Single-Cell RNA-Seq Analysis of Olfactory Mucosal Cells of Alzheimer’s Disease Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethical Considerations
2.2. Patients and OM Cell Cultures
2.3. Enzyme-Linked Immunosorbent Assays
2.4. Cell Hashing and Single Cell RNA Sequencing
2.5. Quality Control and Downstream Analysis
2.6. Differential Expression and Enrichment
2.7. Analysis of Mitochondrial Respiration
2.8. Quantification of the Intracellular Levels of ATP
2.9. Western Blotting
2.10. Statistical Methods and Graphical Illustrations
3. Results
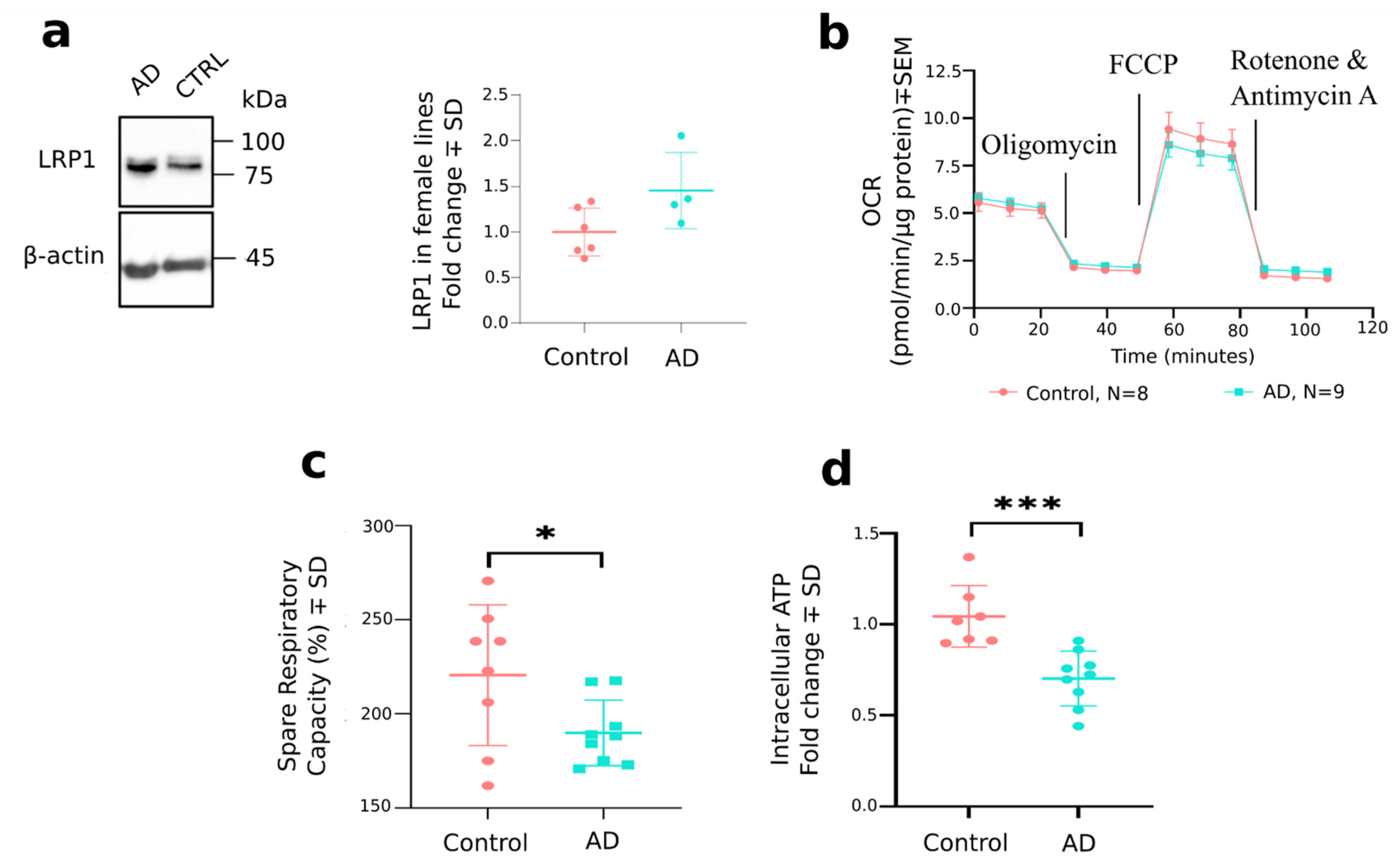
3.1. Secretion of Aβ1–42 Is Increased in AD OM Cells
3.2. Diversity of the Human OM Cells in Health and AD
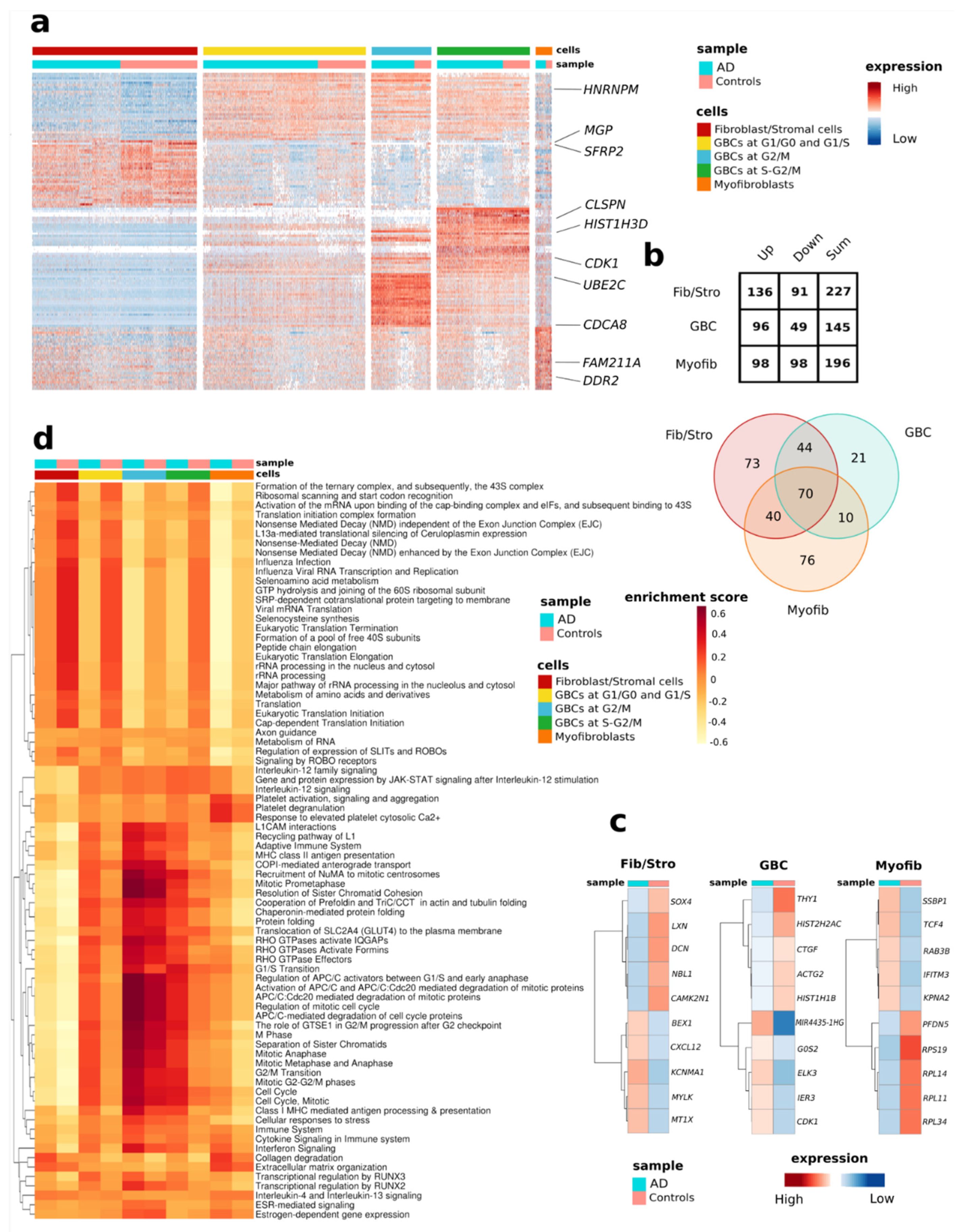
3.3. Differential Expression Analysis Reveals AD-Related Alterations of Gene Expression in the OM Cells
3.4. Analysis of Individual Cell Types Reveals Distinctive AD-Associated Genes and Pathways
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
ScRNA-Seq, Additional Details of the Analysis
References
- Marin, C.; Vilas, D.; Langdon, C.; Alobid, I.; López-Chacón, M.; Haehner, A.; Hummel, T.; Mullol, J. Olfactory Dysfunction in Neurodegenerative Diseases. Curr. Allergy Asthma Rep. 2018, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Féron, F.; Perry, C.; McGrath, J.J.; Mackay-Sim, A. New techniques for biopsy and culture of human olfactory epithelial neurons. Arch. Otolaryngol. Head Neck Surg. 1998, 124, 861–866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.J.; Shin, I.S.; Lee, J.E. Olfactory function in mild cognitive impairment and Alzheimer’s disease: A meta-analysis. Laryngoscope 2019, 129, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, H.R.; Bates, K.A.; Rodrigues, M.; Taddei, K.; Laws, S.M.; Lautenschlager, N.T.; Dhaliwal, S.S.; Johnston, A.N.B.; Mackay-Sim, A.; Gandy, S.; et al. Olfactory dysfunction is associated with subjective memory complaints in community-dwelling elderly individuals. J. Alzheimers Dis. 2009, 17, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, H.R.; Bates, K.A.; Weinborn, M.G.; Johnston, A.N.B.; Bahramian, A.; Taddei, K.; Laws, S.M.; Rodrigues, M.; Morici, M.; Howard, M.; et al. Olfactory discrimination predicts cognitive decline among community-dwelling older adults. Transl. Psychiatry 2012, 2, e118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.J.; Son, G.; Bae, J.; Kim, S.Y.; Yoo, Y.K.; Park, D.; Baek, S.Y.; Chang, K.-A.; Suh, Y.-H.; Lee, Y.-B.; et al. Longitudinal profiling of oligomeric Aβ in human nasal discharge reflecting cognitive decline in probable Alzheimer’s disease. Sci. Rep. 2020, 10, 11234. [Google Scholar] [CrossRef]
- Liu, Z.; Kameshima, N.; Nanjo, T.; Shiino, A.; Kato, T.; Shimizu, S.; Shimizu, T.; Tanaka, S.; Miura, K.; Tooyama, I. Development of a High-Sensitivity Method for the Measurement of Human Nasal Aβ 42, Tau, and Phosphorylated Tau. J. Alzheimers Dis. 2018, 62, 737–744. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.H.; Lee, S.M.; Cho, S.; Kang, J.-H.; Minn, J.-K.; Park, H.; Choi, S.H. Amyloid beta in nasal secretions may be a potential biomarker of Alzheimer’s disease. Sci. Rep. 2019, 9, 4966. [Google Scholar] [CrossRef]
- Arnold, S.E.; Lee, E.B.; Moberg, P.J.; Stutzbach, L.; Kazi, H.; Han, L.-Y.; Lee, V.M.Y.; Trojanowski, J.Q. Olfactory epithelium amyloid-β and paired helical filament-tau pathology in Alzheimer disease. Ann. Neurol. 2010, 67, 462–469. [Google Scholar] [CrossRef]
- Ayala-Grosso, C.A.; Pieruzzini, R.; Diaz-Solano, D.; Wittig, O.; Abrante, L.; Vargas, L.; Cardier, J. Amyloid-Aβ peptide in olfactory mucosa and mesenchymal stromal cells of mild cognitive impairment and Alzheimer’s disease patients. Brain Pathol. 2015, 25, 136–145. [Google Scholar] [CrossRef]
- Talamo, B.R.; Rudel, R.A.; Kosik, K.S.; Lee, V.M.-Y.; Neff, S.; Adelman, L.; Kauer, J.S. Pathological changes in olfactory neurons in patients with Alzheimer’s disease. Nature 1989, 337, 736–739. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Goedert, M.; Hill, W.D.; Lee, V.M.Y.; Trojanowski, J.Q. Tau Proteins Are Abnormally Expressed in Olfactory Epithelium of Alzheimer Patients and Developmentally Regulated in Human Fetal Spinal Cord. Exp. Neurol. 1993, 121, 93–105. [Google Scholar] [CrossRef]
- Ghanbari, H.A.; Ghanbari, K.; Harris, P.L.R.; Jones, P.K.; Kubat, Z.; Castellani, R.J.; Wolozin, R.L.; Smith, M.A.; Perry, G. Oxidative damage in cultured human olfactory neurons from Alzheimer’s disease patients. Aging Cell 2004, 3, 41–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolozin, B.; Lesch, P.; Lebovics, R.; Sunderland, T. Olfactory neuroblasts from Alzheimer donors: Studies on APP processing and cell regulation. Biol. Psychiatry 1993, 34, 824–838. [Google Scholar] [CrossRef]
- Grubman, A.; Chew, G.; Ouyang, J.F.; Sun, G.; Choo, X.Y.; MacLean, C.; Simmons, R.K.; Buckberry, S.; Vargas-Landin, D.B.; Poppe, D.; et al. A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 2019, 22, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Mathys, H.; Davila-Velderrain, J.; Peng, Z.; Gao, F.; Mohammadi, S.; Young, J.Z.; Menon, M.; He, L.; Abdurrob, F.; Jiang, X.; et al. Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 2019, 570, 332–337. [Google Scholar] [CrossRef] [PubMed]
- Olah, M.; Menon, V.; Habib, N.; Taga, M.F.; Ma, Y.; Yung, C.J.; Cimpean, M.; Khairallah, A.; Coronas-Samano, G.; Sankowski, R.; et al. Single cell RNA sequencing of human microglia uncovers a subset associated with Alzheimer’s disease. Nat. Commun. 2020, 11, 6129. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, W.M.; Andhey, P.S.; Swain, A.; Levy, T.; Miller, K.R.; Poliani, P.L.; Cominelli, M.; Grover, S.; Gilfillan, S.; et al. Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 2020, 26, 131–142. [Google Scholar] [CrossRef]
- Kashima, Y.; Sakamoto, Y.; Kaneko, K.; Seki, M.; Suzuki, Y.; Suzuki, A. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med. 2020, 52, 1419–1427. [Google Scholar] [CrossRef]
- Aldridge, S.; Teichmann, S.A. Single cell transcriptomics comes of age. Nat. Commun. 2020, 11, 1–4. [Google Scholar] [CrossRef]
- Martinez-Jimenez, C.P.; Eling, N.; Chen, H.C.; Vallejos, C.A.; Kolodziejczyk, A.A.; Connor, F.; Stojic, L.; Rayner, T.F.; Stubbington, M.J.T.; Teichmann, S.A. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 2017, 355, 1433–1436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, M.P.; Du, J.; Lagoudas, G.; Jiao, Y.; Sawyer, A.; Drummon, D.C.; Lauffenburger, D.A.; Raue, A. Analysis of Single-Cell RNA-Seq Identifies Cell-Cell Communication Associated with Tumor Characteristics. Cell Rep. 2018, 25, 1458–1468.e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vistain, L.F.; Tay, S. Single-Cell Proteomics. Trends Biochem. Sci. 2021, 46, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; George, J.; Wang, J. Deep Profiling of Cellular Heterogeneity by Emerging Single-Cell Proteomic Technologies. Proteomics 2020, 20, e1900226. [Google Scholar] [CrossRef] [PubMed]
- Rubakhin, S.S.; Lanni, E.J.; Sweedler, J.V. Progress toward single cell metabolomics. Curr. Opin. Biotechnol. 2013, 24, 95–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durante, M.A.; Kurtenbach, S.; Sargi, Z.B.; Harbour, J.W.; Choi, R.; Kurtenbach, S.; Goss, G.M.; Matsunami, H.; Goldstein, B.J. Single-cell analysis of olfactory neurogenesis and differentiation in adult humans. Nat. Neurosci. 2020, 23, 323–326. [Google Scholar] [CrossRef]
- Oliva, A.D.; Gupta, R.; Issa, K.; Hachem, R.A.; Jang, D.W.; Wellford, S.A.; Moseman, A.E.; Matsunami, H.; Goldstein, B.J. Aging-related olfactory loss is associated with olfactory stem cell transcriptional alterations in humans. J. Clin. Investig. 2022, e155506. [Google Scholar] [CrossRef]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef]
- Morris, J.C.; Heyman, A.; Mohs, R.C.; Hughes, J.P.; van Belle, G.; Fillenbaum, G.; Mellits, E.D.; Clark, C. The consortium to establish a registry for Alzheimer’s disease (CERAD). Part I. Clinical and neuropsychological assessment of Alzheimer’s disease. Neurology 1989, 39, 1159–1165. [Google Scholar] [CrossRef]
- Mirra, S.S.; Heyman, A.; McKeel, D.; Sumi, S.M.; Crain, B.J.; Brownlee, L.M.; Vogel, F.S.; Hughes, J.P.; van Belle, G.; Berg, L.; et al. The consortium to establish a registry for Alzheimer’s disease (CERAD). Part II. Standardization of the neuropathologic assessment of Alzheimer’s disease. Neurology 1991, 41, 479–486. [Google Scholar] [CrossRef]
- Huovinen, J.; Kastinen, S.; Komulainen, S.; Oinas, M.; Avellan, C.; Frantzen, J.; Rinne, J.; Ronkainen, A.; Kauppinen, M.; Lönnrot, K.; et al. Familial idiopathic normal pressure hydrocephalus. J. Neurol Sci. 2016, 368, 11–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hummel, T.; Konnerth, C.G.; Rosenheim, K.; Kobal, G. Screening of olfactory function with a four-minute odor identification test: Reliability, normative data, and investigations in patients with olfactory loss. Ann. Otol. Rhinol. Laryngol. 2001, 110, 976–981. [Google Scholar] [CrossRef] [PubMed]
- Murrell, W.; Féron, F.; Wetzig, A.; Cameron, N.; Splatt, K.; Bellette, B.; Blanco, J.; Perry, C.; Lee, G.; Mackay-Sim, A. Multipotent stem cells from adult olfactory mucosa. Dev. Dyn. 2005, 233, 496–515. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., III; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Vaz Meirelles, G.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, 90–97. [Google Scholar] [CrossRef] [Green Version]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.J.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Greene, C.S.; Krishnan, A.; Wong, A.K.; Ricciotti, E.; Zelaya, R.A.; Himmelstein, D.S.; Zhang, R.; Hartmann, B.M.; Zaslavsky, E.; Sealfon, S.C.; et al. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015, 47, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Jassal, B.; Matthews, L.; Viteri, G.; Gong, C.; Lorente, P.; Fabregat, A.; Sidiropoulos, K.; Cook, J.; Gillespie, M.; Haw, R.; et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020, 48, 498–503. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’Eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, Y.; Zhang, B. Efficient test and visualization of multi-set intersections. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Rath, S.; Sharma, R.; Gupta, R.; Ast, T.; Chan, C.; Durham, T.J.; Goodman, R.P.; Grabarek, Z.; Haas, M.E.; Hung, W.H.W.; et al. MitoCarta3.0: An updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021, 49, 1541–1547. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Nelson, A.R.; Sagare, A.P.; Zlokovic, B.V. Impaired vascular-mediated clearance of brain amyloid beta in Alzheimer’s disease: The role, regulation and restoration of LRP1. Front. Aging Neurosci. 2015, 7, 136. [Google Scholar] [CrossRef] [Green Version]
- Kameshima, N.; Nanjou, T.; Fukuhara, T.; Yanagisawa, D.; Tooyama, I. Correlation of Aβ deposition in the nasal cavity with the formation of senile plaques in the brain of a transgenic mouse model of Alzheimer’s disease. Neurosci. Lett. 2012, 513, 166–169. [Google Scholar] [CrossRef]
- Johnston, J.A.; Cowburn, R.F.; Norgren, S.; Wiehager, B.; Venizelos, N.; Winblad, B.; Vigo-Pelfrey, C.; Schenk, D.; Lannfelt, L.; O’Neill, C. Increased beta-amyloid release and levels of amyloid precursor protein (APP) in fibroblast cell lines from family members with the Swedish Alzheimer’s disease APP670/671 mutation. FEBS Lett. 1994, 354, 274–278. [Google Scholar] [CrossRef] [Green Version]
- Shinohara, M.; Tachibana, M.; Kanekiyo, T.; Bu, G. Role of LRP1 in the pathogenesis of Alzheimer’s disease: Evidence from clinical and preclinical studies. J. Lipid Res. 2017, 58, 1267–1281. [Google Scholar] [CrossRef] [Green Version]
- Manczak, M.; Park, P.S.; Jung, Y.; Reddy, P.H. Differential expression of oxidative phosphorylation genes in patients with Alzheimer’s disease: Implications for early mitochondrial dysfunction and oxidative damage. Neuromolecular. Med. 2004, 5, 147–162. [Google Scholar] [CrossRef]
- Leake, A.; Morris, C.M.; Whateley, J. Brain matrix metalloproteinase 1 levels are elevated in Alzheimer’s disease. Neurosci. Lett. 2000, 291, 201–203. [Google Scholar] [CrossRef]
- Lanni, C.; Nardinocchi, L.; Puca, R.; Stanga, S.; Uberti, D.; Memo, M.; Govoni, S.; D’Orazi, G.; Racchi, M. Homeodomain Interacting Protein Kinase 2: A Target for Alzheimer’s Beta Amyloid Leading to Misfolded p53 and Inappropriate Cell Survival. PLoS ONE 2010, 5, e10171. [Google Scholar] [CrossRef] [Green Version]
- Lindeque, J.Z.; Levanets, O.; Louw, R.; van der Westhuizen, F.H. The Involvement of Metallothioneins in Mitochondrial Function and Disease. Curr. Protein Pept. Sci. 2010, 11, 292–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bragina, O.; Gurjanova, K.; Krishtal, J.; Kulp, M.; Karro, N.; Tõugu, V.; Palumaa, P. Metallothionein 2A affects the cell respiration by suppressing the expression of mitochondrial protein cytochrome c oxidase subunit II. J. Bioenerg Biomembr. 2015, 47, 209–216. [Google Scholar] [CrossRef]
- Chakravorty, A.; Jetto, C.T.; Manjithaya, R. Dysfunctional Mitochondria and Mitophagy as Drivers of Alzheimer’s Disease Pathogenesis. Front. Aging Neurosci. 2019, 11, 311. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhao, F.; Ma, X.; Perry, G.; Zhu, X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: Recent advances. Mol. Neurodegener. 2020, 15, 30. [Google Scholar] [CrossRef] [PubMed]
- Lunnon, K.; Keohane, A.; Pidsley, R.; Newhouse, S.; Riddoch-Contreras, J.; Thubron, E.B.; Devall, M.; Soininen, H.; Kłoszewska, I.; Mecocci, P.; et al. Mitochondrial genes are altered in blood early in Alzheimer’s disease. Neurobiol. Aging 2017, 53, 36–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, S.M.; de Marco, M.; Barnes, K.; Shaw, P.J.; Ferraiuolo, L.; Blackburn, D.J.; Mortiboys, H.; Venneri, A. Deficits in mitochondrial spare respiratory capacity contribute to the neuropsychological changes of Alzheimer’s disease. J. Pers. Med. 2020, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Lambert, J.C.; Grenier-Boley, B.; Chouraki, V.; Heath, S.; Zelenika, D.; Fievet, N.; Hannequin, D.; Pasquier, F.; Hanon, O.; Brice, A.; et al. Implication of the immune system in Alzheimer’s disease: Evidence from genome-wide pathway analysis. J. Alzheimers Dis. 2010, 20, 1107–1118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, L.; Holmans, P.A.; Hamshere, M.L.; Harold, D.; Moskvina, V.; Ivanov, D.; Pocklington, A.; Abraham, R.; Hollingworth, P.; Sims, R.; et al. Genetic Evidence Implicates the Immune System and Cholesterol Metabolism in the Aetiology of Alzheimer’s Disease. PLoS ONE 2010, 5, e13950. [Google Scholar] [CrossRef]
- Jiang, Q.; Jin, S.; Jiang, Y.; Liao, M.; Feng, R.; Zhang, L.; Liu, G.; Hao, J. Alzheimer’s Disease Variants with the Genome-Wide Significance are Significantly Enriched in Immune Pathways and Active in Immune Cells. Mol. Neurobiol. 2017, 54, 594–600. [Google Scholar] [CrossRef]
- Pillai, J.A.; Maxwell, S.; Bena, J.; Bekris, L.M.; Rao, S.M.; Change, M.; Lamb, B.T.; Leverenz, L.B. Key inflammatory pathway activations in the MCI stage of Alzheimer’s disease. Ann. Clin. Transl. Neurol. 2019, 6, 1248–1262. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Xie, C.; Zhao, Y.; Li, Z.; Xu, P.; Yao, L. Gene expression analysis reveals the dysregulation of immune and metabolic pathways in Alzheimer’s disease. Oncotarget 2016, 7, 72469–72474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef] [PubMed]
- Abdalkader, M.; Lampinen, R.; Kanninen, K.M.; Malm, T.M.; Liddell, J.R. Targeting Nrf2 to Suppress Ferroptosis and Mitochondrial Dysfunction in Neurodegeneration. Front. Neurosci. 2018, 12, 466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Martínez, J.L.; Álvarez-Machancoses, Ó.; de Andrés-Galiana, E.J.; Bea, G.; Kloczkowski, A. Robust Sampling of Defective Pathways in Alzheimer’s Disease. Implications in Drug Repositioning. Int. J. Mol. Sci. 2020, 21, 3594. [Google Scholar] [CrossRef] [PubMed]
- Ming, G.-L.; Song, H. Adult neurogenesis in the mammalian brain: Significant answers and significant questions. Neuron 2011, 70, 687–702. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Jiménez, E.P.; Flor-García, M.; Terreros-Roncal, J.; Rábano, A.; Cafini, F.; Pallas-Bazarra, N.; Ávila, J.; Llorens-Martín, M. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat. Med. 2019, 25, 554–560. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Kuhlwilm, M.; Davierwala, A.; Pääbo, S. Identification of putative target genes of the transcription factor RUNX2. PLoS ONE 2013, 8, e83218. [Google Scholar] [CrossRef] [Green Version]
- Nuutinen, T.; Suuronen, T.; Kauppinen, A.; Salminen, A. Clusterin: A forgotten player in Alzheimer’s disease. Brain Res. Rev. 2009, 61, 89–104. [Google Scholar] [CrossRef]
- Peix, L.; Evans, I.C.; Pearce, D.R.; Simpson, J.K.; Maher, T.M.; McAnulty, R.J. Diverse functions of clusterin promote and protect against the development of pulmonary fibrosis. Sci. Rep. 2018, 8, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Newman, M.P.; Féron, F.; Mackay-Sim, A. Growth factor regulation of neurogenesis in adult olfactory epithelium. Neuroscience 2000, 99, 343–350. [Google Scholar] [CrossRef]
- Kovacs, G.G. Astroglia and Tau: New Perspectives. Front. Aging Neurosci. 2020, 12, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lampinen, R.; Fazaludeen, M.F.; Avesani, S.; Örd, T.; Penttilä, E.; Lehtola, J.-M.; Saari, T.; Hannonen, S.; Saveleva, L.; Kaartinen, E.; et al. Single-Cell RNA-Seq Analysis of Olfactory Mucosal Cells of Alzheimer’s Disease Patients. Cells 2022, 11, 676. https://doi.org/10.3390/cells11040676
Lampinen R, Fazaludeen MF, Avesani S, Örd T, Penttilä E, Lehtola J-M, Saari T, Hannonen S, Saveleva L, Kaartinen E, et al. Single-Cell RNA-Seq Analysis of Olfactory Mucosal Cells of Alzheimer’s Disease Patients. Cells. 2022; 11(4):676. https://doi.org/10.3390/cells11040676
Chicago/Turabian StyleLampinen, Riikka, Mohammad Feroze Fazaludeen, Simone Avesani, Tiit Örd, Elina Penttilä, Juha-Matti Lehtola, Toni Saari, Sanna Hannonen, Liudmila Saveleva, Emma Kaartinen, and et al. 2022. "Single-Cell RNA-Seq Analysis of Olfactory Mucosal Cells of Alzheimer’s Disease Patients" Cells 11, no. 4: 676. https://doi.org/10.3390/cells11040676
APA StyleLampinen, R., Fazaludeen, M. F., Avesani, S., Örd, T., Penttilä, E., Lehtola, J.-M., Saari, T., Hannonen, S., Saveleva, L., Kaartinen, E., Fernández Acosta, F., Cruz-Haces, M., Löppönen, H., Mackay-Sim, A., Kaikkonen, M. U., Koivisto, A. M., Malm, T., White, A. R., Giugno, R., ... Kanninen, K. M. (2022). Single-Cell RNA-Seq Analysis of Olfactory Mucosal Cells of Alzheimer’s Disease Patients. Cells, 11(4), 676. https://doi.org/10.3390/cells11040676








