Chemerin Impact on Alternative mRNA Transcription in the Porcine Luteal Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Samples, In Vitro Cell Culture and Chemerin Administration
2.2. RNA Isolation, Library Preparation and High-Throughput Sequencing Procedure
2.3. In Silico Analyses
2.3.1. Raw Reads Pre-Processing and Mapping to a Reference Genome
2.3.2. Long Non-Coding RNA Analysis
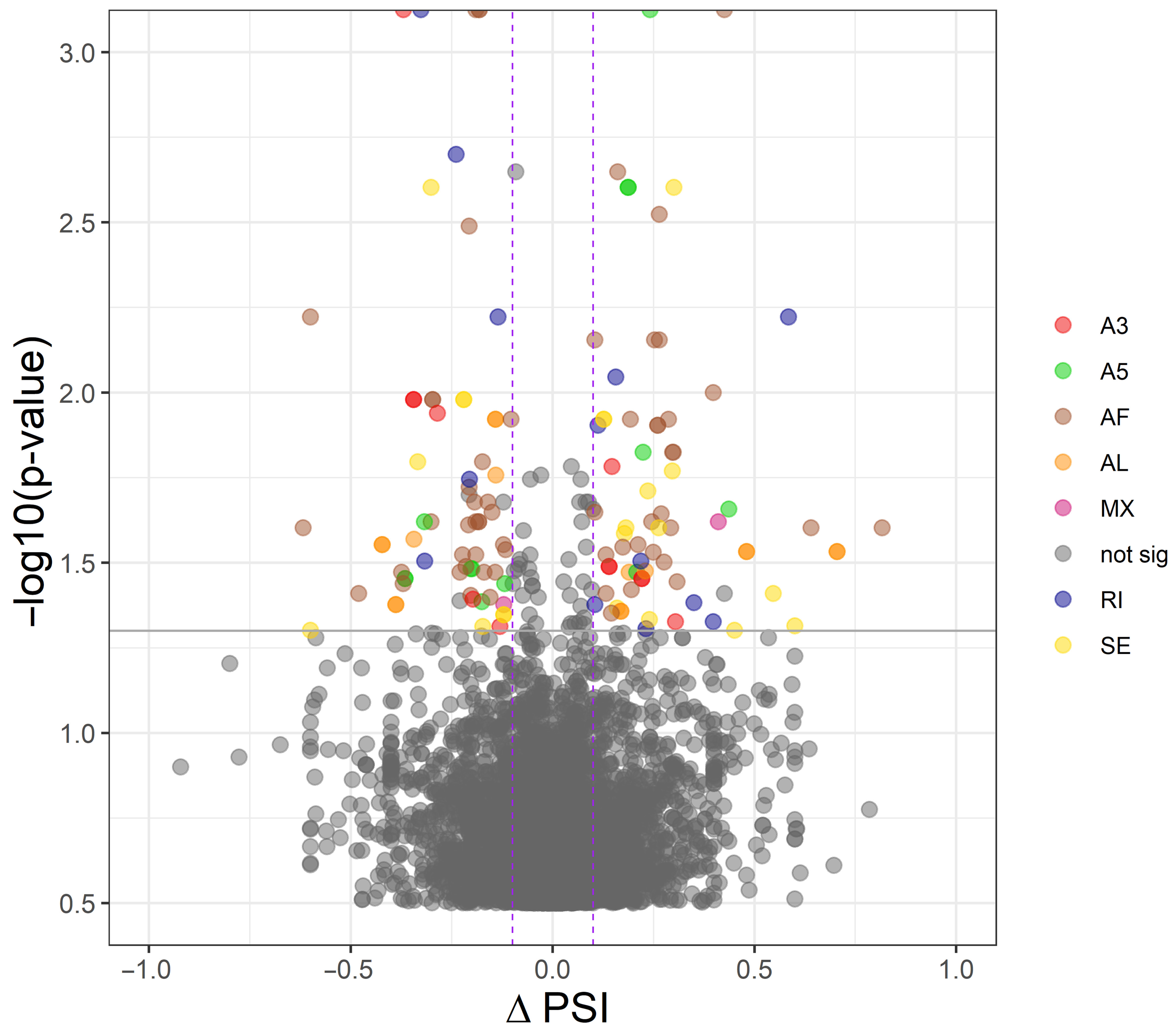
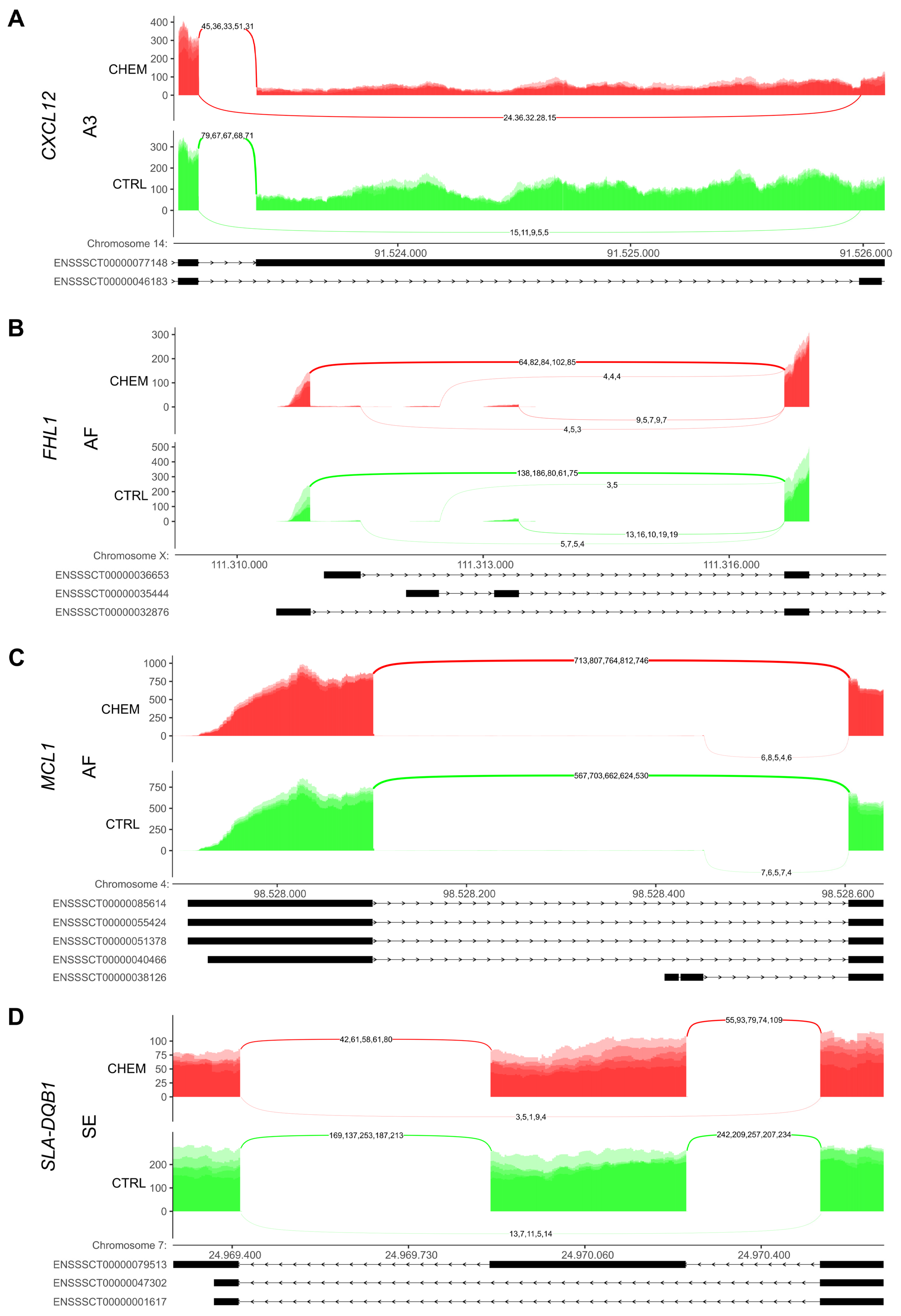
2.3.3. Differential Alternative Splicing Events Analysis
2.3.4. Single Nucleotide Variants Identification and Allele-Specific Expression Variants Analysis
2.3.5. Functional Annotation of Target Genes
2.4. Quantitative Real-Time PCR Validation
| Gene symbol | Primers Sequences | Product Length | Reference |
|---|---|---|---|
| CL.9638.3 | F: GGGGCCCTGTAAGGAAACTC | 141 bp | [The present study] |
| R: TACTTGGCACCAAGCAAGCA | |||
| CL.12742.3 | F: AGCGGGCGCAGATTCAT | 241 bp | [The present study] |
| R: AGCAGAGGGTCATTTCTGGC | |||
| ENSSSCT00000075362 | F: GGGTGTTTCCATGCTCAAGA | 275 bp | [The present study] |
| R: CACAGCCAAGACAGCGAATA | |||
| ENSSSCT00000078829 | F: GTGCTTGGAGGGACATGACA | 186 bp | [The present study] |
| R: TGTCGTTTGAGGGTTCTGGG | |||
| ACTB | F: ACATCAAGGAGAAGCTCTGCTACG | 366 bp | [89] |
| R: GAGGGGCGATGATCTTGATCTTCA | |||
| GAPDH | F: CCTTCATTGACCTCCACTACATGG | 183 bp | [90] |
| R: CCACAACATACGTAGCACCAGCATC |
3. Results
3.1. Overall Statistics of RNA-Seq Data Mapping
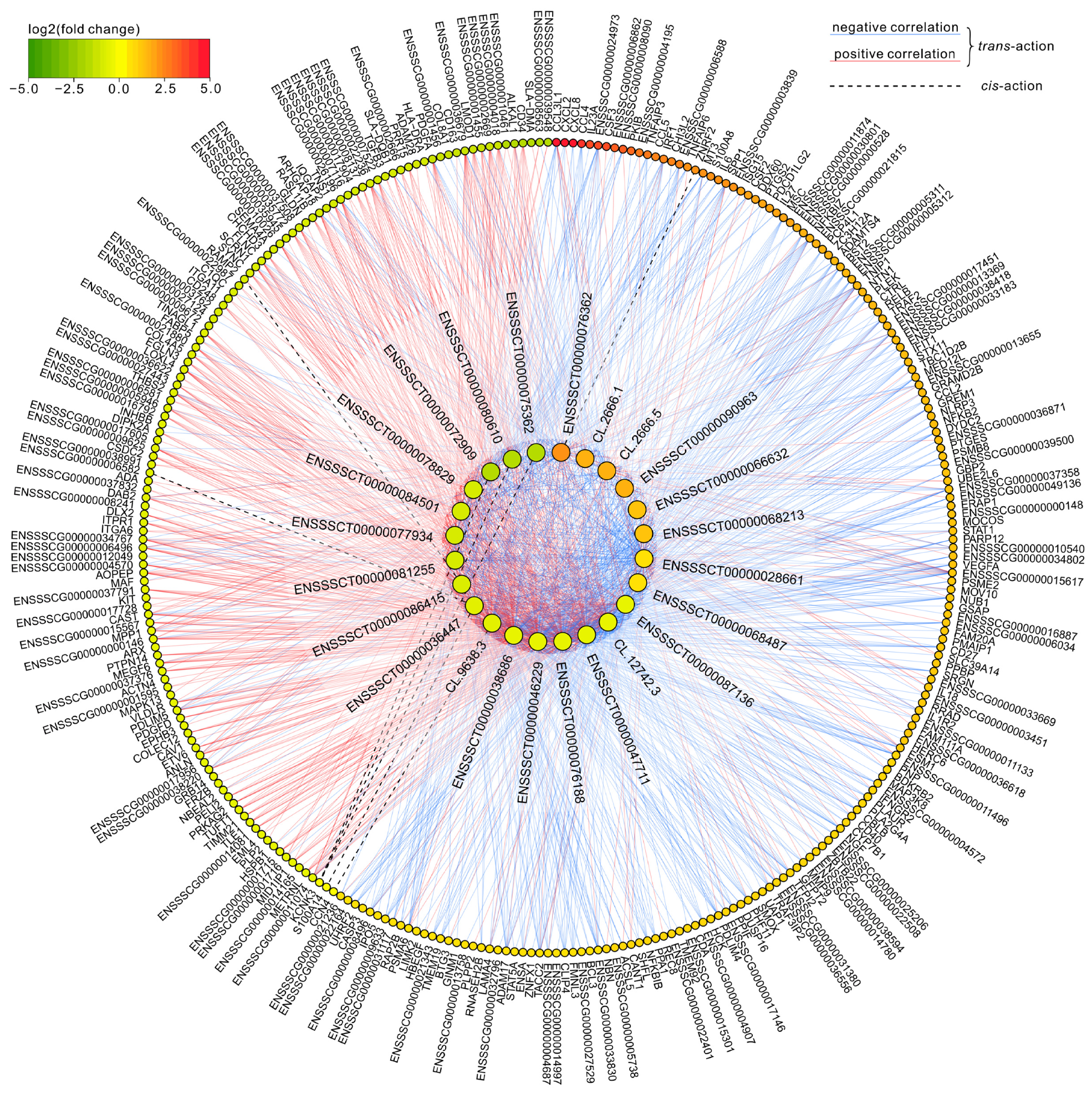
3.2. Long Non-Coding RNA Identification and Cis-/Trans-Acting on Protein-Coding Genes
3.3. Differential Alternative Splicing Events of Differentially Expressed Genes
3.4. Single Nucleotide Variant Calling and Allele-Specific Expression Variants
3.5. Functional Annotation of Target Protein-Coding Genes
3.6. Quantitative Real-Time PCR Validations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACTB | Β-actin |
| Akt | Protein kinase b |
| AMPK | 5′ adenosine monophosphate-activated protein kinase |
| CASP3 | Caspase 3 |
| CCL* | C-C motif chemokine ligand *(2, 4, 5) |
| CCRL2 | C-C chemokine receptor-like 2 |
| ChemR23 | Chemerin receptor 23 |
| CMKLR1 | Chemerin Chemokine-Like Receptor 1 |
| COL6A2 | Collagen type VI α2 chain |
| CTSS | Cathepsin S |
| CXCL* | C-X-C motif chemokine ligand *(2, 8, 12) |
| CXCR4 | C-X-C motif chemokine receptor 4 |
| CYP11A1 | Cytochrome P450 family 11 subfamily A member 1 |
| DUSP4 | Dual specificity phosphatase 4 |
| EIF3F | Eukaryotic translation initiation factor 3 subunit F |
| ERK* | Extracellular signal-regulated kinase *(1/2) |
| FHL1 | Four and a half LIM domains 1 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| GPR1 | G protein-coupled receptor 1 |
| GSTA1 | Glutathione S-transferase α1 |
| HLA-DRA | HLA class II histocompatibility antigen, DRα |
| HPS5 | HPS5 biogenesis of lysosomal organelles complex 2 subunit 2 |
| ICAM1 | Intercellular adhesion molecule 1 |
| IFNγ | Interferon γ |
| IL* | Interleukin *(1A, 6, 8, 17) |
| ITGA2 | Integrin subunit α2 |
| Jak | Janus kinase |
| MAPK | Mitogen-activated protein kinase |
| MCL1 | Mcl1 apoptosis regulator, bcl2 family member |
| MHC* | Major histocompatibility complex *(I, II) |
| MOCS2 | Molybdenum cofactor synthesis 2 |
| NF-κB | Nuclear factor κ-light-chain-enhancer of activated B cells |
| NR5A* | Nuclear receptor subfamily 5 group A *(1) |
| OSBP | Oxysterol binding protein |
| p38 | Mitogen-activated protein kinase 14 |
| P450scc | Cytochrome P450, subfamily XIA (cholesterol side chain cleavage) |
| PA28* | Proteasome activator *(α, β) |
| PBS | Phosphate-buffered saline |
| PGE2 | Prostaglandin E2 |
| PI3K | Phosphoinositide 3-kinases |
| PIK3C2A | Phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2α |
| PSMB* | Proteasome 20S subunit β *(8, 9, 10) |
| PSME* | Proteasome activator subunit *(1, 2) |
| PTGES | Prostaglandin E synthase |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 |
| RARRES2 | Retinoic acid receptor responder 2 |
| RIPK* | Receptor interacting serine/threonine kinase *(1, 3) |
| SAAL1 | Serum amyloid A like 1 |
| SDF-1 | Stromal cell-derived factor 1 |
| SLA-DMA | SLA class II histocompatibility antigen, DMα |
| SLA-DQB1 | SLA class II histocompatibility antigen, DQβ1 |
| SLC25A24 | Solute carrier family 25 member 24 |
| SLC39A14 | Solute carrier family 39 member 14 |
| SORBS3 | Sorbin and SH3 domain containing 3 |
| SPATA2 | Spermatogenesis associated 2 |
| SRSF11 | Serine and arginine rich splicing factor 11 |
| STAT | Signal transducer and activator of transcription |
| TGFB3 | Transforming growth factor β3 |
| TIG2 | Tazarotene-induced gene 2 |
| TNF* | Tumour necrosis factor *(α) |
| TNFAIP3 | TNF α induced protein 3 |
| TRAF3IP2 | TNF receptor associated factor 3 interacting protein 2 |
| VEGFA | Vascular endothelial growth factor A |
References
- Nagpal, S.; Patel, S.; Jacobe, H.; DiSepio, D.; Ghosn, C.; Malhotra, M.; Teng, M.; Duvic, M.; Chandraratna, R.A.S. Tazarotene-induced Gene 2 (TIG2), a Novel Retinoid-Responsive Gene in Skin. J. Investig. Dermatol. 1997, 109, 91–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goralski, K.B.; McCarthy, T.C.; Hanniman, E.A.; Zabel, B.A.; Butcher, E.C.; Parlee, S.D.; Muruganandan, S.; Sinal, C.J. Chemerin, a Novel Adipokine That Regulates Adipogenesis and Adipocyte Metabolism. J. Biol. Chem. 2007, 282, 28175–28188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattern, A.; Zellmann, T.; Beck-Sickinger, A.G. Processing, signaling, and physiological function of chemerin. IUBMB Life 2014, 66, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Wittamer, V.; Franssen, J.-D.D.; Vulcano, M.; Mirjolet, J.-F.F.; Le Poul, E.; Migeotte, I.; Brézillon, S.; Tyldesley, R.; Blanpain, C.; Detheux, M.; et al. Specific Recruitment of Antigen-presenting Cells by Chemerin, a Novel Processed Ligand from Human Inflammatory Fluids. J. Exp. Med. 2003, 198, 977–985. [Google Scholar] [CrossRef]
- Kennedy, A.J.; Davenport, A.P. International Union of Basic and Clinical Pharmacology CIII: Chemerin Receptors CMKLR1 (Chemerin1) and GPR1 (Chemerin2) Nomenclature, Pharmacology, and Function. Pharmacol. Rev. 2018, 70, 174–196. [Google Scholar] [CrossRef] [Green Version]
- De Henau, O.; Degroot, G.-N.N.; Imbault, V.; Robert, V.; De Poorter, C.; Mcheik, S.; Galés, C.; Parmentier, M.; Springael, J.-Y.Y. Signaling Properties of Chemerin Receptors CMKLR1, GPR1 and CCRL2. PLoS ONE 2016, 11, e0164179. [Google Scholar] [CrossRef]
- Zabel, B.A.; Nakae, S.; Zúñiga, L.; Kim, J.-Y.Y.; Ohyama, T.; Alt, C.; Pan, J.; Suto, H.; Soler, D.; Allen, S.J.; et al. Mast cell–expressed orphan receptor CCRL2 binds chemerin and is required for optimal induction of IgE-mediated passive cutaneous anaphylaxis. J. Exp. Med. 2008, 205, 2207–2220. [Google Scholar] [CrossRef]
- Monnier, J.; Lewén, S.; O’Hara, E.; Huang, K.; Tu, H.; Butcher, E.C.; Zabel, B.A. Expression, Regulation, and Function of Atypical Chemerin Receptor CCRL2 on Endothelial Cells. J. Immunol. 2012, 189, 956–967. [Google Scholar] [CrossRef] [Green Version]
- Reverchon, M.; Cornuau, M.; Rame, C.; Guerif, F.; Royere, D.; Dupont, J.; Ram, C.; Guerif, F.; Royre, D.; Dupont, J. Chemerin inhibits IGF-1-induced progesterone and estradiol secretion in human granulosa cells. Hum. Reprod. 2012, 27, 1790–1800. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Kim, J.Y.; Xue, K.; Liu, J.Y.; Leader, A.; Tsang, B.K. Chemerin, a novel regulator of follicular steroidogenesis and its potential involvement in polycystic ovarian syndrome. Endocrinology 2012, 153, 5600–5611. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Leader, A.; Tsang, B.K. Inhibitory roles of prohibitin and chemerin in FSH-induced rat granulosa cell steroidogenesis. Endocrinology 2013, 154, 956–967. [Google Scholar] [CrossRef] [Green Version]
- Garces, M.F.; Sanchez, E.; Acosta, B.J.; Angel, E.; Ruíz, A.I.; Rubio-Romero, J.A.; Diéguez, C.; Nogueiras, R.; Caminos, J.E. Expression and regulation of chemerin during rat pregnancy. Placenta 2012, 33, 373–378. [Google Scholar] [CrossRef]
- Barker, G.; Lim, R.; Rice, G.E.; Lappas, M. Increased chemerin concentrations in fetuses of obese mothers and correlation with maternal insulin sensitivity. J. Matern. Fetal. Neonatal Med. 2012, 25, 2274–2280. [Google Scholar] [CrossRef]
- Smolinska, N.; Kiezun, M.; Dobrzyn, K.; Rytelewska, E.; Kisielewska, K.; Gudelska, M.; Zaobidna, E.; Bogus-Nowakowska, K.; Wyrebek, J.; Bors, K.; et al. Expression of Chemerin and Its Receptors in the Porcine Hypothalamus and Plasma Chemerin Levels during the Oestrous Cycle and Early Pregnancy. Int. J. Mol. Sci. 2019, 20, 3887. [Google Scholar] [CrossRef] [Green Version]
- Kisielewska, K.; Rytelewska, E.; Gudelska, M.; Kiezun, M.; Dobrzyn, K.; Bogus-Nowakowska, K.; Kaminska, B.; Smolinska, N.; Kaminski, T. Relative abundance of chemerin mRNA transcript and protein in pituitaries of pigs during the estrous cycle and early pregnancy and associations with LH and FSH secretion during the estrous cycle. Anim. Reprod. Sci. 2020, 219, 106532. [Google Scholar] [CrossRef]
- Kisielewska, K.; Rytelewska, E.; Gudelska, M.; Kiezun, M.; Dobrzyn, K.; Bogus-Nowakowska, K.; Kaminska, B.; Smolinska, N.; Kaminski, T. Expression of chemerin receptors CMKLR1, GPR1 and CCRL2 in the porcine pituitary during the oestrous cycle and early pregnancy and the effect of chemerin on MAPK/Erk1/2, Akt and AMPK signalling pathways. Theriogenology 2020, 157, 181–198. [Google Scholar] [CrossRef]
- Rytelewska, E.; Kisielewska, K.; Kiezun, M.; Dobrzyn, K.; Gudelska, M.; Rak, A.; Dupont, J.; Kaminska, B.; Kaminski, T.; Smolinska, N. Expression of chemerin and its receptors in the ovaries of prepubertal and mature gilts. Mol. Reprod. Dev. 2020, 87, 739–762. [Google Scholar] [CrossRef]
- Gudelska, M.; Dobrzyn, K.; Kiezun, M.; Rytelewska, E.; Kisielewska, K.; Kaminska, B.; Kaminski, T.; Smolinska, N. The expression of chemerin and its receptors (CMKLR1, GPR1, CCRL2) in the porcine uterus during the oestrous cycle and early pregnancy and in trophoblasts and conceptuses. Animal 2020, 14, 2116–2128. [Google Scholar] [CrossRef]
- Makowczenko, K.G.; Jastrzebski, J.P.; Szeszko, K.; Smolinska, N.; Paukszto, L.; Dobrzyn, K.; Kiezun, M.; Rytelewska, E.; Kaminska, B.; Kaminski, T. Transcription Analysis of the Chemerin Impact on Gene Expression Profile in the Luteal Cells of Gilts. Genes 2020, 11, 651. [Google Scholar] [CrossRef]
- Rytelewska, E.; Kiezun, M.; Kisielewska, K.; Gudelska, M.; Dobrzyn, K.; Kaminska, B.; Kaminski, T.; Smolinska, N. Chemerin as a modulator of ovarian steroidogenesis in pigs: An in vitro study. Theriogenology 2021, 160, 95–101. [Google Scholar] [CrossRef]
- Rytelewska, E.; Kiezun, M.; Zaobidna, E.; Gudelska, M.; Kisielewska, K.; Dobrzyn, K.; Kaminski, T.; Smolinska, N. CHEMERIN as a modulator of angiogenesis and apoptosis processes in the corpus luteum of pigs: An in vitro study. Biol. Reprod. 2021, 105, 1002–1015. [Google Scholar] [CrossRef]
- De Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [Green Version]
- Schwanhäusser, B.; Busse, D.; Li, N.; Dittmar, G.; Schuchhardt, J.; Wolf, J.; Chen, W.; Selbach, M. Global quantification of mammalian gene expression control. Nature 2011, 473, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Li, J.J.; Bickel, P.J.; Biggin, M.D. System wide analyses have underestimated protein abundances and the importance of transcription in mammals. PeerJ 2014, 2014, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Battle, A.; Khan, Z.; Wang, S.H.; Mitrano, A.; Ford, M.J.; Pritchard, J.K.; Gilad, Y. Impact of regulatory variation from RNA to protein. Science 2015, 347, 664–667. [Google Scholar] [CrossRef] [Green Version]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 1259038. [Google Scholar] [CrossRef] [Green Version]
- Kornienko, A.E.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Alessio, E.; Bonadio, R.S.; Buson, L.; Chemello, F.; Cagnin, S. A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 302. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Lin, Y.; Wu, J. Long Non-Coding RNA Expression Profiling of Mouse Testis during Postnatal Development. PLoS ONE 2013, 8, e75750. [Google Scholar] [CrossRef]
- Fraser, L.; Paukszto, Ł.; Mańkowska, A.; Brym, P.; Gilun, P.; Jastrzębski, J.P.; Pareek, C.S.; Kumar, D.; Pierzchała, M. Regulatory potential of long non-coding rnas (LncRNAs) in boar spermatozoa with good and poor freezability. Life 2020, 10, 300. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.H.; Chu, E.T.J.; Spektor, R.; Soloway, P.D. Long non-coding RNA regulation of reproduction and development. Mol. Reprod. Dev. 2015, 82, 932–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.S.; Li, T.P.; Ton, H.; Mao, X.D.; Chen, Y.J. Advances of Long Noncoding RNAs-mediated Regulation in Reproduction. Chin. Med. J. 2018, 131, 226. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Shimada, M.; Yanaka, K.; Mito, M.; Arai, T.; Takahashi, E.; Fujita, Y.; Fujimori, T.; Standaert, L.; Marine, J.C.; et al. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014, 141, 4618–4627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Early, P.; Rogers, J.; Davis, M.; Calame, K.; Bond, M.; Wall, R.; Hood, L. Two mRNAs can be produced from a single immunoglobulin μ gene by alternative RNA processing pathways. Cell 1980, 20, 313–319. [Google Scholar] [CrossRef]
- Saldi, T.; Cortazar, M.A.; Sheridan, R.M.; Bentley, D.L. Coupling of RNA Polymerase II Transcription Elongation with Pre-mRNA Splicing. J. Mol. Biol. 2016, 428, 2623–2635. [Google Scholar] [CrossRef] [Green Version]
- Noble, J.D.; Balmant, K.M.; Dervinis, C.; de los Campos, G.; Resende, M.F.R.; Kirst, M.; Barbazuk, W.B. The Genetic Regulation of Alternative Splicing in Populus deltoides. Front. Plant Sci. 2020, 11, 590. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-Wide Analysis of Alternative Splicing in Zea mays: Landscape and Genetic Regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef] [Green Version]
- Schweingruber, C.; Rufener, S.C.; Zünd, D.; Yamashita, A.; Mühlemann, O. Nonsense-mediated mRNA decay—Mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim. Biophys. Acta 2013, 1829, 612–623. [Google Scholar] [CrossRef]
- Staudt, A.C.; Wenkel, S. Regulation of protein function by ‘microProteins’. EMBO Rep. 2011, 12, 35. [Google Scholar] [CrossRef]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef] [Green Version]
- Madhra, M.; Gay, E.; Fraser, H.M.; Duncan, W.C. Alternative splicing of the human luteal LH receptor during luteolysis and maternal recognition of pregnancy. Mol. Hum. Reprod. 2004, 10, 599–603. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, L.A.; Bacci, M.L.; Seren, E.; Tamanini, C.; Forni, M. Characterization and differential expression of vascular endothelial growth factor isoforms and receptors in swine corpus luteum throughout estrous cycle. Mol. Reprod. Dev. 2007, 74, 163–171. [Google Scholar] [CrossRef]
- Gaur, U.; Li, K.; Mei, S.; Liu, G. Research progress in allele-specific expression and its regulatory mechanisms. J. Appl. Genet. 2013, 54, 271–283. [Google Scholar] [CrossRef]
- Khansefid, M.; Pryce, J.E.; Bolormaa, S.; Chen, Y.; Millen, C.A.; Chamberlain, A.J.; Vander Jagt, C.J.; Goddard, M.E. Comparing allele specific expression and local expression quantitative trait loci and the influence of gene expression on complex trait variation in cattle. BMC Genom. 2018, 19, 793. [Google Scholar] [CrossRef] [Green Version]
- Bozaoglu, K.; Curran, J.E.; Stocker, C.J.; Zaibi, M.S.; Segal, D.; Konstantopoulos, N.; Morrison, S.; Carless, M.; Dyer, T.D.; Cole, S.A.; et al. Chemerin, a novel adipokine in the regulation of angiogenesis. J. Clin. Endocrinol. Metab. 2010, 95, 2476–2485. [Google Scholar] [CrossRef] [Green Version]
- Akins, E.L.; Morrissette, M.C. Gross ovarian changes during estrous cycle of swine. Am. J. Vet. Res. 1968, 29, 1953–1957. [Google Scholar]
- Kaminski, T.; Siawrys, G.; Okrasa, S.; Przala, J. Action of the opioid agonist FK 33-824 on porcine small and large luteal cells from the mid-luteal phase: Effect on progesterone, cAMP, cGMP and inositol phosphate release. Anim. Reprod. Sci. 1999, 56, 245–257. [Google Scholar] [CrossRef]
- Du, X.-Y.; Leung, L.L.K. Proteolytic regulatory mechanism of chemerin bioactivity. Acta Biochim. Biophys. Sin. 2009, 41, 973–979. [Google Scholar] [CrossRef] [Green Version]
- Luangsay, S.; Wittamer, V.; Bondue, B.; Henau, O.D.; Rouger, L.; Brait, M.; Franssen, J.-D.; de Nadai, P.; Huaux, F.; Parmentier, M. Mouse ChemR23 Is Expressed in Dendritic Cell Subsets and Macrophages, and Mediates an Anti-Inflammatory Activity of Chemerin in a Lung Disease Model. J. Immunol. 2009, 183, 6489–6499. [Google Scholar] [CrossRef]
- Shen, Y.; Mao, H.; Huang, M.; Chen, L.; Chen, J.; Cai, Z.; Wang, Y.; Xu, N. Long Noncoding RNA and mRNA Expression Profiles in the Thyroid Gland of Two Phenotypically Extreme Pig Breeds Using Ribo-Zero RNA Sequencing. Genes 2016, 7, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paukszto, L.; Mikolajczyk, A.; Szeszko, K.; Smolinska, N.; Jastrzebski, J.P.; Kaminski, T. Transcription analysis of the response of the porcine adrenal cortex to a single subclinical dose of lipopolysaccharide from Salmonella Enteritidis. Int. J. Biol. Macromol. 2019, 141, 1228–1245. [Google Scholar] [CrossRef]
- Paukszto, L.; Mikolajczyk, A.; Jastrzebski, J.P.; Majewska, M.; Dobrzyn, K.; Kiezun, M.; Smolinska, N.; Kaminski, T. Transcriptome, Spliceosome and Editome Expression Patterns of the Porcine Endometrium in Response to a Single Subclinical Dose of Salmonella Enteritidis Lipopolysaccharide. Int. J. Mol. Sci. 2020, 21, 4217. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 10 September 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Jakobi, T.; Uvarovskii, A.; Dieterich, C. Detect Module—Circtools Documentation. Available online: https://docs.circ.tools/en/latest/Detect.html (accessed on 15 September 2020).
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, G. prepDE.py. Available online: https://github.com/gpertea/stringtie/blob/master/prepDE.py (accessed on 17 September 2021).
- Frazee, A.C.; Pertea, G.; Jaffe, A.E.; Langmead, B.; Salzberg, S.L.; Leek, J.T. Flexible analysis of transcriptome assemblies with Ballgown. bioRxiv 2014, 2014, 003665. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [Green Version]
- Wucher, V.; Legeai, F.; Hédan, B.; Rizk, G.; Lagoutte, L.; Leeb, T.; Jagannathan, V.; Cadieu, E.; David, A.; Lohi, H.; et al. FEELnc: A tool for long non-coding RNA annotation and its application to the dog transcriptome. Nucleic Acids Res. 2017, 45, e57. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Zhang, J.; Zhou, Z. PLEK: A tool for predicting long non-coding RNAs and messenger RNAs based on an improved k-mer scheme. BMC Bioinformatics 2014, 15, 311. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated profile HMM searches. PLoS Comput. Biol. 2011, 7, e1002195. [Google Scholar] [CrossRef] [Green Version]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef]
- Nawrocki, E.P.; Eddy, S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 2013, 29, 2933–2935. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 10 October 2020).
- Ramos, M.; Schiffer, L.; Re, A.; Azhar, R.; Basunia, A.; Rodriguez, C.; Chan, T.; Chapman, P.; Davis, S.R.; Gomez-Cabrero, D.; et al. Software for the integration of multiomics experiments in Bioconductor. Cancer Res. 2017, 77, e39–e42. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ma, W.; Zeng, P.; Wang, J.; Geng, B.; Yang, J.; Cui, Q. LncTar: A tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 2015, 16, 806–812. [Google Scholar] [CrossRef]
- Lu, Q.; Ren, S.; Lu, M.; Zhang, Y.; Zhu, D.; Zhang, X.; Li, T. Computational prediction of associations between long non-coding RNAs and proteins. BMC Genom. 2013, 14, 651. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Trincado, J.L.; Entizne, J.C.; Hysenaj, G.; Singh, B.; Skalic, M.; Elliott, D.J.; Eyras, E. SUPPA2: Fast, accurate, and uncertainty-aware differential splicing analysis across multiple conditions. Genome Biol. 2018, 19, 40. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Martín, D.; Palumbo, E.; Guigó, R.; Breschi, A. ggsashimi: Sashimi plot revised for browser- and annotation-independent splicing visualization. PLOS Comput. Biol. 2018, 14, e1006360. [Google Scholar] [CrossRef]
- Broad Institute. Picard Tools. Available online: https://broadinstitute.github.io/picard/ (accessed on 13 September 2021).
- Wang, J.; Pan, Y.; Shen, S.; Lin, L.; Xing, Y. RMATS-DVR: RMATS discovery of differential variants in RNA. Bioinformatics 2017, 33, 2216–2217. [Google Scholar] [CrossRef]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Carbon, S.; Dietze, H.; Lewis, S.E.; Mungall, C.J.; Munoz-Torres, M.C.; Basu, S.; Chisholm, R.L.; Dodson, R.J.; Fey, P.; Thomas, P.D.; et al. Expansion of the gene ontology knowledgebase and resources: The gene ontology consortium. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef] [Green Version]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Spagnuolo-Weaver, M.; Fuerst, R.; Campbell, S.; Meehan, B.; McNeilly, F.; Adair, B.; Allan, G. A fluorimeter-based RT-PCR method for the detection and quantitation of porcine cytokines. J. Immunol. Methods 1999, 230, 19–27. [Google Scholar] [CrossRef]
- Nitkiewicz, A.; Smolinska, N.; Przala, J.; Kaminski, T. Expression of orexin receptors 1 (OX1R) and 2 (OX2R) in the porcine ovary during the oestrous cycle. Regul. Pept. 2010, 165, 186–190. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef] [Green Version]
- Schioppa, T.; Sozio, F.; Barbazza, I.; Scutera, S.; Bosisio, D.; Sozzani, S.; Del Prete, A. Molecular Basis for CCRL2 Regulation of Leukocyte Migration. Front. Cell Dev. Biol. 2020, 8, 1570. [Google Scholar] [CrossRef]
- Bondue, B.; Wittamer, V.; Parmentier, M. Chemerin and its receptors in leukocyte trafficking, inflammation and metabolism. Cytokine Growth Factor Rev. 2011, 22, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Gonzalvo-Feo, S.; Del Prete, A.; Pruenster, M.; Salvi, V.; Wang, L.; Sironi, M.; Bierschenk, S.; Sperandio, M.; Vecchi, A.; Sozzani, S. Endothelial cell-derived chemerin promotes dendritic cell transmigration. J. Immunol. 2014, 192, 2366–2373. [Google Scholar] [CrossRef] [Green Version]
- Pate, J.; Landis Keyes, P. Immune cells in the corpus luteum: Friends or foes? Reproduction 2001, 122, 665–676. [Google Scholar] [CrossRef] [Green Version]
- Standaert, F.E.; Zamora, C.S.; Chew, B.P. Quantitative and Qualitative Changes in Blood Leukocytes in the Porcine Ovary. Am. J. Reprod. Immunol. 1991, 25, 163–168. [Google Scholar] [CrossRef]
- Ziecik, A.J.; Przygrodzka, E.; Jalali, B.M.; Kaczmarek, M.M. Regulation of the porcine corpus luteum during pregnancy. Reproduction 2018, 156, R57–R67. [Google Scholar] [CrossRef]
- Baggiolini, M. Chemokines and leukocyte traffic. Nature 1998, 392, 565–568. [Google Scholar] [CrossRef]
- Moser, B.; Loetscher, P. Lymphocyte traffic control by chemokines. Nat. Immunol. 2001, 2, 123–128. [Google Scholar] [CrossRef]
- Townson, D.H.; Liptak, A.R. Chemokines in the corpus luteum: Implications of leukocyte chemotaxis. Reprod. Biol. Endocrinol. 2003, 1, 94. [Google Scholar] [CrossRef] [Green Version]
- Witek, K.J.; Ziecik, A.J.; Małysz-Cymborska, I.; Andronowska, A. The presence of CC chemokines and their aberrant role in the porcine corpus luteum. Reprod. Domest. Anim. 2020, 55, 632–646. [Google Scholar] [CrossRef]
- Ito, Y.; Hoare, M.; Narita, M. Spatial and Temporal Control of Senescence. Trends Cell Biol. 2017, 27, 820–832. [Google Scholar] [CrossRef] [Green Version]
- Nacarelli, T.; Lau, L.; Fukumoto, T.; Zundell, J.; Fatkhutdinov, N.; Wu, S.; Aird, K.M.; Iwasaki, O.; Kossenkov, A.V.; Schultz, D.; et al. NAD+ metabolism governs the proinflammatory senescence-associated secretome. Nat. Cell Biol. 2019, 21, 397. [Google Scholar] [CrossRef]
- Sagiv, A.; Krizhanovsky, V. Immunosurveillance of senescent cells: The bright side of the senescence program. Biogerontology 2013, 14, 617–628. [Google Scholar] [CrossRef]
- Gorgoulis, V.G.; Pratsinis, H.; Zacharatos, P.; Demoliou, C.; Sigala, F.; Asimacopoulos, P.J.; Papavassiliou, A.G.; Kletsas, D. p53-Dependent ICAM-1 overexpression in senescent human cells identified in atherosclerotic lesions. Lab. Investig. 2005, 85, 502–511. [Google Scholar] [CrossRef]
- Chien, Y.; Scuoppo, C.; Wang, X.; Fang, X.; Balgley, B.; Bolden, J.E.; Premsrirut, P.; Luo, W.; Chicas, A.; Lee, C.S.; et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011, 25, 2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olson, K.K.; Anderson, L.E.; Wiltbank, M.C.; Townson, D.H. Actions of Prostaglandin F 2 and Prolactin on Intercellular Adhesion Molecule-1 Expression and Monocyte/Macrophage Accumulation in the Rat Corpus Luteum 1. Biol. Reprod. 2001, 64, 890–897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.; Park, J.Y.; Wilson, K.; Rosewell, K.L.; Brännström, M.; Akin, J.W.; Curry, T.E.; Jo, M. The expression of CXCR4 is induced by the luteinizing hormone surge and mediated by progesterone receptors in human preovulatory granulosa cells †. Biol. Reprod. 2017, 96, 1256–1266. [Google Scholar] [CrossRef]
- Zhang, R.-N.; Pang, B.; Xu, S.-R.; Wan, P.-C.; Guo, S.-C.; Ji, H.-Z.; Jia, G.-X.; Hu, L.-Y.; Zhao, X.-Q.; Yang, Q.-E. The CXCL12-CXCR4 signaling promotes oocyte maturation by regulating cumulus expansion in sheep. Theriogenology 2018, 107, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Mcintosh, S.Z.; Quinn, K.E.; Ashley, R.L. CXCL12 May Drive Inflammatory Potential in the Ovine Corpus Luteum During Implantation. Reprod. Sci. 2021, 1, 122–132. [Google Scholar] [CrossRef]
- Kryczek, I.; Frydman, N.; Gaudin, F.; Krzysiek, R.; Fanchin, R.; Emilie, D.; Chouaib, S.; Zou, W.; Machelon, V. The chemokine SDF-1/CXCL12 contributes to T lymphocyte recruitment in human pre-ovulatory follicles and coordinates with lymphocytes to increase granulosa cell survival and embryo quality. Am. J. Reprod. Immunol. 2005, 54, 270–283. [Google Scholar] [CrossRef]
- Nishigaki, A.; Okada, H.; Okamoto, R.; Sugiyama, S.; Miyazaki, K.; Yasuda, K.; Kanzaki, H. Concentrations of stromal cell-derived factor-1 and vascular endothelial growth factor in relation to the diameter of human follicles. Fertil. Steril. 2011, 95, 742–746. [Google Scholar] [CrossRef]
- Nishigaki, A.; Okada, H.; Okamoto, R.; Shimoi, K.; Miyashiro, H.; Yasuda, K.; Kanzaki, H. The concentration of human follicular fluid stromal cell-derived factor-1 is correlated with luteinization in follicles. Gynecol. Endocrinol. 2013, 29, 230–234. [Google Scholar] [CrossRef]
- De Poorter, C.; Baertsoen, K.; Lannoy, V.; Parmentier, M.; Springael, J.-Y. Consequences of ChemR23 Heteromerization with the Chemokine Receptors CXCR4 and CCR7. PLoS ONE 2013, 8, e58075. [Google Scholar] [CrossRef]
- Chardon, P.; Kenard, C.; Vaiman, M.; Renard, C.; Chordon, P. The major histocompatibility complex in swine. Immunol. Rev. 1999, 167, 9–92. [Google Scholar] [CrossRef]
- Bukovský, A.; Caudle, M.R.; Keenan, J.A.; Wimalasena, J.; Upadhyaya, N.B.; Van Meter, S.E. Is Corpus Luteum Regression an Immune-Mediated Event? Localization of Immune System Components and Luteinizing Hormone Receptor in Human Corpora Lutea. Biol. Reprod. 1995, 53, 1373–1384. [Google Scholar] [CrossRef] [Green Version]
- Fairchild Benyo, D.; Haibel, G.K.; Laufman, H.B.; Pate, J.L. Expression of Major Histocompatibility Complex Antigens on the Bovine Corpus Luteum during the Estrous Cycle, Luteolysis, and Early Pregnancy. Biol. Reprod. 1991, 45, 229–234. [Google Scholar] [CrossRef] [Green Version]
- Kenny, N.; Herman, J.R.; Barisas, B.G.; Roess, D.A. Flow Cytometric Analysis of Class I and II MHC Antigens on Ovine Luteal Cell Types. In Signaling Mechanisms and Gene Expression in the Ovary; Springer: New York, NY, USA, 1991; pp. 467–472. [Google Scholar]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, Function, and Antigen Presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Kirk, C.J.; Groettrup, M. The immunoproteasome in antigen processing and other immunological functions. Curr. Opin. Immunol. 2013, 25, 74–80. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Wiltbank, M.C. Luteinization of Bovine Granulosa Cells for 8 Days Increased Major Histocompatibility Complex (MHC) II Molecules, T-Cell Co-stimulatory Ligands, and Prostaglandin F2 Alpha-induced Activation of T Lymphocytes During Cocultures. Biol. Reprod. 2010, 83, 126. [Google Scholar] [CrossRef]
- Cash, J.L.; Hart, R.; Russ, A.; Dixon, J.P.C.; Colledge, W.H.; Doran, J.; Hendrick, A.G.; Carlton, M.B.L.; Greaves, D.R. Synthetic chemerin-derived peptides suppress inflammation through ChemR23. J. Exp. Med. 2008, 205, 767. [Google Scholar] [CrossRef] [Green Version]
- Fimia, G.M.; De Cesare, D.; Sassone-Corsi, P. A Family of LIM-Only Transcriptional Coactivators: Tissue-Specific Expression and Selective Activation of CREB and CREM. Mol. Cell. Biol. 2000, 20, 8613–8622. [Google Scholar] [CrossRef] [Green Version]
- Matulis, C.K.; Mayo, K.E. The LIM Domain Protein FHL2 Interacts with the NR5A Family of Nuclear Receptors and CREB to Activate the Inhibin-α Subunit Gene in Ovarian Granulosa Cells. Mol. Endocrinol. 2012, 26, 1278. [Google Scholar] [CrossRef] [Green Version]
- Fraser, H.M.; Dickson, S.E.; Lunn, S.F.; Wulff, C.; Morris, K.D.; Carroll, V.A.; Bicknell, R. Suppression of luteal angiogenesis in the primate after neutralization of vascular endothelial growth factor. Endocrinology 2000, 141, 995–1000. [Google Scholar] [CrossRef]
- Chouhan, V.; Panda, R.; Yadav, V.; Babitha, V.; Khan, F.; Das, G.; Gupta, M.; Dangi, S.; Singh, G.; Bag, S.; et al. Expression and localization of vascular endothelial growth factor and its receptors in the corpus luteum during oestrous cycle in water buffaloes (Bubalus bubalis). Reprod. Domest. Anim. 2013, 48, 810–818. [Google Scholar] [CrossRef]
- Gram, A.; Hoffmann, B.; Boos, A.; Kowalewski, M.P. Expression and localization of vascular endothelial growth factor A (VEGFA) and its two receptors (VEGFR1/FLT1 and VEGFR2/FLK1/KDR) in the canine corpus luteum and utero-placental compartments during pregnancy and at normal and induced parturition. Gen. Comp. Endocrinol. 2015, 223, 54–65. [Google Scholar] [CrossRef]
- Chen, S.-U.; Chen, R.-J.; Shieh, J.-Y.; Chou, C.-H.; Lin, C.-W.; Lu, H.-F.; Yang, Y.-S. Human Chorionic Gonadotropin Up-Regulates Expression of Myeloid Cell Leukemia-1 Protein in Human Granulosa-Lutein Cells: Implication of Corpus Luteum Rescue and Ovarian Hyperstimulation Syndrome. J. Clin. Endocrinol. Metab. 2010, 95, 3982–3992. [Google Scholar] [CrossRef] [Green Version]
- Patterson, K.I.; Brummer, T.; O’Brien, P.M.; Daly, R.J. Dual-specificity phosphatases: Critical regulators with diverse cellular targets. Biochem. J. 2009, 418, 475–489. [Google Scholar] [CrossRef] [Green Version]
- Barajas-Espinosa, A.; Basye, A.; Angelos, M.G.; Chen, C.-A. Modulation of p38 kinase by DUSP4 is important in regulating cardiovascular function under oxidative stress. Free Radic. Biol. Med. 2015, 89, 170. [Google Scholar] [CrossRef] [Green Version]
- Al-Mutairi, M.; Al-Harthi, S.; Cadalbert, L.; Plevin, R. Over-expression of mitogen-activated protein kinase phosphatase-2 enhances adhesion molecule expression and protects against apoptosis in human endothelial cells. Br. J. Pharmacol. 2010, 161, 782. [Google Scholar] [CrossRef] [Green Version]
- Lawan, A.; Al-Harthi, S.; Cadalbert, L.; McCluskey, A.G.; Shweash, M.; Grassia, G.; Grant, A.; Boyd, M.; Currie, S.; Plevin, R. Deletion of the Dual Specific Phosphatase-4 (DUSP-4) Gene Reveals an Essential Non-redundant Role for MAP Kinase Phosphatase-2 (MKP-2) in Proliferation and Cell Survival. J. Biol. Chem. 2011, 286, 12933–12943. [Google Scholar] [CrossRef] [Green Version]
- Hojo, T.; Piotrowska-Tomala, K.K.; Jonczyk, A.W.; Lukasik, K.; Jankowska, K.; Okuda, K.; Witek, K.J.; Skarzynski, D.J. Receptor interacting protein kinases-dependent necroptosis as a new, potent mechanism for elimination of the endothelial cells during luteolysis in cow. Theriogenology 2019, 128, 193–200. [Google Scholar] [CrossRef]
- Hojo, T.; Siemieniuch, M.J.; Lukasik, K.; Piotrowska-Tomala, K.K.; Jonczyk, A.W.; Okuda, K.; Skarzynski, D.J. Programmed necrosis—A new mechanism of steroidogenic luteal cell death and elimination during luteolysis in cows. Sci. Rep. 2016, 6, 38211. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Treatment | CTRL | CHEM | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | 1_LC | 2_LC | 3_LC | 4_LC | 5_LC | 1_LC | 2_LC | 3_LC | 4_LC | 5_LC |
| Mapped reads | 97.380 | 111.081 | 106.721 | 102.366 | 95.792 | 97.190 | 102.650 | 99.617 | 108.891 | 100.253 |
| Uniquely mapped reads | 94.544 | 106.781 | 103.010 | 98.833 | 92.527 | 93.573 | 99.472 | 95.824 | 104.621 | 96.646 |
| % of uniquely mapped reads | 96.86% | 95.89% | 96.30% | 96.32% | 96.37% | 96.06% | 96.67% | 95.96% | 95.84% | 96.17% |
| Multi-mapped reads | 2.482 | 3.968 | 3.314 | 3.136 | 2.973 | 3.312 | 2.736 | 3.489 | 3.934 | 3.274 |
| % of bases mapped to CDS | 60.70% | 58.88% | 59.83% | 59.31% | 60.05% | 59.22% | 59.38% | 58.87% | 59.15% | 59.83% |
| % of bases mapped to UTR | 22.89% | 23.44% | 23.09% | 23.50% | 23.16% | 23.29% | 23.91% | 23.79% | 23.13% | 22.97% |
| % of bases mapped to introns | 3.31% | 3.21% | 3.35% | 3.43% | 3.21% | 3.49% | 3.32% | 3.25% | 3.47% | 3.43% |
| % of bases mapped to intergenic | 13.10% | 14.48% | 13.73% | 13.76% | 13.59% | 14.00% | 13.38% | 14.08% | 14.24% | 13.76% |
| Transcript ID | Reference Gene ID | log2(FC) | q-Value | Regulation | Chr | Str | Start–End |
|---|---|---|---|---|---|---|---|
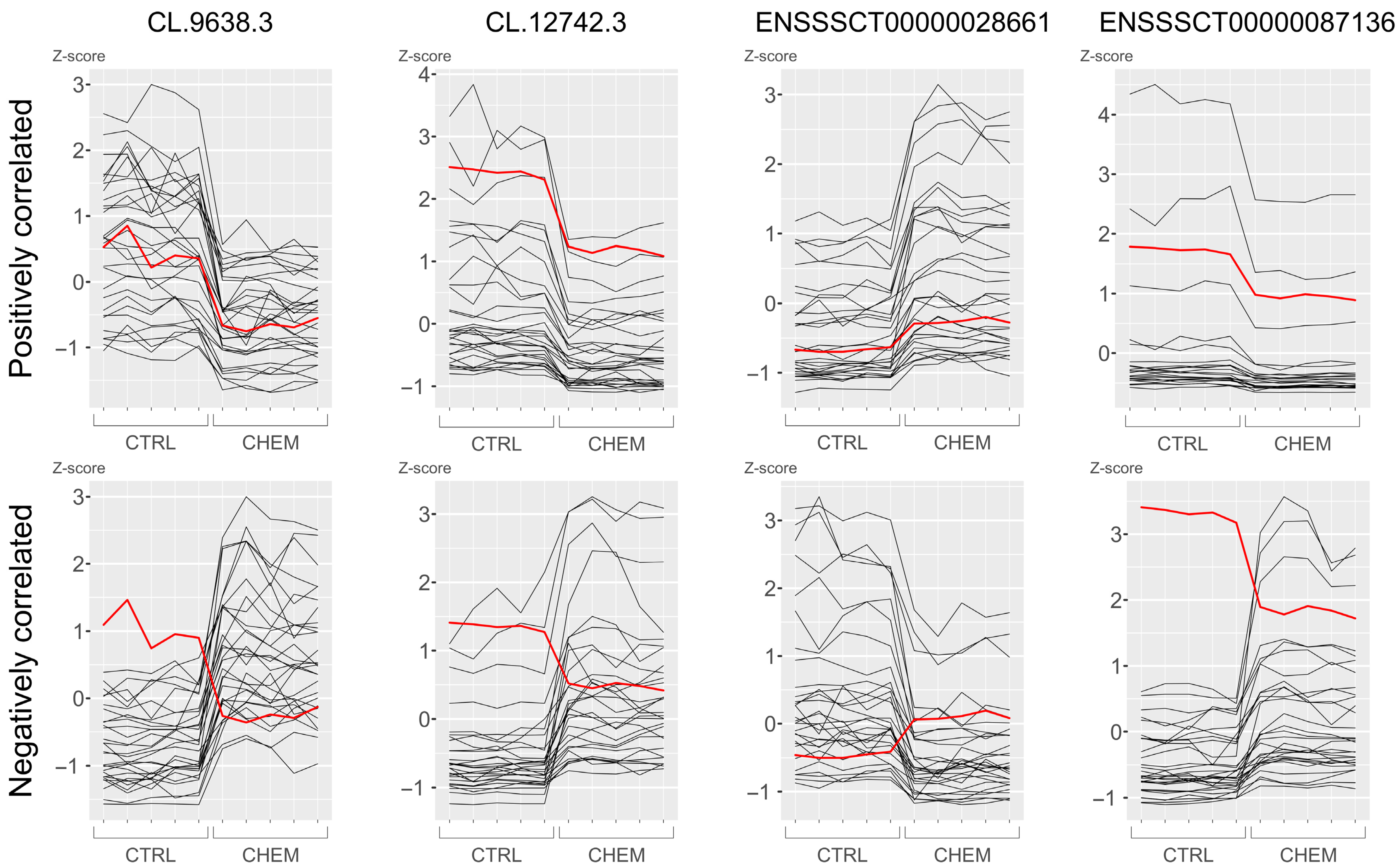
| ENSSSCT00000076362 | ENSSSCG00000047565 | 2.17 | 1.39 × 10−3 | up | 18 | − | 42,660,099–42,669,101 |
| CL.2666.1 | N/A | 1.49 | 2.03 × 10−4 | up | 12 | + | 40,817,836–40,834,552 |
| CL.2666.5 | N/A | 1.49 | 2.03 × 10−4 | up | 12 | + | 40,817,901–40,850,478 |
| ENSSSCT00000090963 | ENSSSCG00000050550 | 1.48 | 2.85 × 10−6 | up | 11 | + | 19,098,312–19,102,389 |
| ENSSSCT00000066632 | ENSSSCG00000042788 | 1.11 | 1.92 × 10−2 | up | 5 | − | 5,752,685–5,756,310 |
| ENSSSCT00000068213 | ENSSSCG00000042788 | 1.11 | 1.92 × 10−2 | up | 5 | − | 5,752,686–5,770,732 |
| ENSSSCT00000028661 | ENSSSCG00000028322 | 0.67 | 1.08 × 10−4 | up | 9 | + | 64,031,854–64,037,769 |
| ENSSSCT00000068487 | ENSSSCG00000050423 | 0.54 | 1.69 × 10−4 | up | 13 | + | 21,429,679–21,433,475 |
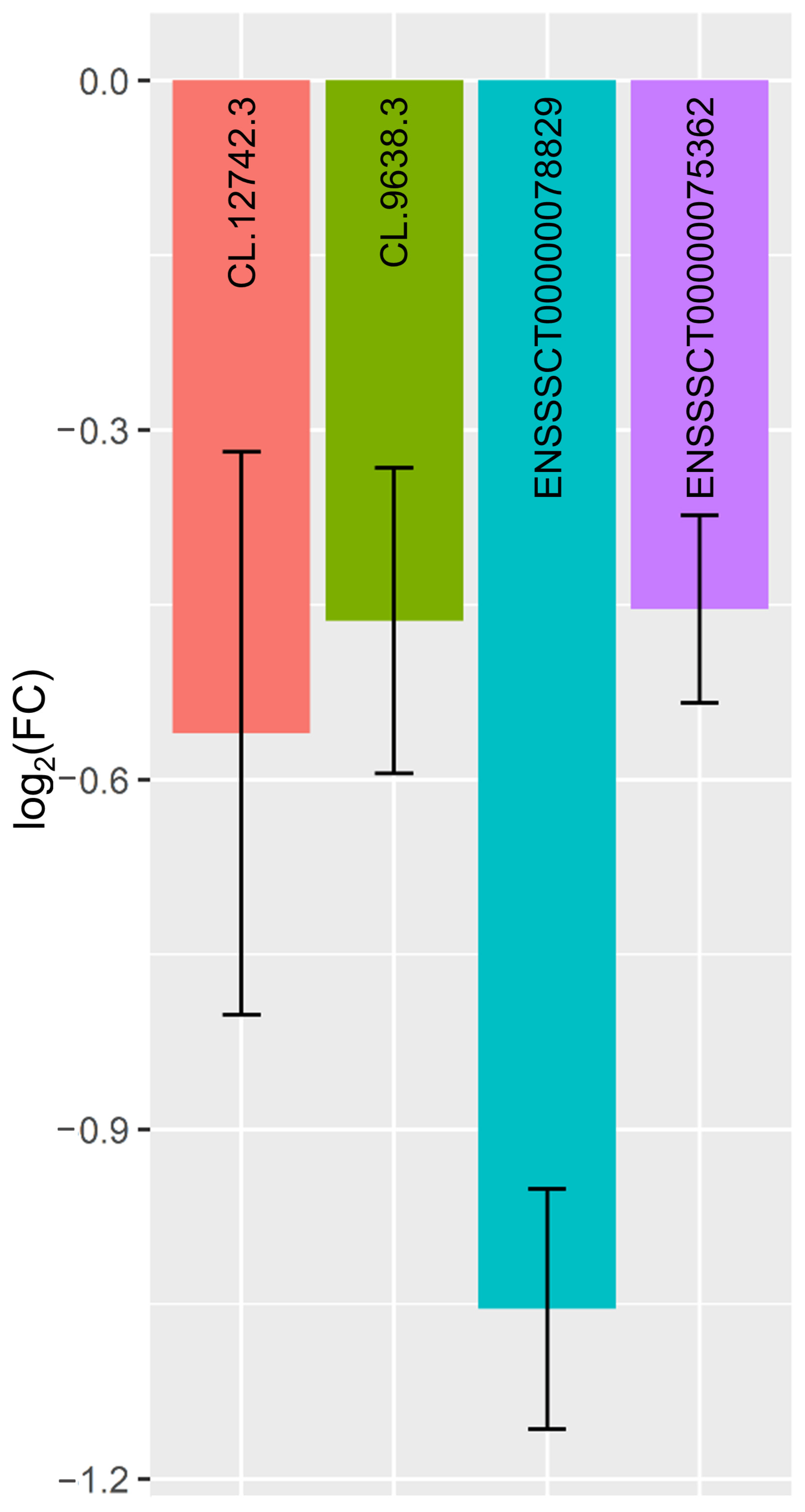
| CL.12742.3 | ENSSSCG00000036505 | −0.58 | 2.29 × 10−5 | down | 6 | + | 144,782,442–144,918,548 |
| ENSSSCT00000038686 | ENSSSCG00000036505 | −0.58 | 2.29 × 10−5 | down | 6 | + | 144,782,503–144,918,549 |
| ENSSSCT00000046229 | ENSSSCG00000036505 | −0.58 | 2.29 × 10−5 | down | 6 | + | 144,782,552–144,918,549 |
| ENSSSCT00000047711 | ENSSSCG00000036505 | −0.58 | 2.29 × 10−5 | down | 6 | + | 144,782,576–144,918,548 |
| ENSSSCT00000076188 | ENSSSCG00000036505 | −0.58 | 2.29 × 10−5 | down | 6 | + | 144,823,167–144,918,548 |
| ENSSSCT00000087136 | ENSSSCG00000036505 | −0.58 | 2.29 × 10−5 | down | 6 | + | 144,781,500–144,912,714 |
| CL.9638.3 | N/A | −0.71 | 8.77 × 10−4 | down | 4 | − | 8,075,901–8,081,556 |
| ENSSSCT00000036447 | ENSSSCG00000006581 | −0.71 | 7.46 × 10−3 | down | 4 | + | 96,035,969–96,037,483 |
| ENSSSCT00000086415 | ENSSSCG00000048436 | −0.85 | 4.04 × 10−2 | down | 12 | − | 20,101,333–20,108,070 |
| ENSSSCT00000077934 | ENSSSCG00000050649 | −0.99 | 2.82 × 10−4 | down | 2 | − | 1,381,838–1,383,221 |
| ENSSSCT00000078829 | ENSSSCG00000050649 | −0.99 | 2.82 × 10−4 | down | 2 | − | 1,381,841–1,384,446 |
| ENSSSCT00000081255 | ENSSSCG00000050649 | −0.99 | 2.82 × 10−4 | down | 2 | − | 1,381,839–1,383,160 |
| ENSSSCT00000084501 | ENSSSCG00000050649 | −0.99 | 2.82 × 10−4 | down | 2 | − | 1,381,841–1,384,446 |
| ENSSSCT00000072909 | ENSSSCG00000048033 | −1.82 | 7.70 × 10−4 | down | 3 | − | 112,308,405–112,318,405 |
| ENSSSCT00000075362 | ENSSSCG00000048033 | −1.82 | 7.70 × 10−4 | down | 3 | − | 112,308,405–112,318,340 |
| ENSSSCT00000080610 | ENSSSCG00000048033 | −1.82 | 7.70 × 10−4 | down | 3 | − | 112,308,405–112,318,450 |
| lncRNA ID | Partner mRNA ID | Partner Gene Name | Dir | Type | Distance | Location |
|---|---|---|---|---|---|---|
| CL.9638.3 | ENSSSCT00000006529 | CCN4 | + | genic—overlapping | 0 | exonic |
| CL.9638.3 | ENSSSCT00000060888 | CCN4 | + | genic—overlapping | 0 | exonic |
| CL.9638.3 | ENSSSCT00000047671 | CCN4 | + | genic—overlapping | 0 | exonic |
| ENSSSCT00000080610 | ENSSSCT00000009368 | KCNK3 | + | intergenic—same_strand | 3996 | downstream |
| ENSSSCT00000072909 | ENSSSCT00000009368 | KCNK3 | + | intergenic—same_strand | 4041 | downstream |
| ENSSSCT00000075362 | ENSSSCT00000009368 | KCNK3 | + | intergenic—same_strand | 4106 | downstream |
| ENSSSCT00000036447 | ENSSSCT00000042276 | ENSSSCG00000038991 | + | intergenic—same_strand | 6100 | downstream |
| ENSSSCT00000036447 | ENSSSCT00000007211 | S100A14 | + | intergenic—same_strand | 6100 | downstream |
| ENSSSCT00000086415 | ENSSSCT00000076358 | RAMP2 | + | intergenic—same_strand | 6859 | upstream |
| ENSSSCT00000086415 | ENSSSCT00000018929 | RAMP2 | + | intergenic—same_strand | 6864 | upstream |
| ENSSSCT00000076362 | ENSSSCT00000048117 | ZNRF2 | + | intergenic—same_strand | 8292 | upstream |
| Pathway Name | Pathway ID | Input (Background) Number | FDR | Visualization |
|---|---|---|---|---|
| Cytokine-cytokine receptor interaction | ssc04060 | 25 (259) | 4.74 × 10−11 | Figure S1 |
| TNF signalling pathway | ssc04668 | 17 (107) | 2.94 × 10−10 | Figure S2 |
| IL-17 signalling pathway | ssc04657 | 15 (88) | 1.96 × 10−9 | Figure S3 |
| C-type lectin receptor signalling pathway | ssc04625 | 15 (103) | 1.13 × 10−8 | - |
| NOD-like receptor signalling pathway | ssc04621 | 17 (148) | 1.56 × 10−8 | Figure S4 |
| NF-kappa B signalling pathway | ssc04064 | 13 (99) | 3.89 × 10−7 | Figure S5 |
| Focal adhesion | ssc04510 | 16 (194) | 1.98 × 10−6 | - |
| MAPK signalling pathway | ssc04010 | 19 (286) | 2.43 × 10−6 | Figure S6 |
| PI3K-Akt signalling pathway | ssc04151 | 20 (335) | 4.28 × 10−6 | - |
| Hematopoietic cell lineage | ssc04640 | 11 (88) | 4.93 × 10−6 | - |
| Toll-like receptor signalling pathway | ssc04620 | 11 (95) | 8.69 × 10−6 | Figure S7 |
| Necroptosis | ssc04217 | 13 (147) | 1.11 × 10−5 | Figure S8 |
| Jak-STAT signalling pathway | ssc04630 | 13 (151) | 1.41 × 10−5 | Figure S9 |
| ECM-receptor interaction | ssc04512 | 10 (84) | 2.08 × 10−5 | - |
| Antigen processing and presentation | ssc04612 | 9 (45) | 3.78 × 10−5 | Figure S10 |
| Cell adhesion molecules (CAMs) | ssc04514 | 12 (143) | 4.46 × 10−5 | Figure S11 |
| Th17 cell differentiation | ssc04659 | 10 (107) | 1.20 × 10−4 | - |
| Apoptosis | ssc04210 | 11 (134) | 1.23 × 10−4 | Figure S12 |
| Chemokine signalling pathway | ssc04062 | 11 (177) | 1.03 × 10−3 | - |
| Th1 and Th2 cell differentiation | ssc04658 | 8 (91) | 1.18 × 10−3 | - |
| Regulation of actin cytoskeleton | ssc04810 | 11 (207) | 3.04 × 10−3 | Figure S13 |
| Leukocyte transendothelial migration | ssc04670 | 8 (110) | 3.21 × 10−3 | Figure S14 |
| Phospholipase D signalling pathway | ssc04072 | 9 (146) | 4.00 × 10−3 | - |
| Cellular senescence | ssc04218 | 9 (154) | 5.43 × 10−3 | Figure S15 |
| HIF-1 signalling pathway | ssc04066 | 7 (108) | 1.10 × 10−2 | - |
| Cytosolic DNA-sensing pathway | ssc04623 | 5 (55) | 1.39 × 10−2 | - |
| VEGF signalling pathway | ssc04370 | 5 (55) | 1.39 × 10−2 | Figure S16 |
| RIG-I-like receptor signalling pathway | ssc04622 | 5 (63) | 2.08 × 10−2 | - |
| Rap1 signalling pathway | ssc04015 | 9 (211) | 2.59 × 10−2 | - |
| Adipocytokine signalling pathway | ssc04920 | 5 (71) | 2.97 × 10−2 | - |
| Proteasome | ssc03050 | 4 (44) | 3.11 × 10−2 | Figure S17 |
| Ras signalling pathway | ssc04014 | 9 (229) | 3.53 × 10−2 | - |
| Phagosome | ssc04145 | 8 (126) | 3.87 × 10−2 | Figure S18 |
| Folate biosynthesis | ssc00790 | 3 (24) | 3.92 × 10−2 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makowczenko, K.G.; Jastrzebski, J.P.; Paukszto, L.; Dobrzyn, K.; Kiezun, M.; Smolinska, N.; Kaminski, T. Chemerin Impact on Alternative mRNA Transcription in the Porcine Luteal Cells. Cells 2022, 11, 715. https://doi.org/10.3390/cells11040715
Makowczenko KG, Jastrzebski JP, Paukszto L, Dobrzyn K, Kiezun M, Smolinska N, Kaminski T. Chemerin Impact on Alternative mRNA Transcription in the Porcine Luteal Cells. Cells. 2022; 11(4):715. https://doi.org/10.3390/cells11040715
Chicago/Turabian StyleMakowczenko, Karol G., Jan P. Jastrzebski, Lukasz Paukszto, Kamil Dobrzyn, Marta Kiezun, Nina Smolinska, and Tadeusz Kaminski. 2022. "Chemerin Impact on Alternative mRNA Transcription in the Porcine Luteal Cells" Cells 11, no. 4: 715. https://doi.org/10.3390/cells11040715
APA StyleMakowczenko, K. G., Jastrzebski, J. P., Paukszto, L., Dobrzyn, K., Kiezun, M., Smolinska, N., & Kaminski, T. (2022). Chemerin Impact on Alternative mRNA Transcription in the Porcine Luteal Cells. Cells, 11(4), 715. https://doi.org/10.3390/cells11040715









