Application of Rapid Fluorescence Lifetime Imaging Microscopy (RapidFLIM) to Examine Dynamics of Nanoparticle Uptake in Live Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Nanoparticles
2.2. Sample Preparation for 2D Lifetime Imaging of Nanoparticles in Glioblastoma
2.3. Acquisition of Nanoparticle Fluorescence Lifetimes Using RapidFLIM
2.4. Phasor Analysis
2.5. Temporal Tracking Analysis
3. Results
3.1. RapidFLIM Demonstrates Faster Photon Detection Rates to Resolve Fluorescence Lifetimes
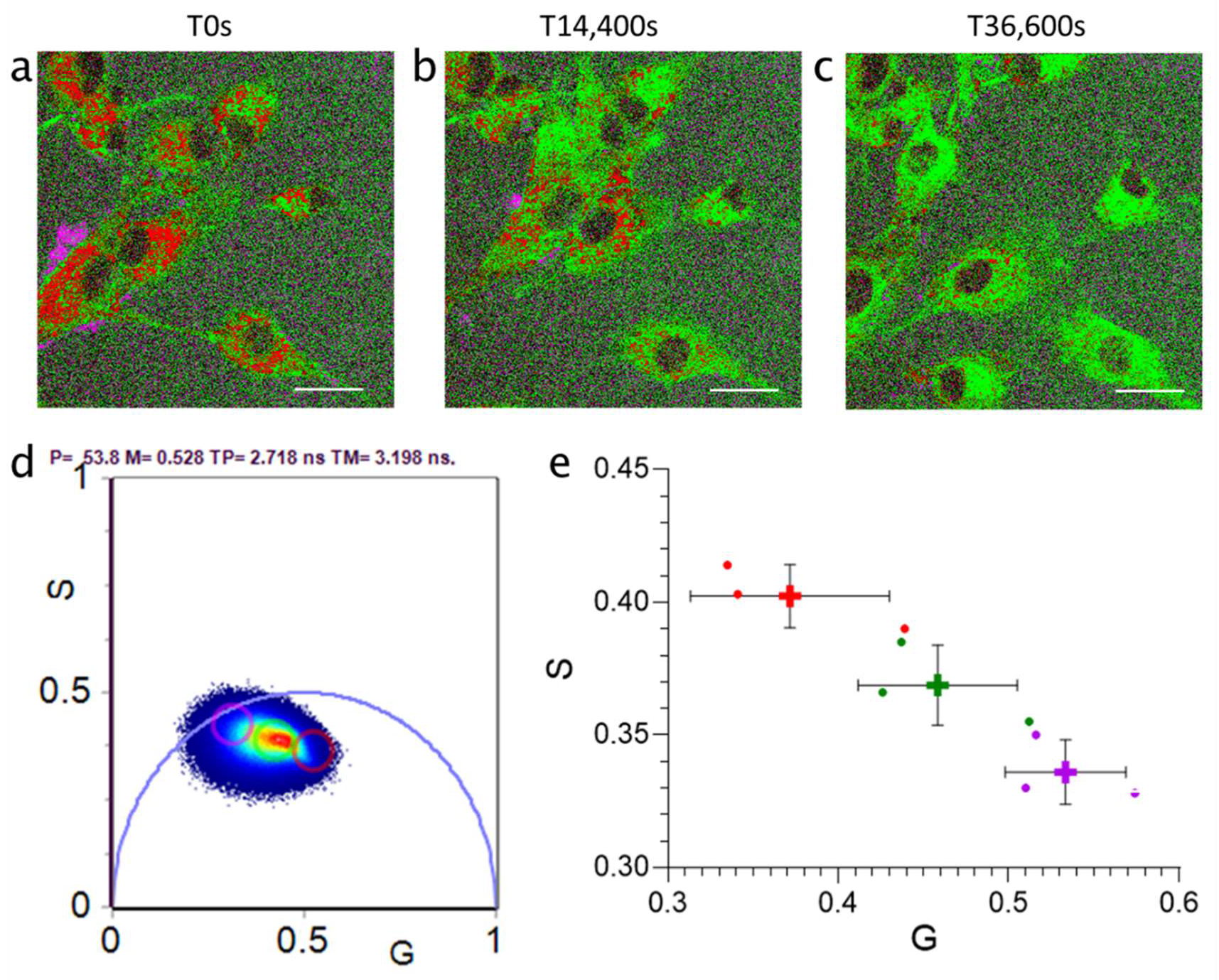
3.2. Particle Size Affected Silica Nanoparticle Lifetimes Acquired during Cellular Uptake
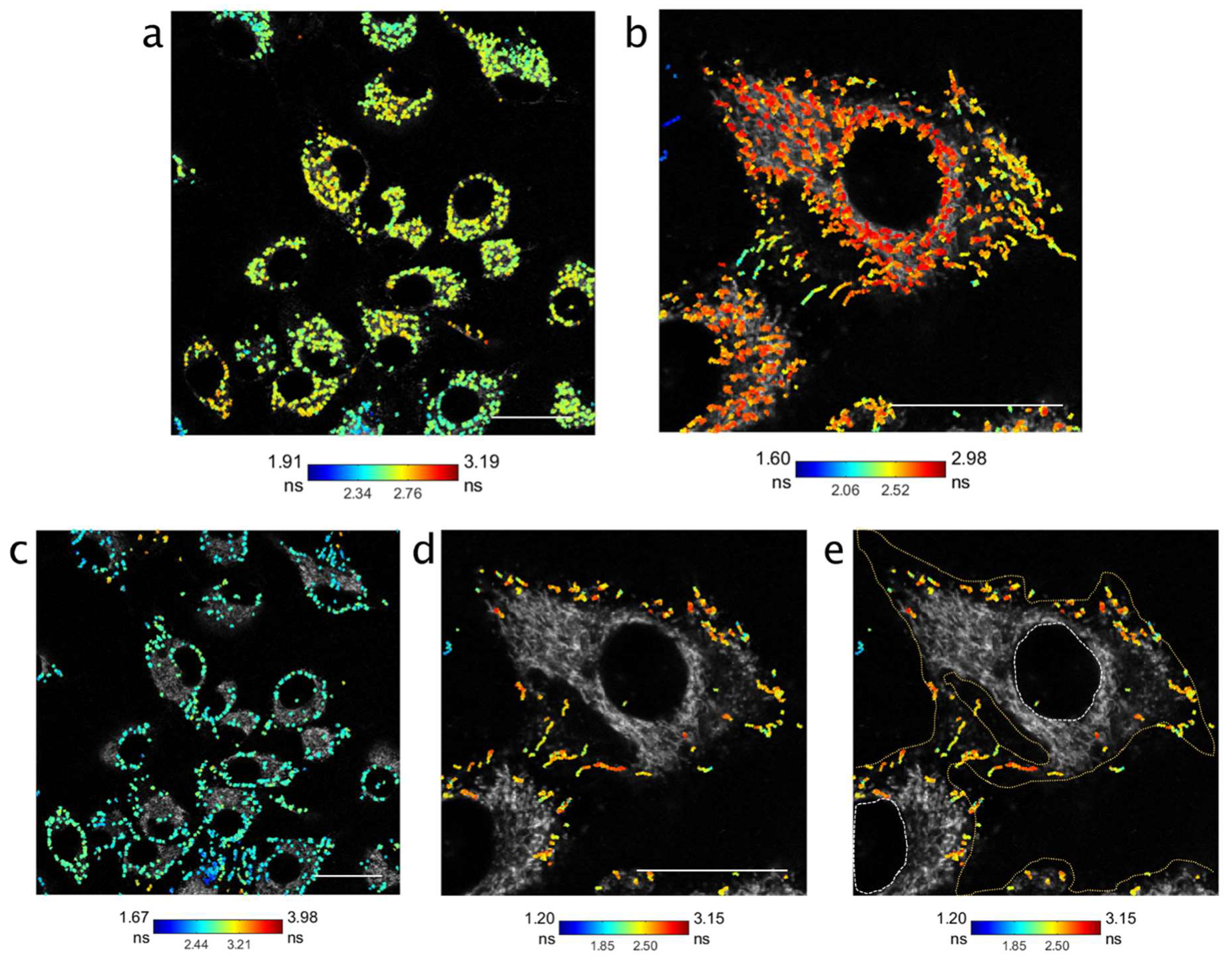
3.3. RapidFLIM Trajectories of Nanoparticles Identifies Significant Changes in Lifetimes Proximal to Membrane Boundaries
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sagnella, S.M.; McCarroll, J.A.; Kavallaris, M. Drug delivery: Beyond active tumour targeting. Nanomed. Nanotechnol. Biol. Med. 2014, 10, 1131–1137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-Nogales, C.; Gonzalez-Fernandez, Y.; Aldaz, A.; Couvreur, P.; Blanco-Prieto, M.J. Nanomedicines for pediatric cancers. ACS Nano 2018, 12, 7482–7496. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Kantoff, P.W.; Wooster, R.; Farokhzad, O.C. Cancer nanomedicine: Progress, challenges and opportunities. Nat. Rev. Cancer 2017, 17, 20–37. [Google Scholar] [CrossRef] [PubMed]
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941. [Google Scholar] [CrossRef]
- Sindhwani, S.; Syed, A.M.; Ngai, J.; Kingston, B.R.; Maiorino, L.; Rothschild, J.; MacMillan, P.; Zhang, Y.; Rajesh, N.U.; Hoang, T. The entry of nanoparticles into solid tumours. Nat. Mater. 2020, 19, 566–575. [Google Scholar] [CrossRef]
- Bastiaens, P.I.; Squire, A. Fluorescence lifetime imaging microscopy: Spatial resolution of biochemical processes in the cell. Trends Cell Biol. 1999, 9, 48–52. [Google Scholar] [CrossRef]
- Pliss, A.; Zhao, L.; Ohulchanskyy, T.Y.; Qu, J.; Prasad, P.N. Fluorescence lifetime of fluorescent proteins as an intracellular environment probe sensing the cell cycle progression. ACS Chem. Biol. 2012, 7, 1385–1392. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, K.; Moriya, D.; Nakabayashi, T.; Li, L.; Ohta, N. Sensitive detection of intracellular environment of normal and cancer cells by autofluorescence lifetime imaging. J. Photochem. Photobiol. B Biol. 2016, 165, 256–265. [Google Scholar] [CrossRef]
- Okabe, K.; Inada, N.; Gota, C.; Harada, Y.; Funatsu, T.; Uchiyama, S. Intracellular temperature mapping with a fluorescent polymeric thermometer and fluorescence lifetime imaging microscopy. Nat. Commun. 2012, 3, 705. [Google Scholar] [CrossRef] [Green Version]
- Basuki, J.S.; Duong, H.T.; Macmillan, A.; Erlich, R.B.; Esser, L.; Akerfeldt, M.C.; Whan, R.M.; Kavallaris, M.; Boyer, C.; Davis, T.P. Using fluorescence lifetime imaging microscopy to monitor theranostic nanoparticle uptake and intracellular doxorubicin release. ACS Nano 2013, 7, 10175–10189. [Google Scholar] [CrossRef]
- Romero, G.; Qiu, Y.; Murray, R.A.; Moya, S.E. Study of Intracellular Delivery of Doxorubicin from Poly (lactide-co-glycolide) Nanoparticles by Means of Fluorescence Lifetime Imaging and Confocal Raman Microscopy. Macromol. Biosci. 2013, 13, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Yue, Z.; Eccleston, M.E.; Swartling, J.; Slater, N.K.; Kaminski, C.F. Fluorescence intensity and lifetime imaging of free and micellar-encapsulated doxorubicin in living cells. Nanomed. Nanotechnol. Biol. Med. 2008, 4, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Nam, H.Y.; Kwon, S.M.; Chung, H.; Lee, S.; Kwon, S.; Jeon, H.; Kim, Y.; Park, J.H.; Kim, J.; Her, S. Cellular uptake mechanism and intracellular fate of hydrophobically modified glycol chitosan nanoparticles. J. Control. Release 2009, 135, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Chithrani, B.D.; Chan, W.C. Elucidating the mechanism of cellular uptake and removal of protein-coated gold nanoparticles of different sizes and shapes. Nano Lett. 2007, 7, 1542–1550. [Google Scholar] [CrossRef] [PubMed]
- Pelras, T.; Duong, H.; Kim, B.J.; Hawkett, B.S.; Mullner, M. A’grafting from’approach to polymer nanorods for pH-triggered intracellular drug delivery. Polymer 2017, 112, 244–251. [Google Scholar] [CrossRef]
- O’Connor, D. Time-Correlated Single Photon Counting; Academic Press: Orlando, FL, USA, 2012. [Google Scholar]
- Birch, D.J.; Imhof, R.E. Time-domain fluorescence spectroscopy using time-correlated single-photon counting. In Anonymous Topics in Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2002; pp. 1–95. [Google Scholar]
- Orthaus-Mueller, S.; Kraemer, B.; Dowler, R.; Devaux, A.; Tannert, A.; Roehlicke, T.; Wahl, M.; Rahn, H.; Erdmann, R. rapidFLIM: The new and innovative method for ultra fast FLIM imaging. PicoQuant Appl. Note 2016. Available online: https://www.picoquant.com/rapidFLIM (accessed on 10 January 2022).
- Faria, A.R.; Silvestre, O.F.; Maibohm, C.; Adão, R.M.; Silva, B.F.; Nieder, J.B. Cubosome nanoparticles for enhanced delivery of mitochondria anticancer drug elesclomol and therapeutic monitoring via sub-cellular NAD (P) H multi-photon fluorescence lifetime imaging. Nano Res. 2019, 12, 991–998. [Google Scholar] [CrossRef]
- Sandin, P.; Fitzpatrick, L.W.; Simpson, J.C.; Dawson, K.A. High-speed imaging of Rab family small GTPases reveals rare events in nanoparticle trafficking in living cells. ACS Nano 2012, 6, 1513–1521. [Google Scholar] [CrossRef]
- Poudel, C.; Mela, I.; Kaminski, C.F. High-throughput, multi-parametric, and correlative fluorescence lifetime imaging. Methods Appl. Fluoresc. 2020, 8, 024005. [Google Scholar] [CrossRef]
- Davis, M.E.; Chen, Z.; Shin, D.M. Nanoparticle therapeutics: An emerging treatment modality for cancer. Nat. Rev. Drug Discov. 2008, 7, 771–782. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, X.; Li, P.; Nguyen, K.T.; Wang, X.; Luo, Z.; Zhang, H.; Tan, N.S.; Zhao, Y. Biocompatible, uniform, and redispersible mesoporous silica nanoparticles for cancer-targeted drug delivery in vivo. Adv. Funct. Mater. 2014, 24, 2450–2461. [Google Scholar] [CrossRef]
- Digman, M.A.; Gratton, E.; Marcu, L.; French, P.; Elson, D.S. The phasor approach to fluorescence lifetime imaging: Exploiting phasor linear properties. In Florescence Lifetime Spectroscopy and Imaging; CRC Press: Boca Raton, FL, USA, 2014; pp. 235–248. [Google Scholar]
- Colyer, R.; Siegmund, O.; Tremsin, A.; Vallerga, J.; Weiss, S.; Michalet, X. Phasor-based single-molecule fluorescence lifetime imaging using a wide-field photon-counting detector. In Proceedings of the Single Molecule Spectroscopy and Imaging II, San Jose, CA, USA, 24 February 2009; Volume 7185, p. 71850T. [Google Scholar]
- Stringari, C.; Cinquin, A.; Cinquin, O.; Digman, M.A.; Donovan, P.J.; Gratton, E. Phasor approach to fluorescence lifetime microscopy distinguishes different metabolic states of germ cells in a live tissue. Proc. Natl. Acad. Sci. USA 2011, 108, 13582–13587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malacrida, L.; Jameson, D.M.; Gratton, E. A multidimensional phasor approach reveals LAURDAN photophysics in NIH-3T3 cell membranes. Sci. Rep. 2017, 7, 9215. [Google Scholar] [CrossRef]
- Vallotton, P.; van Oijen, A.M.; Whitchurch, C.B.; Gelfand, V.; Yeo, L.; Tsiavaliaris, G.; Heinrich, S.; Dultz, E.; Weis, K. Grünwald, Diatrack particle tracking software: Review of applications and performance evaluation. Traffic 2017, 18, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, J.; Xiong, J.; Liu, X. Determination of agarose gel pore size: Absorbance measurements vis a vis other techniques. J. Phys. Conf. Ser. 2006, 28, 017. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, X.; Chen, J.; Chen, J.; Kuznetsova, L.; Wong, S.S.; Ojima, I. Mechanism-based tumor-targeting drug delivery system. Validation of efficient vitamin receptor-mediated endocytosis and drug release. Bioconjug. Chem. 2010, 21, 979–987. [Google Scholar] [CrossRef] [Green Version]
- Vu, M.N.; Rajasekhar, P.; Poole, D.P.; Khor, S.Y.; Truong, N.P.; Nowell, C.J.; Quinn, J.F.; Whittaker, M.; Veldhuis, N.A.; Davis, T.P. Rapid Assessment of Nanoparticle Extravasation in a Microfluidic Tumor Model. ACS Appl. Nano Mater. 2019, 2, 1844–1856. [Google Scholar] [CrossRef] [Green Version]
- Ahmed-Cox, A.; Pandzic, E.; Johnston, S.T.; Heu, C.; McGhee, J.; Mansfeld, F.M.; Crampin, E.J.; Davis, T.P.; Whan, R.M.; Kavallaris, M. Spatio-temporal analysis of nanoparticles in live tumor spheroids impacted by cell origin and density. J. Control. Release 2021, 341, 661–675. [Google Scholar] [CrossRef]
- Feiner-Gracia, N.; Beck, M.; Pujals, S.; Tosi, S.; Mandal, T.; Buske, C.; Linden, M.; Albertazzi, L. Super-Resolution Microscopy Unveils Dynamic Heterogeneities in Nanoparticle Protein Corona. Small 2017, 13, 1701631. [Google Scholar] [CrossRef]
- Zhang, L.; Feng, Q.; Wang, J.; Zhang, S.; Ding, B.; Wei, Y.; Dong, M.; Ryu, J.Y.; Yoon, T.Y.; Shi, X.; et al. Microfluidic Synthesis of Hybrid Nanoparticles with Controlled Lipid Layers: Understanding Flexibility-Regulated Cell-Nanoparticle Interaction. ACS Nano 2015, 9, 9912–9921. [Google Scholar] [CrossRef] [Green Version]
- Goodarzi, S.; Prunet, A.; Rossetti, F.; Bort, G.; Tillement, O.; Porcel, E.; Lacombe, S.; Wu, T.; Guerquin-Kern, J.; Delanoë-Ayari, H. Quantifying nanotherapeutic penetration using a hydrogel-based microsystem as a new 3D in vitro platform. Lab A Chip. 2021, 21, 2495–2510. [Google Scholar] [CrossRef] [PubMed]
- Digman, M.A.; Caiolfa, V.R.; Zamai, M.; Gratton, E. The phasor approach to fluorescence lifetime imaging analysis. Biophys. J. 2008, 94, L14–L16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinde, E.; Thammasiraphop, K.; Duong, H.T.; Yeow, J.; Karagoz, B.; Boyer, C.; Gooding, J.J.; Gaus, K. Pair correlation microscopy reveals the role of nanoparticle shape in intracellular transport and site of drug release. Nat. Nanotechnol. 2017, 12, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Samukaite-Bubniene, U.; Mazetyte-Stasinskiene, R.; Chernyakova, K.; Karpicz, R.; Ramanavicius, A. Time-resolved fluorescence spectroscopy based evaluation of stability of glucose oxidase. Int. J. Biol. Macromol. 2020, 163, 676–682. [Google Scholar] [CrossRef]
- Wright, B.K.; Andrews, L.M.; Markham, J.; Jones, M.R.; Stringari, C.; Digman, M.A.; Gratton, E. NADH distribution in live progenitor stem cells by phasor-fluorescence lifetime image microscopy. Biophys. J. 2012, 103, L7–L9. [Google Scholar] [CrossRef] [Green Version]
- Ung, T.P.L.; Lim, S.; Solinas, X.; Mahou, P.; Chessel, A.; Marionnet, C.; Bornschlögl, T.; Beaurepaire, E.; Bernerd, F.; Pena, A. Simultaneous NAD (P) H and FAD fluorescence lifetime microscopy of long UVA–induced metabolic stress in reconstructed human skin. Sci. Rep. 2021, 11, 22171. [Google Scholar] [CrossRef]
- Ramanavicius, A.; Ryskevic, N.; Oztekin, Y.; Kausaite-Minkstimiene, A.; Jursenas, S.; Baniukevic, J.; Kirlyte, J.; Bubniene, U.; Ramanaviciene, A. Immunosensor based on fluorescence quenching matrix of the conducting polymer polypyrrole. Anal. Bioanal. Chem. 2010, 398, 3105–3113. [Google Scholar] [CrossRef]
- Wilhelm, S.; del Barrio, M.; Heiland, J.; Himmelstoß, S.F.; Galbán, J.; Wolfbeis, O.S.; Hirsch, T. Spectrally matched upconverting luminescent nanoparticles for monitoring enzymatic reactions. ACS Appl. Mater. Interfaces 2014, 6, 15427–15433. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed-Cox, A.; Macmillan, A.M.; Pandzic, E.; Whan, R.M.; Kavallaris, M. Application of Rapid Fluorescence Lifetime Imaging Microscopy (RapidFLIM) to Examine Dynamics of Nanoparticle Uptake in Live Cells. Cells 2022, 11, 642. https://doi.org/10.3390/cells11040642
Ahmed-Cox A, Macmillan AM, Pandzic E, Whan RM, Kavallaris M. Application of Rapid Fluorescence Lifetime Imaging Microscopy (RapidFLIM) to Examine Dynamics of Nanoparticle Uptake in Live Cells. Cells. 2022; 11(4):642. https://doi.org/10.3390/cells11040642
Chicago/Turabian StyleAhmed-Cox, Aria, Alexander M. Macmillan, Elvis Pandzic, Renee M. Whan, and Maria Kavallaris. 2022. "Application of Rapid Fluorescence Lifetime Imaging Microscopy (RapidFLIM) to Examine Dynamics of Nanoparticle Uptake in Live Cells" Cells 11, no. 4: 642. https://doi.org/10.3390/cells11040642






