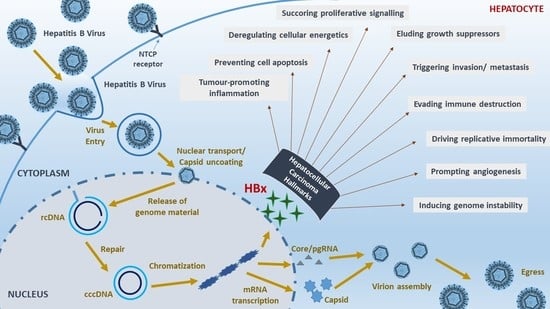
Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review
Abstract
1. Introduction
2. Sustaining Proliferative Signalling
3. Evading Growth Suppressors
4. Resisting Cell Death
5. Enabling Replicative Immortality
6. Prompting Angiogenesis
7. Triggering Invasion and Metastasis
8. Evading Immune Destruction
9. Tumour-Promoting Inflammation
10. Genome Instability and Mutation
11. Deregulating Cellular Energetics
12. Therapeutic Potential of HBx
13. Conclusions and Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections. 2021. Available online: https://www.who.int/publications/i/item/9789240027077 (accessed on 29 December 2021).
- Wong, V.W.-S.; Janssen, H.L.A. Can we use hcc risk scores to individualize surveillance in chronic hepatitis b infection? J. Hepatol. 2015, 63, 722–732. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Kgatle, M. Recent advancement in hepatitis b virus, epigenetics alterations and related complications. In Advances in Treatment of Hepatitis C and B; IntechOpen: London, UK, 2017. [Google Scholar]
- Seeger, C.; Mason, W.S. Hepatitis b virus biology. Microbiol. Mol. Biol. Rev. MMBR 2000, 64, 51–68. [Google Scholar] [CrossRef] [PubMed]
- Feitelson, M.A.; Lee, J. Hepatitis b virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007, 252, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Berasain, C.; Castillo, J.; Perugorria, M.J.; Latasa, M.U.; Prieto, J.; Avila, M.A. Inflammation and liver cancer. Ann. N. Y. Acad. Sci. 2009, 1155, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Abdel-Hafiz, H.; Suhail, M.; Al-Mars, A.; Zakaria, M.K.; Fatima, K.; Ahmad, S.; Azhar, E.; Chaudhary, A.; Qadri, I. Hepatitis b virus, hbx mutants and their role in hepatocellular carcinoma. World J. Gastroenterol. 2014, 20, 10238–10248. [Google Scholar] [CrossRef]
- Mathew, M.A.; Kurian, S.C.; Varghese, A.P.; Oommen, S.; Manoj, G. Hbx gene mutations in hepatitis b virus and hepatocellular carcinoma. Gastroenterol. Res. 2014, 7, 1–4. [Google Scholar]
- Miller, R.H.; Robinson, W.S. Common evolutionary origin of hepatitis b virus and retroviruses. Proc. Natl. Acad. Sci. USA 1986, 83, 2531–2535. [Google Scholar] [CrossRef]
- Martin-Vilchez, S.; Lara-Pezzi, E.; Trapero-Marugán, M.; Moreno-Otero, R.; Sanz-Cameno, P. The molecular and pathophysiological implications of hepatitis b x antigen in chronic hepatitis b virus infection. Rev. Med. Virol. 2011, 21, 315–329. [Google Scholar] [CrossRef]
- Henkler, F.; Hoare, J.; Waseem, N.; Goldin, R.D.; McGarvey, M.J.; Koshy, R.; King, I.A. Intracellular localization of the hepatitis b virus hbx protein. J. Gen. Virol. 2001, 82, 871–882. [Google Scholar] [CrossRef]
- Bouchard Michael, J.; Schneider Robert, J. The enigmatic x gene of hepatitis b virus. J. Virol. 2004, 78, 12725–12734. [Google Scholar] [CrossRef] [PubMed]
- Faktor, O.; Shaul, Y. The identification of hepatitis b virus x gene responsive elements reveals functional similarity of x and htlv-i tax. Oncogene 1990, 5, 867–872. [Google Scholar] [PubMed]
- Murakami, S. Hepatitis b virus x protein: A multifunctional viral regulator. J. Gastroenterol. 2001, 36, 651–660. [Google Scholar] [CrossRef] [PubMed]
- Caselmann, W.H.; Koshy, R. Transactivators of hbv, signal transduction and tumorigenesis. In Hepatitis B Virus; Imperial College Press: London, UK, 1998; pp. 161–181. [Google Scholar]
- Rawat, S.; Clippinger, A.J.; Bouchard, M.J. Modulation of apoptotic signaling by the hepatitis b virus x protein. Viruses 2012, 4, 2945–2972. [Google Scholar] [CrossRef]
- Robinson, W.S. Molecular events in the pathogenesis of hepadnavirus-associated hepatocellular carcinoma. Annu. Rev. Med. 1994, 45, 297–323. [Google Scholar] [CrossRef]
- Seifer, M.; Höhne, M.; Schaefer, S.; Gerlich, W.H. In vitro tumorigenicity of hepatitis b virus DNA and hbx protein. J. Hepatol. 1991, 13, S61–S65. [Google Scholar] [CrossRef]
- Paterlini, P.; Poussin, K.; Kew, M.; Franco, D.; Brechot, C. Selective accumulation of the x transcript of hepatitis b virus in patients negative for hepatitis b surface antigen with hepatocellular carcinoma. Hepatology 1995, 21, 313–321. [Google Scholar]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Guo, G.H.; Tan, D.M.; Zhu, P.A.; Liu, F. Hepatitis b virus x protein promotes proliferation and upregulates tgf-beta1 and ctgf in human hepatic stellate cell line, lx-2. Hepatobiliary Pancreat. Dis. Int. HBPD INT 2009, 8, 59–64. [Google Scholar]
- Shukla, S.K.; Kumar, V. Hepatitis b virus x protein and c-myc cooperate in the upregulation of ribosome biogenesis and in cellular transformation. FEBS J. 2012, 279, 3859–3871. [Google Scholar] [CrossRef] [PubMed]
- Terradillos, O.; Billet, O.; Renard, C.A.; Levy, R.; Molina, T.; Briand, P.; Buendia, M.A. The hepatitis b virus x gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene 1997, 14, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; He, J.; Chen, L.; Wang, G. Hepatitis b virus x protein upregulates expression of smyd3 and c-myc in hepg2 cells. Med. Oncol. 2009, 26, 445–451. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.L.; Guo, Y.; Factor, V.M.; Thorgeirsson, S.S.; Bell, D.W.; Testa, J.R.; Peifley, K.A.; Winkles, J.A. The fn14 immediate-early response gene is induced during liver regeneration and highly expressed in both human and murine hepatocellular carcinomas. Am. J. Pathol. 2000, 156, 1253–1261. [Google Scholar] [CrossRef]
- Qiao, L.; Leach, K.; McKinstry, R.; Gilfor, D.; Yacoub, A.; Park, J.S.; Grant, S.; Hylemon, P.B.; Fisher, P.B.; Dent, P. Hepatitis b virus x protein increases expression of p21(cip-1/waf1/mda6) and p27(kip-1) in primary mouse hepatocytes, leading to reduced cell cycle progression. Hepatology 2001, 34, 906–917. [Google Scholar] [CrossRef]
- Xu, J.; Liu, H.; Chen, L.; Wang, S.; Zhou, L.; Yun, X.; Sun, L.; Wen, Y.; Gu, J. Hepatitis b virus x protein confers resistance of hepatoma cells to anoikis by up-regulating and activating p21-activated kinase 1. Gastroenterology 2012, 143, 199–212.e4. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, W.; Liu, Q.; Zhang, X.; Lv, N.; Ye, L.; Zhang, X. A mutant of hepatitis b virus x protein (hbxdelta127) promotes cell growth through a positive feedback loop involving 5-lipoxygenase and fatty acid synthase. Neoplasia 2010, 12, 103–115. [Google Scholar] [CrossRef]
- Shan, C.; Xu, F.; Zhang, S.; You, J.; You, X.; Qiu, L.; Zheng, J.; Ye, L.; Zhang, X. Hepatitis b virus x protein promotes liver cell proliferation via a positive cascade loop involving arachidonic acid metabolism and p-erk1/2. Cell Res. 2010, 20, 563–575. [Google Scholar] [CrossRef]
- Lee, Y.I.; Kang-Park, S.; Do, S.I.; Lee, Y.I. The hepatitis b virus-x protein activates a phosphatidylinositol 3-kinase-dependent survival signaling cascade. J. Biol. Chem. 2001, 276, 16969–16977. [Google Scholar] [CrossRef]
- Noh, E.J.; Jung, H.J.; Jeong, G.; Choi, K.S.; Park, H.J.; Lee, C.H.; Lee, J.S. Subcellular localization and transcriptional repressor activity of hbx on p21(waf1/cip1) promoter is regulated by erk-mediated phosphorylation. Biochem. Biophys. Res. Commun. 2004, 319, 738–745. [Google Scholar] [CrossRef]
- Yang, B.; Bouchard, M.J. The hepatitis b virus x protein elevates cytosolic calcium signals by modulating mitochondrial calcium uptake. J. Virol. 2012, 86, 313–327. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Kong, F.; Hu, L.; You, H.; Zhang, P.; Du, W.; Zheng, K. Role of hepatitis b virus x protein in regulating lim and sh3 protein 1 (lasp-1) expression to mediate proliferation and migration of hepatoma cells. Virol. J. 2012, 9, 163. [Google Scholar] [CrossRef] [PubMed]
- Elmore, L.W.; Hancock, A.R.; Chang, S.F.; Wang, X.W.; Chang, S.; Callahan, C.P.; Geller, D.A.; Will, H.; Harris, C.C. Hepatitis b virus x protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. USA 1997, 94, 14707–14712. [Google Scholar] [CrossRef]
- Truant, R.; Antunovic, J.; Greenblatt, J.; Prives, C.; Cromlish, J.A. Direct interaction of the hepatitis b virus hbx protein with p53 leads to inhibition by hbx of p53 response element-directed transactivation. J. Virol. 1995, 69, 1851–1859. [Google Scholar] [CrossRef]
- Choi, B.H.; Choi, M.; Jeon, H.Y.; Rho, H.M. Hepatitis b viral x protein overcomes inhibition of e2f1 activity by prb on the human rb gene promoter. DNA Cell Biol. 2001, 20, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Kumar, V. Hbx protein of hepatitis b virus promotes reinitiation of DNA replication by regulating expression and intracellular stability of replication licensing factor cdc6. J. Biol. Chem. 2012, 287, 20545–20554. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, J.; Cui, M.; Liu, F.; You, X.; Du, Y.; Gao, Y.; Zhang, S.; Lu, Z.; Ye, L.; et al. Hepatitis b virus x protein inhibits tumor suppressor mir-205 through inducing hypermethylation of mir-205 promoter to enhance carcinogenesis. Neoplasia 2013, 15, 1282–1291. [Google Scholar] [CrossRef]
- Zhang, W.; Lu, Z.; Kong, G.; Gao, Y.; Wang, T.; Wang, Q.; Cai, N.; Wang, H.; Liu, F.; Ye, L.; et al. Hepatitis b virus x protein accelerates hepatocarcinogenesis with partner survivin through modulating mir-520b and hbxip. Mol. Cancer 2014, 13, 128. [Google Scholar] [CrossRef]
- Zhao, Z.; Hu, Y.; Shen, X.; Lao, Y.; Zhang, L.; Qiu, X.; Hu, J.; Gong, P.; Cui, H.; Lu, S.; et al. Hbx represses riz1 expression by DNA methyltransferase 1 involvement in decreased mir-152 in hepatocellular carcinoma. Oncol. Rep. 2017, 37, 2811–2818. [Google Scholar] [CrossRef]
- Lee, J.O.; Kwun, H.J.; Jung, J.K.; Choi, K.H.; Min, D.S.; Jang, K.L. Hepatitis b virus x protein represses e-cadherin expression via activation of DNA methyltransferase 1. Oncogene 2005, 24, 6617–6625. [Google Scholar] [CrossRef]
- Zhu, Y.Z.; Zhu, R.; Fan, J.; Pan, Q.; Li, H.; Chen, Q.; Zhu, H.G. Hepatitis b virus x protein induces hypermethylation of p16(ink4a) promoter via DNA methyltransferases in the early stage of hbv-associated hepatocarcinogenesis. J. Viral Hepat. 2010, 17, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.K.; Park, S.H.; Jang, K.L. Hepatitis b virus x protein overcomes the growth-inhibitory potential of retinoic acid by downregulating retinoic acid receptor-beta2 expression via DNA methylation. J. Gen. Virol. 2010, 91, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; You, H.; Zhao, J.; Liu, W.; Hu, L.; Luo, W.; Hu, W.; Tang, R.; Zheng, K. The enhanced expression of death receptor 5 (dr5) mediated by hbv x protein through nf-kappab pathway is associated with cell apoptosis induced by (tnf-α related apoptosis inducing ligand) trail in hepatoma cells. Virol. J. 2015, 12, 192. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, C.; Wang, Y.; Lu, Z.; Zhuang, N.; Zhao, D.; He, J.; Shi, L. Hepatitis b virus x protein sensitizes trail-induced hepatocyte apoptosis by inhibiting the e3 ubiquitin ligase a20. PLoS ONE 2015, 10, e0127329. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, X.; Hu, D.; Feng, T.; Li, H.; Lu, Y.; Huang, J. Hepatitis b virus x (hbx) play an anti-apoptosis role in hepatic progenitor cells by activating wnt/β-catenin pathway. Mol. Cell. Biochem. 2013, 383, 213–222. [Google Scholar] [CrossRef]
- Liu, H.; Yuan, Y.; Guo, H.; Mitchelson, K.; Zhang, K.; Xie, L.; Qin, W.; Lu, Y.; Wang, J.; Guo, Y.; et al. Hepatitis b virus encoded x protein suppresses apoptosis by inhibition of the caspase-independent pathway. J. Proteome Res. 2012, 11, 4803–4813. [Google Scholar] [CrossRef]
- Liu, B.; Fang, M.; Hu, Y.; Huang, B.; Li, N.; Chang, C.; Huang, R.; Xu, X.; Yang, Z.; Chen, Z.; et al. Hepatitis b virus x protein inhibits autophagic degradation by impairing lysosomal maturation. Autophagy 2014, 10, 416–430. [Google Scholar] [CrossRef]
- Son, J.; Kim, M.J.; Lee, J.S.; Kim, J.Y.; Chun, E.; Lee, K.Y. Hepatitis b virus x protein promotes liver cancer progression through autophagy induction in response to tlr4 stimulation. Immune Netw. 2021, 21, e37. [Google Scholar] [CrossRef]
- Wang, P.; Guo, Q.S.; Wang, Z.W.; Qian, H.X. Hbx induces hepg-2 cells autophagy through pi3k/akt-mtor pathway. Mol. Cell. Biochem. 2013, 372, 161–168. [Google Scholar] [CrossRef]
- Qu, Z.L.; Zou, S.Q.; Cui, N.Q.; Wu, X.Z.; Qin, M.F.; Kong, D.; Zhou, Z.L. Upregulation of human telomerase reverse transcriptase mrna expression by in vitro transfection of hepatitis b virus x gene into human hepatocarcinoma and cholangiocarcinoma cells. World J. Gastroenterol. 2005, 11, 5627–5632. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, N.; Zhang, H.; You, J.; Wang, H.; Ye, L. Effects of hepatitis b virus x protein on human telomerase reverse transcriptase expression and activity in hepatoma cells. J. Lab. Clin. Med. 2005, 145, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Su, J.-M.; Lai, X.-M.; Lan, K.-H.; Li, C.-P.; Chao, Y.; Yen, S.-H.; Chang, F.-Y.; Lee, S.-D.; Lee, W.-P. X protein of hepatitis b virus functions as a transcriptional corepressor on the human telomerase promoter. Hepatology 2007, 46, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.W.; Lee, Y.M.; Bae, S.K.; Murakami, S.; Yun, Y.; Kim, K.W. Human hepatitis b virus x protein is a possible mediator of hypoxia-induced angiogenesis in hepatocarcinogenesis. Biochem. Biophys. Res. Commun. 2000, 268, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Yun, C.; Lee, J.H.; Wang, J.H.; Seong, J.K.; Oh, S.H.; Yu, D.Y.; Cho, H. Expression of hepatitis b virus x (hbx) gene is up-regulated by adriamycin at the post-transcriptional level. Biochem. Biophys. Res. Commun. 2002, 296, 1157–1163. [Google Scholar] [CrossRef]
- Moon, E.J.; Jeong, C.H.; Jeong, J.W.; Kim, K.R.; Yu, D.Y.; Murakami, S.; Kim, C.W.; Kim, K.W. Hepatitis b virus x protein induces angiogenesis by stabilizing hypoxia-inducible factor-1alpha. FASEB J. 2004, 18, 382–384. [Google Scholar] [CrossRef]
- Yoo, Y.G.; Oh, S.H.; Park, E.S.; Cho, H.; Lee, N.; Park, H.; Kim, D.K.; Yu, D.Y.; Seong, J.K.; Lee, M.O. Hepatitis b virus x protein enhances transcriptional activity of hypoxia-inducible factor-1alpha through activation of mitogen-activated protein kinase pathway. J. Biol. Chem. 2003, 278, 39076–39084. [Google Scholar] [CrossRef]
- Yoo, Y.G.; Na, T.Y.; Seo, H.W.; Seong, J.K.; Park, C.K.; Shin, Y.K.; Lee, M.O. Hepatitis b virus x protein induces the expression of mta1 and hdac1, which enhances hypoxia signaling in hepatocellular carcinoma cells. Oncogene 2008, 27, 3405–3413. [Google Scholar] [CrossRef]
- Lara-Pezzi, E.; Gómez-Gaviro, M.V.; Gálvez, B.G.; Mira, E.; Iñiguez, M.A.; Fresno, M.; Martínez-A, C.; Arroyo, A.G.; López-Cabrera, M. The hepatitis b virus x protein promotes tumor cell invasion by inducing membrane-type matrix metalloproteinase-1 and cyclooxygenase-2 expression. J. Clin. Investig. 2002, 110, 1831–1838. [Google Scholar] [CrossRef]
- Yu, F.L.; Liu, H.J.; Lee, J.W.; Liao, M.H.; Shih, W.L. Hepatitis b virus x protein promotes cell migration by inducing matrix metalloproteinase-3. J. Hepatol. 2005, 42, 520–527. [Google Scholar] [CrossRef]
- Liu, L.P.; Liang, H.F.; Chen, X.P.; Zhang, W.G.; Yang, S.L.; Xu, T.; Ren, L. The role of nf-kappab in hepatitis b virus x protein-mediated upregulation of vegf and mmps. Cancer Investig. 2010, 28, 443–451. [Google Scholar] [CrossRef]
- Cheng, A.S.L.; Chan, H.L.Y.; Leung, W.K.; To, K.F.; Go, M.Y.Y.; Chan, J.Y.H.; Liew, C.T.; Sung, J.J.Y. Expression of hbx and cox-2 in chronic hepatitis b, cirrhosis and hepatocellular carcinoma: Implication of hbx in upregulation of cox-2. Mod. Pathol. 2004, 17, 1169–1179. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kongkavitoon, P.; Tangkijvanich, P.; Hirankarn, N.; Palaga, T. Hepatitis b virus hbx activates notch signaling via delta-like 4/notch1 in hepatocellular carcinoma. PLoS ONE 2016, 11, e0146696. [Google Scholar] [CrossRef] [PubMed]
- Sanz-Cameno, P.; Martín-Vílchez, S.; Lara-Pezzi, E.; Borque, M.J.; Salmerón, J.; Muñoz de Rueda, P.; Solís, J.A.; López-Cabrera, M.; Moreno-Otero, R. Hepatitis b virus promotes angiopoietin-2 expression in liver tissue: Role of hbv x protein. Am. J. Pathol. 2006, 169, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Majano, P.L.; García-Monzón, C.; López-Cabrera, M.; Lara-Pezzi, E.; Fernández-Ruiz, E.; García-Iglesias, C.; Borque, M.J.; Moreno-Otero, R. Inducible nitric oxide synthase expression in chronic viral hepatitis. Evidence for a virus-induced gene upregulation. J. Clin. Investig. 1998, 101, 1343–1352. [Google Scholar] [CrossRef]
- Giannelli, G.; Bergamini, C.; Marinosci, F.; Fransvea, E.; Quaranta, M.; Lupo, L.; Schiraldi, O.; Antonaci, S. Clinical role of mmp-2/timp-2 imbalance in hepatocellular carcinoma. Int. J. Cancer 2002, 97, 425–431. [Google Scholar] [CrossRef]
- Lara-Pezzi, E.; Majano, P.L.; Yáñez-Mó, M.; Gómez-Gonzalo, M.; Carretero, M.; Moreno-Otero, R.; Sánchez-Madrid, F.; López-Cabrera, M. Effect of the hepatitis b virus hbx protein on integrin-mediated adhesion to and migration on extracellular matrix. J. Hepatol. 2001, 34, 409–415. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, Q.; Ye, L.; Feng, Y.; Zhang, X. Hepatitis b virus x protein upregulates expression of calpain small subunit 1 via nuclear facter-κb/p65 in hepatoma cells. J. Med. Virol. 2010, 82, 920–928. [Google Scholar] [CrossRef]
- Lin, X.; Zuo, S.; Luo, R.; Li, Y.; Yu, G.; Zou, Y.; Zhou, Y.; Liu, Z.; Liu, Y.; Hu, Y.; et al. Hbx-induced mir-5188 impairs foxo1 to stimulate β-catenin nuclear translocation and promotes tumor stemness in hepatocellular carcinoma. Theranostics 2019, 9, 7583–7598. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, S.; Hu, T.; Liu, S.; He, Y.; Sun, S. Up-regulated microrna-143 transcribed by nuclear factor kappa b enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009, 50, 490–499. [Google Scholar] [CrossRef]
- Lara-Pezzi, E.; Serrador, J.M.; Montoya, M.C.; Zamora, D.; Yáñez-Mó, M.; Carretero, M.; Furthmayr, H.; Sánchez-Madrid, F.; López-Cabrera, M. The hepatitis b virus x protein (hbx) induces a migratory phenotype in a cd44-dependent manner: Possible role of hbx in invasion and metastasis. Hepatology 2001, 33, 1270–1281. [Google Scholar] [CrossRef]
- Zhang, X.; Ye, L.-h.; Zhang, X.-d. A mutant of hepatitis b virus x protein (hbxδ127) enhances hepatoma cell migration via osteopontin involving 5-lipoxygenase. Acta Pharmacol. Sin. 2010, 31, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.K.; Man, K.; Poon, R.T.; Lo, C.M.; Yuen, A.P.; Ng, I.O.; Ng, K.T.; Leonard, W.; Fan, S.T. Signal transducers and activators of transcription 5b activation enhances hepatocellular carcinoma aggressiveness through induction of epithelial-mesenchymal transition. Cancer Res. 2006, 66, 9948–9956. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.Z.; Zhang, L.D.; Zhang, Y.; Xiong, Y.; Zhang, Y.J.; Li, H.L.; Li, X.W.; Dong, J.H. Hbx protein induces emt through c-src activation in smmc-7721 hepatoma cell line. Biochem. Biophys. Res. Commun. 2009, 382, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, L.; He, H.; Zhu, Y.; Liu, J.; Wang, S.; Chen, L.; Wu, Q.; Xu, J.; Gu, J. Hepatitis b virus x protein promotes hepatoma cell invasion and metastasis by stabilizing snail protein. Cancer Sci. 2012, 103, 2072–2081. [Google Scholar] [CrossRef]
- You, H.; Yuan, D.; Bi, Y.; Zhang, N.; Li, Q.; Tu, T.; Wei, X.; Lian, Q.; Yu, T.; Kong, D.; et al. Hepatitis b virus x protein promotes vimentin expression via lim and sh3 domain protein 1 to facilitate epithelial-mesenchymal transition and hepatocarcinogenesis. Cell Commun. Signal. 2021, 19, 33. [Google Scholar] [CrossRef]
- Arzumanyan, A.; Friedman, T.; Kotei, E.; Ng, I.O.L.; Lian, Z.; Feitelson, M.A. Epigenetic repression of e-cadherin expression by hepatitis b virus x antigen in liver cancer. Oncogene 2012, 31, 563–572. [Google Scholar] [CrossRef]
- Lee, M.J.; Jin, Y.H.; Kim, K.; Choi, Y.; Kim, H.C.; Park, S. Expression of hepatitis b virus x protein in hepatocytes suppresses cd8 t cell activity. Immune Netw. 2010, 10, 126–134. [Google Scholar] [CrossRef]
- Kumar, M.; Jung, S.Y.; Hodgson, A.J.; Madden, C.R.; Qin, J.; Slagle, B.L. Hepatitis b virus regulatory hbx protein binds to adaptor protein ips-1 and inhibits the activation of beta interferon. J. Virol. 2011, 85, 987–995. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Mao, A.; Li, C.; Li, Y.; Tien, P. Hepatitis b virus x protein suppresses virus-triggered irf3 activation and ifn-beta induction by disrupting the visa-associated complex. Cell. Mol. Immunol. 2010, 7, 341–348. [Google Scholar] [CrossRef]
- Wei, C.; Ni, C.; Song, T.; Liu, Y.; Yang, X.; Zheng, Z.; Jia, Y.; Yuan, Y.; Guan, K.; Xu, Y.; et al. The hepatitis b virus x protein disrupts innate immunity by downregulating mitochondrial antiviral signaling protein. J. Immunol. 2010, 185, 1158–1168. [Google Scholar] [CrossRef]
- Wang, L.; Sun, Y.; Song, X.; Wang, Z.; Zhang, Y.; Zhao, Y.; Peng, X.; Zhang, X.; Li, C.; Gao, C.; et al. Hepatitis b virus evades immune recognition via rna adenosine deaminase adar1-mediated viral rna editing in hepatocytes. Cell. Mol. Immunol. 2021, 18, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Huang, Y. Antagonism of rip1 using necrostatin-1 (nec-1) ameliorated damage and inflammation of hbv x protein (hbx) in human normal hepatocytes. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1194–1199. [Google Scholar] [CrossRef] [PubMed]
- Duan, L.; Wu, R.; Zhang, X.; Wang, D.; You, Y.; Zhang, Y.; Zhou, L.; Chen, W. Hbx-induced s100a9 in nf-κb dependent manner promotes growth and metastasis of hepatocellular carcinoma cells. Cell Death Dis. 2018, 9, 629. [Google Scholar] [CrossRef]
- Xiang, W.-Q.; Feng, W.-F.; Ke, W.; Sun, Z.; Chen, Z.; Liu, W. Hepatitis b virus x protein stimulates il-6 expression in hepatocytes via a myd88-dependent pathway. J. Hepatol. 2011, 54, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Tian, D.; Huang, W.; Zhu, H.; Wang, J.; Zhang, Y.; Hu, H.; Nie, Y.; Fan, D.; Wu, K. Upregulation of il-23 expression in patients with chronic hepatitis b is mediated by the hbx/erk/nf-κb pathway. J. Immunol. 2012, 188, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.-J.; Yang, S.-T.; Chen, R.-Y.; Huang, W.; Chayama, K.; Lee, M.-H.; Yang, S.-J.; Lai, H.-S.; Yen, H.-Y.; Hsiao, Y.-W.; et al. Hepatitis b virus x protein (hbx) enhances centrosomal p4.1-associated protein (cpap) expression to promote hepatocarcinogenesis. J. Biomed. Sci. 2019, 26, 44. [Google Scholar] [CrossRef]
- Fujii, R.; Zhu, C.; Wen, Y.; Marusawa, H.; Bailly-Maitre, B.; Matsuzawa, S.; Zhang, H.; Kim, Y.; Bennett, C.F.; Jiang, W.; et al. Hbxip, cellular target of hepatitis b virus oncoprotein, is a regulator of centrosome dynamics and cytokinesis. Cancer Res. 2006, 66, 9099–9107. [Google Scholar] [CrossRef]
- Forgues, M.; Marrogi, A.J.; Spillare, E.A.; Wu, C.-G.; Yang, Q.; Yoshida, M.; Wang, X.W. Interaction of the hepatitis b virus x protein with the crm1-dependent nuclear export pathway. J. Biol. Chem. 2001, 276, 22797–22803. [Google Scholar] [CrossRef]
- Becker Sherry, A.; Lee, T.-H.; Butel Janet, S.; Slagle Betty, L. Hepatitis b virus x protein interferes with cellular DNA repair. J. Virol. 1998, 72, 266–272. [Google Scholar] [CrossRef]
- Oishi, N.; Shilagardi, K.; Nakamoto, Y.; Honda, M.; Kaneko, S.; Murakami, S. Hepatitis b virus x protein overcomes oncogenic ras-induced senescence in human immortalized cells. Cancer Sci. 2007, 98, 1540–1548. [Google Scholar] [CrossRef]
- Qadri, I.; Conaway, J.W.; Conaway, R.C.; Schaack, J.; Siddiqui, A. Hepatitis b virus transactivator protein, hbx, associates with the components of tfiih and stimulates the DNA helicase activity of tfiih. Proc. Natl. Acad. Sci. USA 1996, 93, 10578–10583. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.M.; Xu, Y.; Li, F.; Nio, K.; Reszka-Blanco, N.; Li, X.; Wu, Y.; Yu, Y.; Xiong, Y.; Su, L. Hepatitis b virus x protein promotes degradation of smc5/6 to enhance hbv replication. Cell Rep. 2016, 16, 2846–2854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yan, Q.; Gong, L.; Xu, H.; Liu, B.; Fang, X.; Yu, D.; Li, L.; Wei, T.; Wang, Y.; et al. C-terminal truncated hbx initiates hepatocarcinogenesis by downregulating txnip and reprogramming glucose metabolism. Oncogene 2021, 40, 1147–1161. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, Y.; Li, D.; Liu, Z.; Zhang, J. Oncoprotein lamtor5 activates glut1 via upregulating nf-κb in liver cancer. Open Med. 2019, 14, 264–270. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.-J.; Park, Y.-H.; Kim, S.-U.; Moon, H.-B.; Park, D.S.; Han, Y.-H.; Lee, C.-H.; Lee, D.-S.; Song, I.-S.; Lee, D.H.; et al. Hepatitis b virus x protein regulates hepatic glucose homeostasis via activation of inducible nitric oxide synthase. J. Biol. Chem. 2011, 286, 29872–29881. [Google Scholar] [CrossRef]
- Jung, S.-Y.; Kim, Y.-J. C-terminal region of hbx is crucial for mitochondrial DNA damage. Cancer Lett. 2013, 331, 76–83. [Google Scholar] [CrossRef]
- Klein, N.P.; Bouchard, M.J.; Wang, L.H.; Kobarg, C.; Schneider, R.J. Src kinases involved in hepatitis b virus replication. EMBO J. 1999, 18, 5019–5027. [Google Scholar] [CrossRef]
- Benn, J.; Schneider, R.J. Hepatitis b virus hbx protein deregulates cell cycle checkpoint controls. Proc. Natl. Acad. Sci. USA 1995, 92, 11215–11219. [Google Scholar] [CrossRef]
- Feitelson, M.A.; Bonamassa, B.; Arzumanyan, A. The roles of hepatitis b virus-encoded x protein in virus replication and the pathogenesis of chronic liver disease. Expert Opin. Ther. Targets 2014, 18, 293–306. [Google Scholar] [CrossRef]
- Wang, L.H.; Wu, C.F.; Rajasekaran, N.; Shin, Y.K. Loss of tumor suppressor gene function in human cancer: An overview. Cell. Physiol. Biochem. 2018, 51, 2647–2693. [Google Scholar] [CrossRef]
- Kim, M.P.; Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 2018, 25, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ko, L.J.; Jayaraman, L.; Prives, C. P53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996, 10, 2438–2451. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.T.; Olson, E.N. Micrornas in stress signaling and human disease. Cell 2012, 148, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kong, G.; Zhang, J.; Wang, T.; Ye, L.; Zhang, X. Microrna-520b inhibits growth of hepatoma cells by targeting mekk2 and cyclin d1. PLoS ONE 2012, 7, e31450. [Google Scholar]
- Yokota, T.; Suda, T.; Igarashi, M.; Kuroiwa, T.; Waguri, N.; Kawai, H.; Mita, Y.; Aoyagi, Y. Telomere length variation and maintenance in hepatocarcinogenesis. Cancer 2003, 98, 110–118. [Google Scholar] [CrossRef]
- Riedl, S.J.; Shi, Y. Molecular mechanisms of caspase regulation during apoptosis. Nat. Rev. Mol. Cell Biol. 2004, 5, 897–907. [Google Scholar] [CrossRef]
- Niller, H.H.; Ay, E.; Banati, F.; Demcsák, A.; Takacs, M.; Minarovits, J. Wild type hbx and truncated hbx: Pleiotropic regulators driving sequential genetic and epigenetic steps of hepatocarcinogenesis and progression of hbv-associated neoplasms. Rev. Med. Virol. 2016, 26, 57–73. [Google Scholar] [CrossRef]
- An, P.; Xu, J.; Yu, Y.; Winkler, C.A. Host and viral genetic variation in hbv-related hepatocellular carcinoma. Front. Genet. 2018, 9, 261. [Google Scholar] [CrossRef]
- Sirma, H.; Giannini, C.; Poussin, K.; Paterlini, P.; Kremsdorf, D.; Bréchot, C. Hepatitis b virus x mutants, present in hepatocellular carcinoma tissue abrogate both the antiproliferative and transactivation effects of hbx. Oncogene 1999, 18, 4848–4859. [Google Scholar] [CrossRef]
- Bock, C.T.; Toan, N.L.; Koeberlein, B.; Song, L.H.; Chin, R.; Zentgraf, H.; Kandolf, R.; Torresi, J. Subcellular mislocalization of mutant hepatitis b x proteins contributes to modulation of stat/socs signaling in hepatocellular carcinoma. Intervirology 2008, 51, 432–443. [Google Scholar] [CrossRef]
- Hoare, J.; Henkler, F.; Dowling, J.J.; Errington, W.; Goldin, R.D.; Fish, D.; McGarvey, M.J. Subcellular localisation of the x protein in hbv infected hepatocytes. J. Med. Virol. 2001, 64, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef] [PubMed]
- White, E.; DiPaola, R.S. The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 2009, 15, 5308–5316. [Google Scholar] [CrossRef] [PubMed]
- Salimi-Jeda, A.; Badrzadeh, F.; Esghaei, M.; Abdoli, A. The role of telomerase and viruses interaction in cancer development, and telomerase-dependent therapeutic approaches. Cancer Treat. Res. Commun. 2021, 27, 100323. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.Q.; Qu, Z.L.; Li, Z.F.; Wang, X. Hepatitis b virus x gene induces human telomerase reverse transcriptase mrna expression in cultured normal human cholangiocytes. World J. Gastroenterol. 2004, 10, 2259–2262. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Kaita, K.D.; Xu, Z.; Ou, J.H.; Gong, Y.; Zhang, M.; Minuk, G.Y. The absence of up-regulation of telomerase activity during regeneration after partial hepatectomy in hepatitis b virus x gene transgenic mice. J. Hepatol. 2003, 39, 262–268. [Google Scholar] [CrossRef]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef]
- Hanahan, D.; Folkman, J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell 1996, 86, 353–364. [Google Scholar] [CrossRef]
- Vrancken, K.; Paeshuyse, J.; Liekens, S. Angiogenic activity of hepatitis b and c viruses. Antivir. Chem. Chemother. 2012, 22, 159–170. [Google Scholar] [CrossRef]
- Park, Y.N.; Kim, Y.B.; Yang, K.M.; Park, C. Increased expression of vascular endothelial growth factor and angiogenesis in the early stage of multistep hepatocarcinogenesis. Arch. Pathol. Lab. Med. 2000, 124, 1061–1065. [Google Scholar] [CrossRef]
- Dai, C.X.; Gao, Q.; Qiu, S.J.; Ju, M.J.; Cai, M.Y.; Xu, Y.F.; Zhou, J.; Zhang, B.H.; Fan, J. Hypoxia-inducible factor-1 alpha, in association with inflammation, angiogenesis and myc, is a critical prognostic factor in patients with hcc after surgery. BMC Cancer 2009, 9, 418. [Google Scholar] [CrossRef]
- Meirson, T.; Gil-Henn, H.; Samson, A.O. Invasion and metastasis: The elusive hallmark of cancer. Oncogene 2020, 39, 2024–2026. [Google Scholar] [CrossRef] [PubMed]
- Elkington, P.T.G.; O’Kane, C.M.; Friedland, J.S. The paradox of matrix metalloproteinases in infectious disease. Clin. Exp. Immunol. 2005, 142, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.-S.; Dai, Z.; Zhou, J.; Liu, Y.-K.; Qiu, S.-J.; Tan, C.-J.; Shi, Y.-H.; Huang, C.; Wang, Z.; He, Y.-F.; et al. Capn4 overexpression underlies tumor invasion and metastasis after liver transplantation for hepatocellular carcinoma. Hepatology 2009, 49, 460–470. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.-F.; Lau, S.H.; Hu, L.; Xie, D.; Wu, J.; Yang, J.; Wang, Y.; Wu, M.-C.; Fung, J.; Bai, X.; et al. Cooh-terminal truncated hbv x protein plays key role in hepatocarcinogenesis. Clin. Cancer Res. 2008, 14, 5061–5068. [Google Scholar] [CrossRef] [PubMed]
- van Zijl, F.; Zulehner, G.; Petz, M.; Schneller, D.; Kornauth, C.; Hau, M.; Machat, G.; Grubinger, M.; Huber, H.; Mikulits, W. Epithelial-mesenchymal transition in hepatocellular carcinoma. Future Oncol. 2009, 5, 1169–1179. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, N.C. Recognition of human oncogenic viruses by host pattern-recognition receptors. Front. Immunol. 2014, 5, 353. [Google Scholar] [CrossRef] [PubMed]
- Thimme, R.; Wieland, S.; Steiger, C.; Ghrayeb, J.; Reimann, K.A.; Purcell, R.H.; Chisari, F.V. Cd8(+) t cells mediate viral clearance and disease pathogenesis during acute hepatitis b virus infection. J. Virol. 2003, 77, 68–76. [Google Scholar] [CrossRef]
- Masson, J.J.R.; Billings, H.W.W.; Palmer, C.S. Metabolic reprogramming during hepatitis b disease progression offers novel diagnostic and therapeutic opportunities. Antivir. Chem. Chemother. 2017, 25, 53–57. [Google Scholar] [CrossRef]
- Sartorius, K.; An, P.; Winkler, C.; Chuturgoon, A.; Li, X.; Makarova, J.; Kramvis, A. The epigenetic modulation of cancer and immune pathways in hepatitis b virus-associated hepatocellular carcinoma: The influence of hbx and mirna dysregulation. Front. Immunol. 2021, 12, 661204. [Google Scholar] [CrossRef]
- Wei, X.; Tang, C.; Lu, X.; Liu, R.; Zhou, M.; He, D.; Zheng, D.; Sun, C.; Wu, Z. Mir-101 targets dusp1 to regulate the tgf-β secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget 2015, 6, 18389–18405. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-X.; Ling, Y.; Wang, H.-Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis. Oncol. 2018, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Duan, L.; Cui, F.; Cao, J.; Xiang, Y.; Tang, Y.; Zhou, L. S100a9 promotes human hepatocellular carcinoma cell growth and invasion through rage-mediated erk1/2 and p38 mapk pathways. Exp. Cell Res. 2015, 334, 228–238. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Nagaraj, S. Myeloid-derived suppressor cells as regulators of the immune system. Nat. Rev. Immunol. 2009, 9, 162–174. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.X.; Taraboulos, A.; Ou, J.H.; Yen, T.S. Activation of class i major histocompatibility complex gene expression by hepatitis b virus. J. Virol. 1990, 64, 4025–4028. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, J.K.; Kim, H.J.; Ahn, J.K. Hepatitis b virus x protein sensitizes uv-induced apoptosis by transcriptional transactivation of fas ligand gene expression. IUBMB Life 2005, 57, 651–658. [Google Scholar] [CrossRef]
- Fu, S.; Zhou, R.-R.; Li, N.; Huang, Y.; Fan, X.-G. Hepatitis b virus x protein in liver tumor microenvironment. Tumour Biol. 2016, 37, 15371–15381. [Google Scholar] [CrossRef]
- Wang, D.Y.; Zou, L.P.; Liu, X.J.; Zhu, H.G.; Zhu, R. Chemokine expression profiles of human hepatoma cell lines mediated by hepatitis b virus x protein. Pathol. Oncol. Res. POR 2016, 22, 393–399. [Google Scholar] [CrossRef]
- Gómez-Moreno, A.; Garaigorta, U. Hepatitis b virus and DNA damage response: Interactions and consequences for the infection. Viruses 2017, 9, 304. [Google Scholar] [CrossRef]
- Forgues, M.; Difilippantonio, M.J.; Linke, S.P.; Ried, T.; Nagashima, K.; Feden, J.; Valerie, K.; Fukasawa, K.; Wang, X.W. Involvement of crm1 in hepatitis b virus x protein-induced aberrant centriole replication and abnormal mitotic spindles. Mol. Cell. Biol. 2003, 23, 5282–5292. [Google Scholar] [CrossRef]
- Ahodantin, J.; Bou-Nader, M.; Cordier, C.; Mégret, J.; Soussan, P.; Desdouets, C.; Kremsdorf, D. Hepatitis b virus x protein promotes DNA damage propagation through disruption of liver polyploidization and enhances hepatocellular carcinoma initiation. Oncogene 2019, 38, 2645–2657. [Google Scholar] [CrossRef] [PubMed]
- Groisman, I.J.; Koshy, R.; Henkler, F.; Groopman, J.D.; Alaoui-Jamali, M.A. Downregulation of DNA excision repair by the hepatitis b virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis 1999, 20, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Königer, C.; Wingert, I.; Marsmann, M.; Rösler, C.; Beck, J.; Nassal, M. Involvement of the host DNA-repair enzyme tdp2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis b viruses. Proc. Natl. Acad. Sci. USA 2014, 111, E4244–E4253. [Google Scholar] [CrossRef] [PubMed]
- Decorsière, A.; Mueller, H.; van Breugel, P.C.; Abdul, F.; Gerossier, L.; Beran, R.K.; Livingston, C.M.; Niu, C.; Fletcher, S.P.; Hantz, O.; et al. Hepatitis b virus x protein identifies the smc5/6 complex as a host restriction factor. Nature 2016, 531, 386–389. [Google Scholar] [CrossRef]
- Chen, Y.; Ning, J.; Cao, W.; Wang, S.; Du, T.; Jiang, J.; Feng, X.; Zhang, B. Research progress of txnip as a tumor suppressor gene participating in the metabolic reprogramming and oxidative stress of cancer cells in various cancers. Front. Oncol. 2020, 10, 1861. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.I.; Hwang, J.M.; Im, J.H.; Lee, Y.I.; Kim, N.S.; Kim, D.G.; Yu, D.Y.; Moon, H.B.; Park, S.K. Human hepatitis b virus-x protein alters mitochondrial function and physiology in human liver cells. J. Biol. Chem. 2004, 279, 15460–15471. [Google Scholar] [CrossRef]
- Xie, N.; Chen, X.; Zhang, T.; Liu, B.; Huang, C. Using proteomics to identify the hbx interactome in hepatitis b virus: How can this inform the clinic? Expert Rev. Proteom. 2014, 11, 59–74. [Google Scholar] [CrossRef]
- Lee, S.-H.; Cha, E.-J.; Lim, J.-E.; Kwon, S.-H.; Kim, D.-H.; Cho, H.; Han, K.-H. Structural characterization of an intrinsically unfolded mini-hbx protein from hepatitis b virus. Mol. Cells 2012, 34, 165–169. [Google Scholar] [CrossRef]
- Slagle, B.L.; Andrisani, O.M.; Bouchard, M.J.; Lee, C.G.; Ou, J.H.; Siddiqui, A. Technical standards for hepatitis b virus x protein (hbx) research. Hepatology 2015, 61, 1416–1424. [Google Scholar] [CrossRef]
- Horng, J.-H.; Lin, W.-H.; Wu, C.-R.; Lin, Y.-Y.; Wu, L.-L.; Chen, D.-S.; Chen, P.-J. Hbv x protein-based therapeutic vaccine accelerates viral antigen clearance by mobilizing monocyte infiltration into the liver in hbv carrier mice. J. Biomed. Sci. 2020, 27, 70. [Google Scholar] [CrossRef]
- Shin, D.; Kim, S.I.; Kim, M.; Park, M. Efficient inhibition of hepatitis b virus replication by small interfering rnas targeted to the viral x gene in mice. Virus Res. 2006, 119, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.W.; Ng, I.O. Knock-down of hepatitis b virus x protein reduces the tumorigenicity of hepatocellular carcinoma cells. J. Pathol. 2006, 208, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Xiong, H.L.; Cao, J.L.; Wang, S.J.; Guo, X.R.; Lin, B.Y.; Zhang, Y.; Zhao, J.H.; Wang, Y.B.; Zhang, T.Y.; et al. A cell-penetrating whole molecule antibody targeting intracellular hbx suppresses hepatitis b virus via trim21-dependent pathway. Theranostics 2018, 8, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.T.; Hu, J.L.; Ren, J.H.; Yu, H.B.; Zhong, S.; Wai Wong, V.K.; Kwan Law, B.Y.; Chen, W.X.; Xu, H.M.; Zhang, Z.Z.; et al. Dicoumarol, an nqo1 inhibitor, blocks cccdna transcription by promoting degradation of hbx. J. Hepatol. 2021, 74, 522–534. [Google Scholar] [CrossRef]
- Sekiba, K.; Otsuka, M.; Ohno, M.; Yamagami, M.; Kishikawa, T.; Suzuki, T.; Ishibashi, R.; Seimiya, T.; Tanaka, E.; Koike, K. Inhibition of hbv transcription from cccdna with nitazoxanide by targeting the hbx-ddb1 interaction. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 297–312. [Google Scholar] [CrossRef]
- van de Klundert, M.A.A.; Zaaijer, H.L.; Kootstra, N.A. Identification of fda-approved drugs that target hepatitis b virus transcription. J. Viral Hepat. 2016, 23, 191–201. [Google Scholar] [CrossRef]
- Lok, A.S.; Pan, C.Q.; Han, S.H.; Trinh, H.N.; Fessel, W.J.; Rodell, T.; Massetto, B.; Lin, L.; Gaggar, A.; Subramanian, G.M.; et al. Randomized phase ii study of gs-4774 as a therapeutic vaccine in virally suppressed patients with chronic hepatitis b. J. Hepatol. 2016, 65, 509–516. [Google Scholar] [CrossRef]
- Boni, C.; Janssen, H.L.A.; Rossi, M.; Yoon, S.K.; Vecchi, A.; Barili, V.; Yoshida, E.M.; Trinh, H.; Rodell, T.C.; Laccabue, D.; et al. Combined gs-4774 and tenofovir therapy can improve hbv-specific t-cell responses in patients with chronic hepatitis. Gastroenterology 2019, 157, 227–241.e227. [Google Scholar] [CrossRef]
- Bartoli, A.; Gabrielli, F.; Tassi, A.; Cursaro, C.; Pinelli, A.; Andreone, P. Treatments for hbv: A glimpse into the future. Viruses 2021, 13, 1767. [Google Scholar] [CrossRef]
- Miri, S.M.; Alavian, S.M. Risk factors of hepatitis b infection: Health policy makers should be aware of their importance in each community. Hepat. Mon. 2011, 11, 238–239. [Google Scholar]
- Sivasudhan, E.; Blake, N.; Lu, Z.-L.; Meng, J.; Rong, R. Dynamics of m6a rna methylome on the hallmarks of hepatocellular carcinoma. Front. Cell Dev. Biol. 2021, 9, 642443. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Wong, C.-M. The emerging roles of n6-methyladenosine (m6a) deregulation in liver carcinogenesis. Mol. Cancer 2020, 19, 44. [Google Scholar] [CrossRef] [PubMed]
- Qu, N.; Bo, X.; Li, B.; Ma, L.; Wang, F.; Zheng, Q.; Xiao, X.; Huang, F.; Shi, Y.; Zhang, X. Role of n6-methyladenosine (m6a) methylation regulators in hepatocellular carcinoma. Front. Oncol. 2021, 11, 4134. [Google Scholar] [CrossRef]
- Kim, G.-W.; Siddiqui, A. The role of n6-methyladenosine modification in the life cycle and disease pathogenesis of hepatitis b and c viruses. Exp. Mol. Med. 2021, 53, 339–345. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.-W.; Siddiqui, A. Hepatitis b virus x protein recruits methyltransferases to affect cotranscriptional n6-methyladenosine modification of viral/host rnas. Proc. Natl. Acad. Sci. USA 2021, 118, e2019455118. [Google Scholar] [CrossRef]
- Kim, G.W.; Siddiqui, A. Hepatitis b virus x (hbx) protein expression is tightly regulated by n6-methyladenosine modification of its mrna. J. Virol. 2021, Jvi0165521. [Google Scholar] [CrossRef]
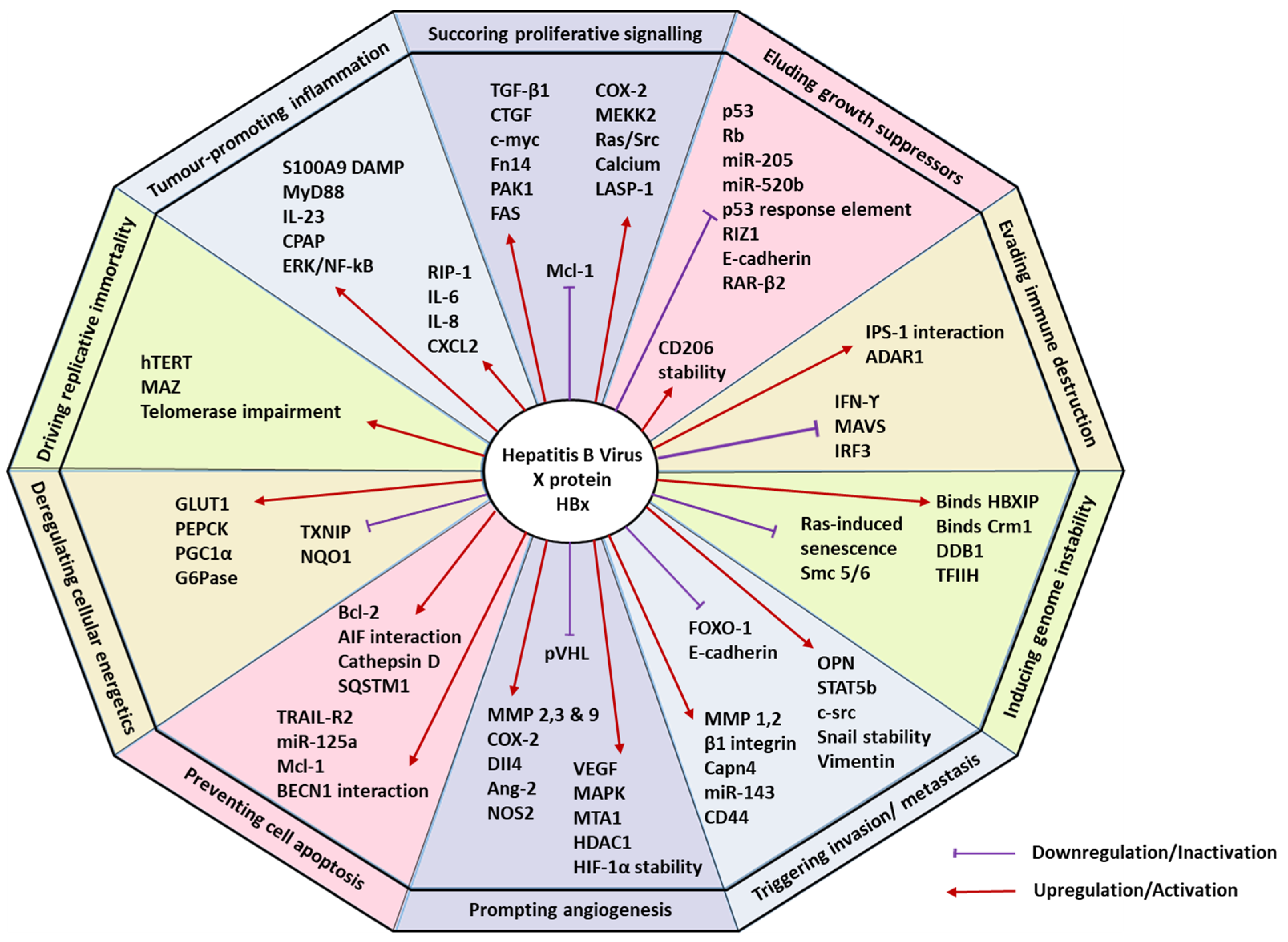
| HCC Hallmark | HBx Activity | Study Design | Ref. |
|---|---|---|---|
| Sustaining proliferative signalling | Activates stellate cells and elevates transforming-growth factor β1 (TGF-β1) and connective tissue growth factor (CTGF) | In vitro co-culture with LX-2 cells and stable QSG7701-HBx cell line | Guo et al. [23] |
| Five-fold elevated expression of c-myc | In vitro human hepatoma cell lines Huh7 and IHHs and in vivo X15–myc transgenic mouse model In vivo HBx-transgenic mice with c-myc driven by woodchuck hepatitis virus (WHV) | Shukla & Kumar et al. Terradillos et al. [24,25] | |
| HBx-SMYD3 interaction, guided by the downstream target gene c-myc | In vitro HBx-expressing HepG2 cells and HBV containing HepG2.2.15 cells | Yang et al. [26] | |
| Enhanced expression of fibroblast growth factor-inducible 14 (fn14) | In vitro human fibroblasts and HCC cells and In vivo HBx-transgenic mice. | Feng et al. [27] | |
| Disrupts cell cycle progression by upregulating p21 and p27 | In vivo pX expressing primary mouse hepatocytes | Qiao et al. [28] | |
| Elevates serine/threonine p21 activated kinase 1 (PAK1) | In vivo tumour xenografts in mice and in vitro human hepatoma cells with pHBV1.3 | Xu et al. [29] | |
| Upregulates transcription of fatty acid synthase (FAS), mediated by 5-lipoxygenase (5-LOX) | In vitro HBx-expressing human hepatoma HepG2 and H7402 cells | Wang et al. [30] | |
| Upregulates cyclooxygenase (COX-2) and MERK/ERK kinase 2 (MEKK2) | In vitro HBx-expressing L-O2 and H7402 cell lines. | Shan et al. [31] | |
| Suppressed anti-apoptotic protein Mcl-1 | In vitro Chang liver cells transiently transfected with HBx (CHL-X) | Lee et al. [32] | |
| Activates Ras and Src kinase | In vitro human hepatoma Hep3B cells with transiently transfected HA-tagged HBx | Noh et al. [33] | |
| Enhances cytosolic calcium levels | In vitro HepG2 cells transfected with full length HBx | Yang & Bouchard, 2012 [34] | |
| Elevates adhesion protein LASP-1 via PI3K pathway | In vitro HBx stably transfected HepG2 and Huh-7 cells | Tang et al. [35] | |
| Eluding growth suppressors | Partial sequestration of p53 causing G1 arrest | In vitro HBx expressing human fibroblasts, HepG2 cells and liver tissue from patients | Elmore et al. [36] |
| Inhibition of p53 response element | In vitro HBx transiently transfected human Calu-6 cells | Truant et al. [37] | |
| Inactivates Rb gene promoter | In vitro HepG2 and Hela cells | Choi et al. [38] | |
| Confers stability to replication initiator CDC6 | In vitro HBx-expressing human hepatoma cells Huh7, HepG2. In vivo X15-myc transgenic mouse model | Pandey & Kumar, 2012 [39] | |
| miR-205 inhibition via promoter hypermethylation | In vitro HBx-expressing hepatoma cell lines and In vivo HBx- transgenic mice and patient samples | Zhang et al. [40] | |
| Controls miR-520b and hepatitis B X-interacting protein (HBXIP) | In vitro HBx-expressing human hepatoma cells and In vivo nude mice transplantation and patient samples | Zhang et al. [41] | |
| Represses RIZ1 via hypermethylation | In vitro HBx-expressing human hepatoma cells and patient samples | Zhao et al. [42] | |
| Suppresses E-cadherin tumour suppressor | In vitro HBx-expressing HepG2 cell line | Lee et al. [43] | |
| Hypermethylates p16 via pRb-E2f pathway | HBV-HCC patient and tissue specimens | Zhu et al. [44] | |
| Downregulates retinoic acid receptor-beta 2 (RAR-β2) | In vitro HBx-expressing HepG2 cells | Jung et al. [45] | |
| Resisting cell death | Pro-apoptosis | ||
| Induces the expression of TRAIL-R2 (DR5) | In vitro HBx-expressing Huh-7 cells | Kong et al. [46] | |
| Upregulation of miR-125a | In vitro HBx-expressing HepG2 and LO-2 liver cells | Zhang et al. [47] | |
| Anti-apoptosis | |||
| Induces myeloid cell leukemia-1 (Mcl-1) and B cell lymphoma 2 (Bcl-2) | In vitro HBx-expressing HPCs (HP14.5) cells | Shen et al. [48] | |
| Interacts with apoptosis-inducing factor (AIF) and AIF-homologue mitochondrian-associated inducer of death (AMID) | In vitro HBx-expressing HepG2 cells | Liu et al. [49] | |
| Autophagy | |||
| Upregulating SQSTM1 and lysosomal aspartic protease cathepsin D | In vitro HBx-expressing Huh-7 cells and human tissue specimens | Liu et al. [50] | |
| Interacts with BECN1 (Beclin 1) | In vitro HBx-expressing HepG2 and SK-Hep-1 | Son et al. [51] | |
| PI3K-Akt-mTOR pathway | In vitro HBx-expressing HepG2 cells | Wang et al. [52] | |
| Facilitating replicative immortality | Activates human telomerase reverse transcriptase (hTERT) | In vitro HBx-expressing HepG2 and QBC939 cell lines | Zhang et al. Qu et al. [53,54] |
| MAZ binding aided telomerase impairment | In vitro HBx-expressing H7402 hepatoma cells | Su et al. [55] | |
| Prompting angiogenesis | Upregulates VEGF mRNA expression and stabilizes HIF-1α | In vitro HBx-expressing human HepG2 and mouse Hepa 1–6 HCC cell lines In vitro ChangX-34 and HBx transgenic mice model In vitro HBx-expressing HEK293 cells | Lee et al. Moon et al. Yun et al. [56,57,58] |
| Mitigates binding of von Hippel-Lindau (pVHL) | In vitro HBx-expressing HEK293 cells | Moon et al. [58] | |
| Activates p42/44 mitogen-activated protein kinases (MAPK) | In vitro HBx-expressing human hepatoma cell lines and HBx transgenic mice model | Yoo et al. [59] | |
| Upregulates metastasis-associated protein 1 (MTA1) and histone deacetylase (HDAC1) | In vitro Chang X-34 cells, HBx transgenic mice and patient samples | Yoo et al. [60] | |
| Overexpresses matrix metalloproteinases (MMP) 2,3 and 9 | In vitro HBx-expressing human Chang cell lines and murine AML-12 liver cell line In vitro HBx-expressing human hepatoma cell lines In vitro HBx-expressing HepG2 cell line and xenograft mice model | Lara-Pezzi et al. [61] Yu et al. [62] Liu et al. [63] | |
| Induces COX-2 enzyme | In vitro HBx-expressing Hep3B cell line and patient samples | Cheng et al. [64] | |
| Mediates Dll4 upregulation | In vitro HBx-expressing human hepatoma cell lines and HCC patient samples | Kongkavitoon et al. [65] | |
| Stimulates Ang-2 isoform | In vitro HBx-expressing Chang cell line and rat hepatic stellate cells CFSC-2G, THP1 promonocyte cell line and patient and tissue specimens | Sanz-Cameno et al. [66] | |
| Induces nitrogen oxide synthase 2 (NOS2) | In vitro HBV-expressing HepG2 and HepG2.2.15 cell lines and patient and tissue specimens | Majano et al. [67] | |
| Triggering invasion and metastasis | Promotes production of MMPs 1 and 2 and disrupts adherens junctions | In vitro HBx-expressing human Chang cell lines and murine AML-12 liver cell line HCC patient tissue specimens | Lara-Pezzi et al. [61] Giannelli et al. [68] |
| Modifies α integrin subunits and activates β1 integrin subunits | In vitro HBx-expressing Chang cell line | Lara-Pezzi et al. [69] | |
| Upregulation of Capn4 via nuclear factor-kB/p65 | In vitro HBx-expressing HepG2 and H7402 cell lines | Zhang et al. [70] | |
| Promotes tumour stemness via impaired FOXO1 and β-catenin nuclear translocation | In vitro HBx-expressing cell lines and In vivo tumour xenograft mice model | Lin et al. [71] | |
| Elevates miRNA-143 (miR-143) | In vitro HepG2 and Huh7 cell lines and In vivo HBx transgenic mice and patient tissue samples | Zhang et al. [72] | |
| Activates cell-surface adhesion molecule CD44 | In vitro HBx expressing Chang cell line | Lara-Pezzi et al. [73] | |
| Activates ossteopontin (OPN) through 5-LOX | In vitro HBx-expressing HepG2 cell line | Zhang et al. [74] | |
| Activates (STAT5b) and c-Src proto-oncogene | In vitro HBx-expressing Huh7 and HCC patient samples In vitro HBx-expressing SMMC-7721 cell line | Lee et al. [75] Yang et al. [76] | |
| Stabilizes Snail protein | In vitro human hepatoma Huh7 and Chang cell lines and patient samples | Liu et al. [77] | |
| Induces expression of vimentin | In vitro HBx-expressing HepG2 and Huh7 cell lines and patient samples | You et al. [78] | |
| Represses E-cadherin | In vitro HBx-expressing HepG2 cell line and patient samples | Arzumanyan et al. [79] | |
| Evading immune destruction | Induces apoptosis in HBV-specific CD8+ T cells | In vitro HBx-expressing primary hepatocytes | Lee et al. [80] |
| Interacts with IPS-1 and inhibits interferon-ϒ | In vitro HBx-expressing HepG2 and In vivo HBx transgenic mice | Kumar et al. [81] | |
| Inhibits IRF3 and associations between VISA and RIG-1/MDA5 | In vitro BHK and HEK 293 cell lines | Wang et al. [82] | |
| Degradation of MAVS via Lys(136) ubiquitination | In vitro human hepatoma cell lines, In vivo HBx knock-in mice model and liver tumour samples | Wei et al. [83] | |
| Promotes RNA adenosine deaminase ADAR1 | In vitro HepG2.2.15 and NTCP-expressing HepG2 and Huh7, In vivo mice model | Wang et al. [84] | |
| Tumour-promoting inflammation | Induces RIP-1 and aids in activation of cytokines IL-6, IL-8 and CXCL2 | In vitro HBx-expressing LO-2 hepatocytes | Xie & Huang [85] |
| Induces S100A9 DAMP protein | In vitro HBx-expressing human hepatoma cell lines, In vivo HBx transgenic mice model and patient samples | Duan et al. [86] | |
| Activates signal transduction adaptor MyD88, including, IRAK-1, NF-kB and ERKs/p38 | In vitro HBx-expressing human hepatic L02 cells and human hepatoma SMMC-7721 | Xiang et al. [87] | |
| Activates ERK/NF-kB pathway and IL-23 subunits | In vitro HepG2 and Huh7, normal hepatocyte Chang liver and HL-7702, and HepG2.2.15 cells lines and patient samples | Xia et al. [88] | |
| Interacts with CPAP regulator | In vitro HBx- and NTCP-expressing human hepatoma cell lines and In vivo xenograft mice model | Yen et al. [89] | |
| Inducing genomic instability | Binds HBXIP | In vitro Hela and mouse embryonic fibroblast (MEF) cell lines and In vivo liver regeneration mice model | Fujii et al. [90] |
| Binds Crm1 with the NES domain on HBx | In vitro HBx-expressing Hep3B and primary human fibroblast cell lines | Forgues et al. [91] | |
| Binds DDB proteins | In vitro wild-type or mutant HBx-expressing HepG2 cell lines | Becker et al. [92] | |
| Disrupts Ras-induced senescence | In vitro HBx-expressing human primary fibroblasts BJ and TIG3 cell lines and In vivo mice model | Oishi et al. [93] | |
| Stimulates DNA helicase catalytic activity of TFIIH subunits | In vitro HBx-expressing Hela cells and yeast model | Qadri et al. [94] | |
| Degradation of Smc 5/6 | In vitro wild type and mutant HBx-expressing HepG2, HepAD38, HepG2-NTCP cell lines | Murphy et al. [95] | |
| Deregulating cellular energetics | Downregulates TXNIP protein | In vitro HBx-expressing MIHA and LO-2 cell lines, In vivo mice model and HCC patient samples | Zhang et al. [96] |
| Elevates expression of GLUT1 | In vitro human hepatoma HepG2 cell line and HCC patient samples | Zhou et al. [97] | |
| Overexpression of PEPCK, PGC1α, and G6Pase | In vitro HBx-expressing HepG2 cell line, In vivo HBx transgenic mice and HCC patient samples | Shin et al. [98] | |
| Downregulates NQO1 enzyme | In vitro HBx-expressing Huh7 cell line | Jung et al. [99] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sivasudhan, E.; Blake, N.; Lu, Z.; Meng, J.; Rong, R. Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review. Cells 2022, 11, 741. https://doi.org/10.3390/cells11040741
Sivasudhan E, Blake N, Lu Z, Meng J, Rong R. Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review. Cells. 2022; 11(4):741. https://doi.org/10.3390/cells11040741
Chicago/Turabian StyleSivasudhan, Enakshi, Neil Blake, Zhiliang Lu, Jia Meng, and Rong Rong. 2022. "Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review" Cells 11, no. 4: 741. https://doi.org/10.3390/cells11040741
APA StyleSivasudhan, E., Blake, N., Lu, Z., Meng, J., & Rong, R. (2022). Hepatitis B Viral Protein HBx and the Molecular Mechanisms Modulating the Hallmarks of Hepatocellular Carcinoma: A Comprehensive Review. Cells, 11(4), 741. https://doi.org/10.3390/cells11040741