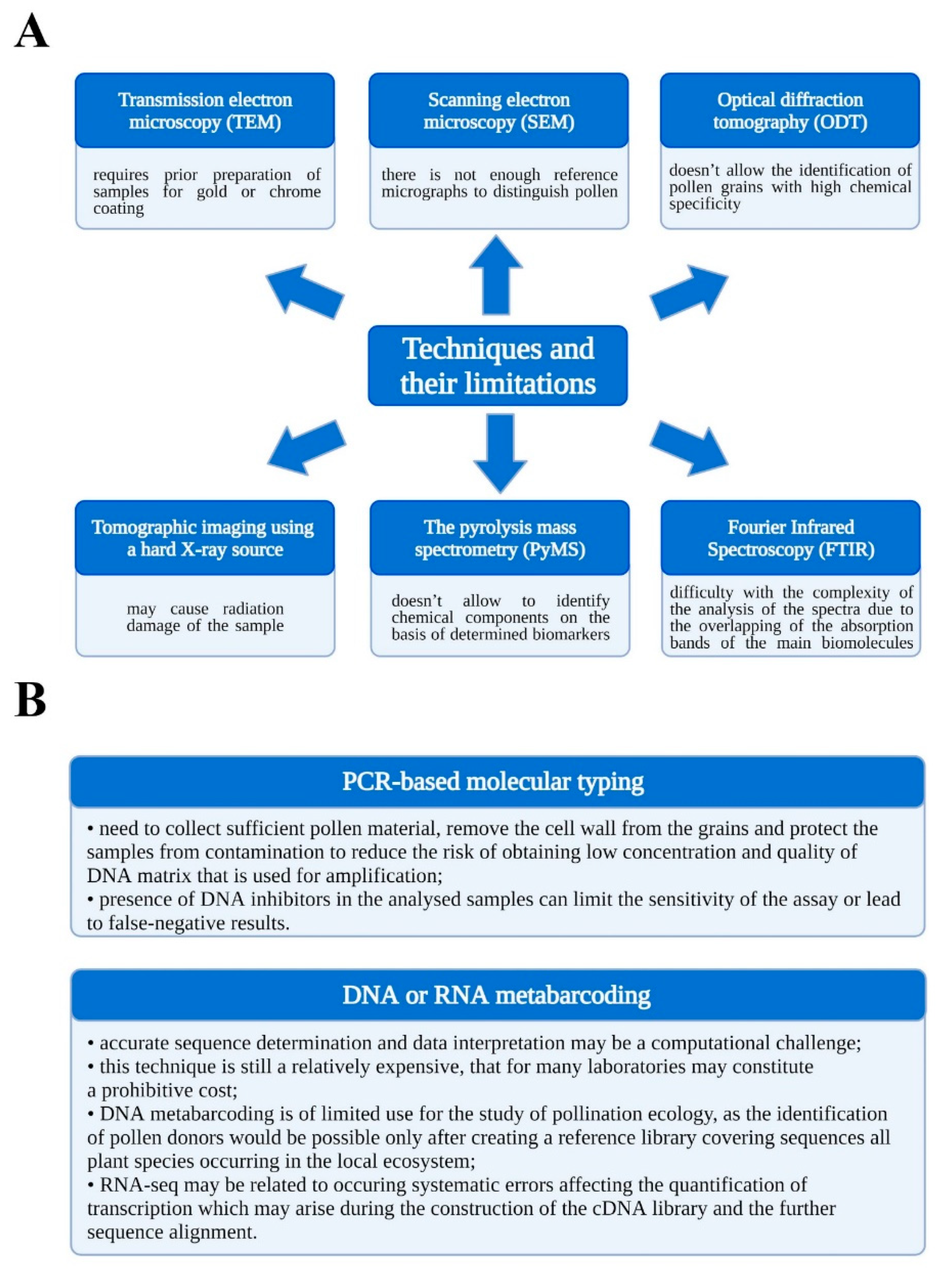
:1. Introduction
2. Materials and Methods
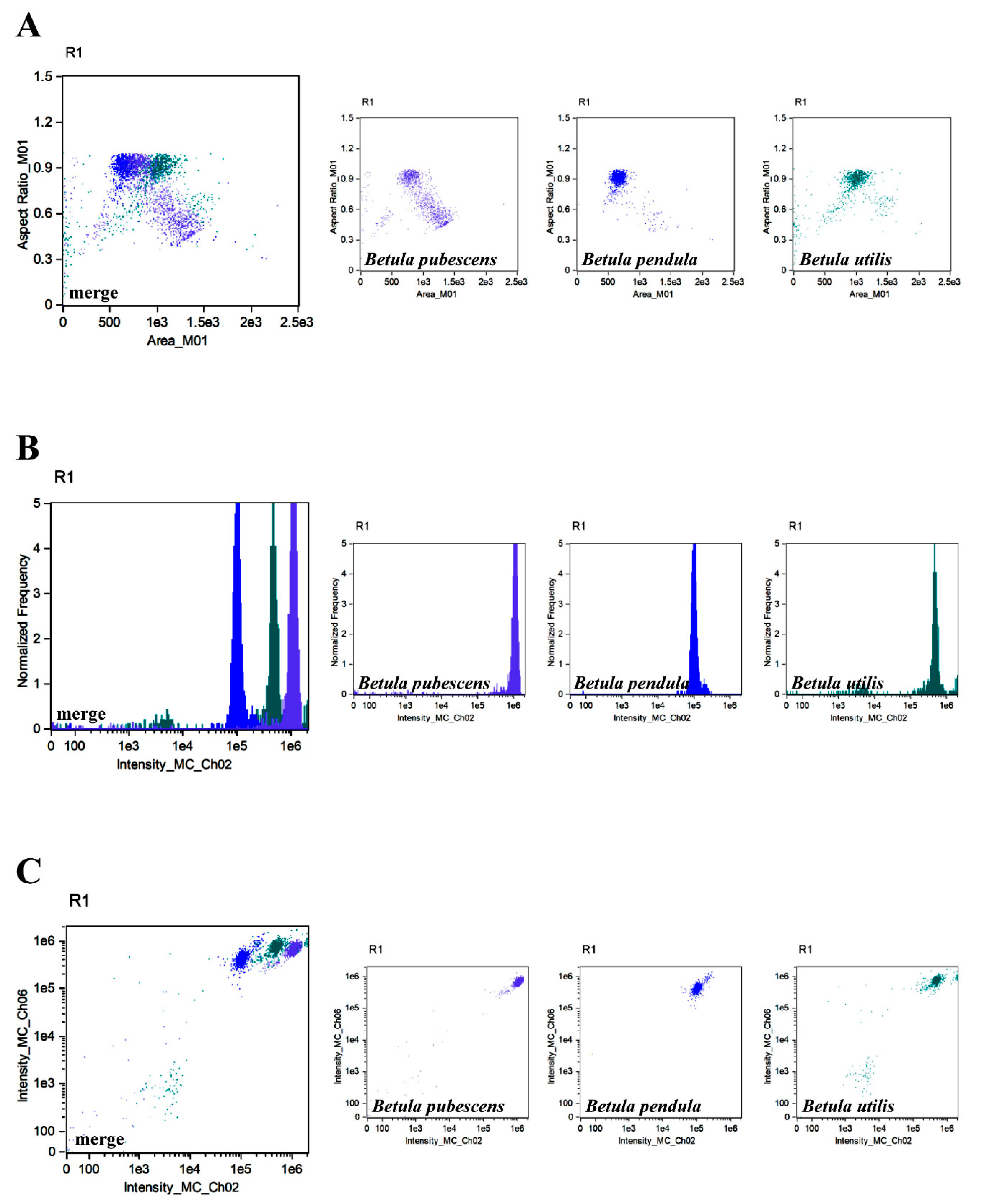
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beggs, P.J.; Šikoparija, B.; Smith, M. Aerobiology in the International Journal of Biometeorology, 1957–2017. Int. J. Biometeorol. 2017, 61, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Lancia, A.; Capone, P.; Vonesch, N.; Pelliccioni, A.; Grandi, C.; Magri, D.; D’Ovidio, M.C. Research Progress on Aerobiology in the Last 30 Years: A Focus on Methodology and Occupational Health. Sustainability 2021, 13, 4337. [Google Scholar] [CrossRef]
- Erdtman, G. Pollen Morphology and Plant Taxonomy: Angiosperms. An Introduction to the Study of Pollen Grains and Spores; Brill: Leiden, The Netherlands, 1986; ISBN 978-90-04-08122-2. [Google Scholar]
- Blackmore, S.; Steinman, J.A.J.; Hoen, P.P.; Punt, W. Betulaceae and Corylaceae. Rev. Palaeobot. Palynol. 2003, 123, 71–98. [Google Scholar] [CrossRef]
- Frenguelli, G.; Kasprzyk, I. Description of Pollen Grains. In Manual for Aerobiology; Kasprzyk, I., Smith, M., Eds.; Wyd. Univ. Rzeszow: Rzeszów, Poland, 2015; ISBN 978-83-7996-146-7. [Google Scholar]
- Dunker, S.; Motivans, E.; Rakosy, D.; Boho, D.; Mäder, P.; Hornick, T.; Knight, T.M. Pollen Analysis Using Multispectral Imaging Flow Cytometry and Deep Learning. New Phytol. 2021, 229, 593–606. [Google Scholar] [CrossRef]
- Holt, K.A.; Bennett, K.D. Principles and Methods for Automated Palynology. New Phytol. 2014, 203, 735–742. [Google Scholar] [CrossRef] [PubMed]
- Sevillano, V.; Holt, K.; Aznarte, J.L. Precise Automatic Classification of 46 Different Pollen Types with Convolutional Neural Networks. PLoS ONE 2020, 15, e0229751. [Google Scholar] [CrossRef]
- Šaulienė, I.; Šukienė, L.; Daunys, G.; Valiulis, G.; Vaitkevičius, L.; Matavulj, P.; Brdar, S.; Panic, M.; Sikoparija, B.; Clot, B.; et al. Automatic Pollen Recognition with the Rapid-E Particle Counter: The First-Level Procedure, Experience and next Steps. Atmos. Meas. Tech. 2019, 12, 3435–3452. [Google Scholar] [CrossRef] [Green Version]
- Oteros, J.; Pusch, G.; Weichenmeier, I.; Heimann, U.; Möller, R.; Röseler, S.; Traidl-Hoffmann, C.; Schmidt-Weber, C.; Buters, J.T.M. Automatic and Online Pollen Monitoring. IAA 2015, 167, 158–166. [Google Scholar] [CrossRef]
- Schaefer, J.; Milling, M.; Schuller, B.W.; Bauer, B.; Brunner, J.O.; Traidl-Hoffmann, C.; Damialis, A. Towards Automatic Airborne Pollen Monitoring: From Commercial Devices to Operational by Mitigating Class-Imbalance in a Deep Learning Approach. Sci. Total Environ. 2021, 796, 148932. [Google Scholar] [CrossRef]
- Kim, G.; Lee, S.; Shin, S.; Park, Y. Three-Dimensional Label-Free Imaging and Analysis of Pinus Pollen Grains Using Optical Diffraction Tomography. Sci. Rep. 2018, 8, 1782. [Google Scholar] [CrossRef] [Green Version]
- de Win, A.H.N.; Knuiman, B.; Pierson, E.S.; Geurts, H.; Kengen, H.M.P.; Derksen, J. Development and Cellular Organization of Pinus Sylvestris Pollen Tubes. Sex. Plant Reprod. 1996, 9, 93–101. [Google Scholar] [CrossRef]
- Derksen, J.; Li, Y.; Knuiman, B.; Geurts, H. Wall of Pinus sylvestris L. Pollen Tubes. Protoplasma 1999, 208, 26–36. [Google Scholar] [CrossRef]
- Ami, D.; Mereghetti, P.; Doglia, S.M. Multivariate Analysis for Fourier Transform Infrared Spectra of Complex Biological Systems and Processes; IntechOpen: London, UK, 2013; ISBN 978-953-51-0921-1. [Google Scholar]
- Chang, H.; Guo, J.; Fu, X.; Liu, Y.; Wyckhuys, K.A.G.; Hou, Y.; Wu, K. Molecular-Assisted Pollen Grain Analysis Reveals Spatiotemporal Origin of Long-Distance Migrants of a Noctuid Moth. Int. J. Mol. Sci. 2018, 19, 567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR Inhibitors—Occurrence, Properties and Removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Bell, K.L.; Burgess, K.S.; Botsch, J.C.; Dobbs, E.K.; Read, T.D.; Brosi, B.J. Quantitative and Qualitative Assessment of Pollen DNA Metabarcoding Using Constructed Species Mixtures. Mol. Ecol. 2019, 28, 431–455. [Google Scholar] [CrossRef]
- Bell, K.L.; Loeffler, V.M.; Brosi, B.J. An RbcL Reference Library to Aid in the Identification of Plant Species Mixtures by DNA Metabarcoding. Appl. Plant Sci. 2017, 5, 1600110. [Google Scholar] [CrossRef]
- Baksay, S.; Pornon, A.; Burrus, M.; Mariette, J.; Andalo, C.; Escaravage, N. Experimental Quantification of Pollen with DNA Metabarcoding Using ITS1 and TrnL. Sci. Rep. 2020, 10, 4202. [Google Scholar] [CrossRef] [Green Version]
- Pérez-de-Castro, A.M.; Vilanova, S.; Cañizares, J.; Pascual, L.; Blanca, J.M.; Díez, M.J.; Prohens, J.; Picó, B. Application of Genomic Tools in Plant Breeding. Curr. Genom. 2012, 13, 179–195. [Google Scholar] [CrossRef] [Green Version]
- Bell, K.L.; de Vere, N.; Keller, A.; Richardson, R.T.; Gous, A.; Burgess, K.S.; Brosi, B.J. Pollen DNA Barcoding: Current Applications and Future Prospects. Genome 2016, 59, 629–640. [Google Scholar] [CrossRef] [Green Version]
- Lobaton, J.; Andrew, R.; Duitama, J.; Kirkland, L.; Macfadyen, S.; Rader, R. Using RNA-Seq to Characterize Pollen–Stigma Interactions for Pollination Studies. Sci. Rep. 2021, 11, 6635. [Google Scholar] [CrossRef]
- Wnuk, M.; Lewinska, A. Imaging Flow Cytometry-Based Analysis of Bacterial Profiles in Milk Samples. Food Bioprod. Process. 2021, 128, 102–108. [Google Scholar] [CrossRef]
- Whitley, S.K.; Horne, W.T.; Kolls, J.K. Research Techniques Made Simple: Methodology and Clinical Applications of RNA Sequencing. J. Investig. Dermatol. 2016, 136, e77–e82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, Y.; Wang, Y.; Jin, F.; Xing, L.; Fang, Y.; Zhang, Z.; Zou, J.; Wang, L.; Xu, M. Differentially Expressed MiRNAs in Anthers May Contribute to the Fertility of a Novel Brassica Napus Genic Male Sterile Line CN12A. J. Integr. Agric. 2020, 19, 1731–1742. [Google Scholar] [CrossRef]
- Ding, X.; Chen, L.; Guo, J.; Gai, J.; Yang, S. A Small RNA of MiR2119b from Soybean CMS Line Acts as a Negative Regulator of Male Fertility in Transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 167, 210–221. [Google Scholar] [CrossRef]
- Potocki, L.; Karbarz, M.; Adamczyk-Grochala, J.; Kasprzyk, I.; Pawlina-Tyszko, K.; Lewinska, A.; Wnuk, M. Silver Birch Pollen-Derived MicroRNAs Promote NF-ΚB-Mediated Inflammation in Human Lung Cells. Sci. Total Environ. 2021, 800, 149531. [Google Scholar] [CrossRef] [PubMed]
- Macey, M.G. Principles of Flow Cytometry. In Flow Cytometry—Principles and Applications; Macey, M.G., Ed.; Humana Press: Totova, NJ, USA, 2007; ISBN 978-1-58829-691-7. [Google Scholar]
- Burchardt, D.; Machowska, L.; Derwich, K.; Samara, H.; Dworacki, G. Applications of Flow Cytometry in Clinical—Its Applications to Diagnose and Recognize Pathological Changes in Oral Cavity. Now. Lek. 2008, 77, 324–329. [Google Scholar]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow Cytometry: Basic Principles and Applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- McKinnon, K.M. Flow Cytometry: An Overview. Curr. Protoc. Immunol. 2018, 120, 5.1.1–5.1.11. [Google Scholar] [CrossRef]
- Dewitte, A.; Eeckhaut, T.; Van Huylenbroeck, J.; Van Bockstaele, E. Occurrence of Viable Unreduced Pollen in a Begonia Collection. Euphytica 2009, 168, 81–94. [Google Scholar] [CrossRef]
- Moon, H.S.; Eda, S.; Saxton, A.M.; Ow, D.W.; Stewart, C.N., Jr. An Efficient and Rapid Transgenic Pollen Screening and Detection Method Using Flow Cytometry. Biotechnol. J. 2011, 6, 118–123. [Google Scholar] [CrossRef]
- Kron, P.; Kwok, A.; Husband, B.C. Flow Cytometric Analysis of Pollen Grains Collected from Individual Bees Provides Information about Pollen Load Composition and Foraging Behaviour. Ann. Bot. 2014, 113, 191–197. [Google Scholar] [CrossRef] [Green Version]
- Tennant, R.K.; Jones, R.T.; Brock, F.; Cook, C.; Turney, C.S.M.; Love, J.; Lee, R. A New Flow Cytometry Method Enabling Rapid Purification of Fossil Pollen from Terrestrial Sediments for AMS Radiocarbon Dating. J. Quat. Sci. 2013, 28, 229–236. [Google Scholar] [CrossRef]
- Barteneva, N.S.; Fasler-Kan, E.; Vorobjev, I.A. Imaging Flow Cytometry. J. Histochem. Cytochem. 2012, 60, 723–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleiber, A.; Kraus, D.; Henkel, T.; Fritzsche, W. Review: Tomographic Imaging Flow Cytometry. Lab Chip 2021, 21, 3655–3666. [Google Scholar] [CrossRef]
- Veal, D.A.; Deere, D.; Ferrari, B.; Piper, J.; Attfield, P.V. Fluorescence Staining and Flow Cytometry for Monitoring Microbial Cells. J. Immunol. Methods 2000, 243, 191–210. [Google Scholar] [CrossRef]
- Díaz, M.; Herrero, M.; García, L.A.; Quirós, C. Application of Flow Cytometry to Industrial Microbial Bioprocesses. Biochem. Eng. J. 2010, 48, 385–407. [Google Scholar] [CrossRef]
- Pöhlker, C.; Huffman, J.A.; Förster, J.-D.; Pöschl, U. Autofluorescence of Atmospheric Bioaerosols: Spectral Fingerprints and Taxonomic Trends of Pollen. Atmos. Meas. Tech. 2013, 6, 3369–3392. [Google Scholar] [CrossRef] [Green Version]
- Antoniak, M.A.; Pązik, R.; Bazylińska, U.; Wiwatowski, K.; Tomaszewska, A.; Kulpa-Greszta, M.; Adamczyk-Grochala, J.; Wnuk, M.; Maćkowski, S.; Lewińska, A.; et al. Multimodal Polymer Encapsulated CdSe/Fe3O4 Nanoplatform with Improved Biocompatibility for Two-Photon and Temperature Stimulated Bioapplications. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 127, 112224. [Google Scholar] [CrossRef]
- Potocki, L.; Kuna, E.; Filip, K.; Kasprzyk, B.; Lewinska, A.; Wnuk, M. Activation of Transposable Elements and Genetic Instability during Long-Term Culture of the Human Fungal Pathogen Candida Albicans. Biogerontology 2019, 20, 457–474. [Google Scholar] [CrossRef] [Green Version]
- Szpyrka, E.; Broda, D.; Oklejewicz, B.; Podbielska, M.; Slowik-Borowiec, M.; Jagusztyn, B.; Chrzanowski, G.; Kus-Liskiewicz, M.; Duda, M.; Zuczek, J.; et al. A Non-Vector Approach to Increase Lipid Levels in the Microalga Planktochlorella Nurekis. Molecules 2020, 25, 270. [Google Scholar] [CrossRef] [Green Version]
- Depciuch, J.; Kasprzyk, I.; Drzymała, E.; Parlinska-Wojtan, M. Identification of Birch Pollen Species Using FTIR Spectroscopy. Aerobiologia 2018, 34, 525–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowosad, J. Forecasting of Corylus, Alnus, and Betula Pollen Concentration in the Air in Poland. Ph.D. Thesis, Adam Mickiewicz University, Poznan, Poland, 2016. [Google Scholar]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gierlicka, I.; Kasprzyk, I.; Wnuk, M. Imaging Flow Cytometry as a Quick and Effective Identification Technique of Pollen Grains from Betulaceae, Oleaceae, Urticaceae and Asteraceae. Cells 2022, 11, 598. https://doi.org/10.3390/cells11040598
Gierlicka I, Kasprzyk I, Wnuk M. Imaging Flow Cytometry as a Quick and Effective Identification Technique of Pollen Grains from Betulaceae, Oleaceae, Urticaceae and Asteraceae. Cells. 2022; 11(4):598. https://doi.org/10.3390/cells11040598
Chicago/Turabian StyleGierlicka, Iwona, Idalia Kasprzyk, and Maciej Wnuk. 2022. "Imaging Flow Cytometry as a Quick and Effective Identification Technique of Pollen Grains from Betulaceae, Oleaceae, Urticaceae and Asteraceae" Cells 11, no. 4: 598. https://doi.org/10.3390/cells11040598
APA StyleGierlicka, I., Kasprzyk, I., & Wnuk, M. (2022). Imaging Flow Cytometry as a Quick and Effective Identification Technique of Pollen Grains from Betulaceae, Oleaceae, Urticaceae and Asteraceae. Cells, 11(4), 598. https://doi.org/10.3390/cells11040598