Divalent Metal Ions Boost Effect of Nucleic Acids Delivered by Cell-Penetrating Peptides
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Cultivation
2.3. Nanoparticle Formation
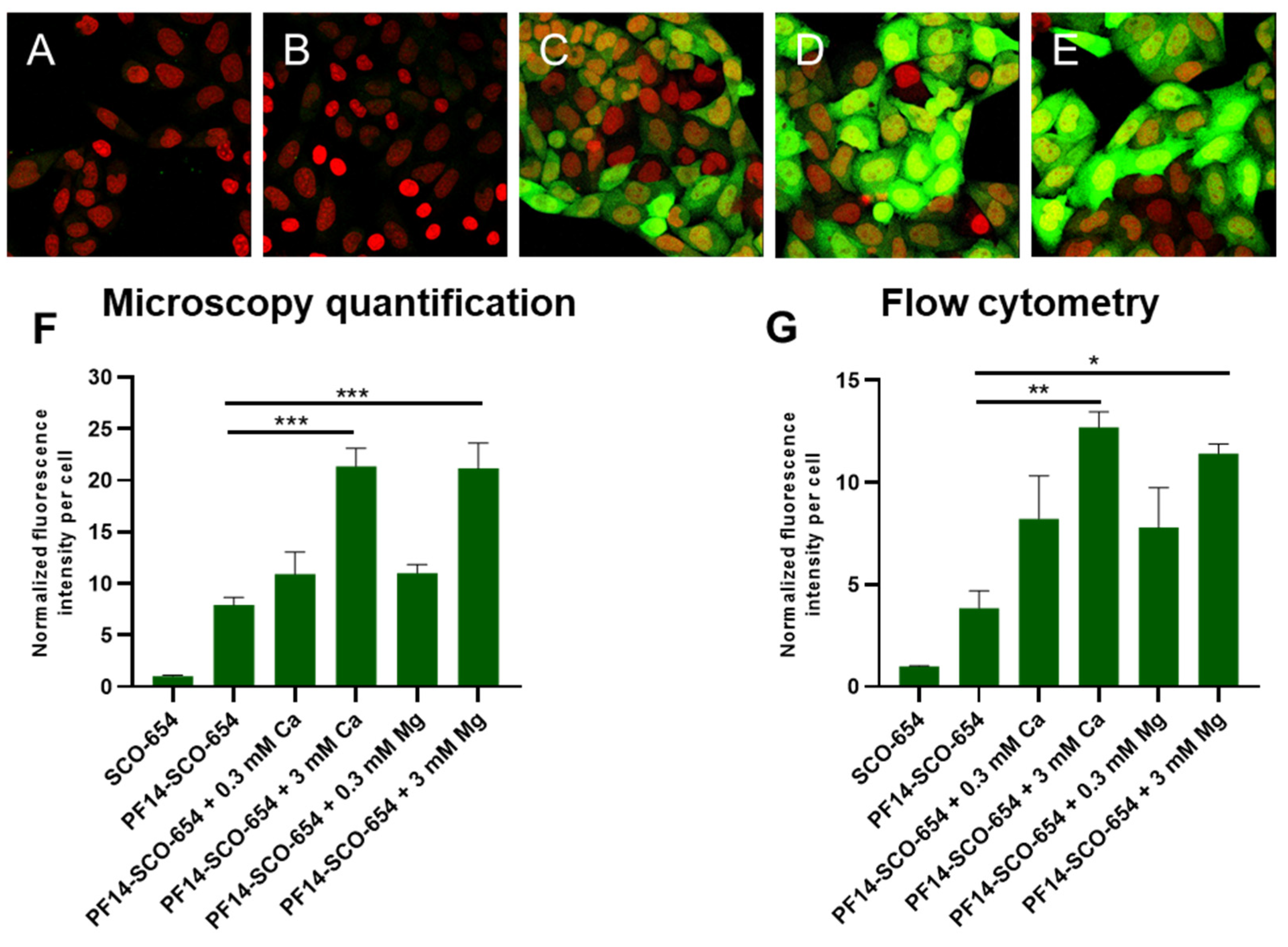
2.4. Confocal Microscopy
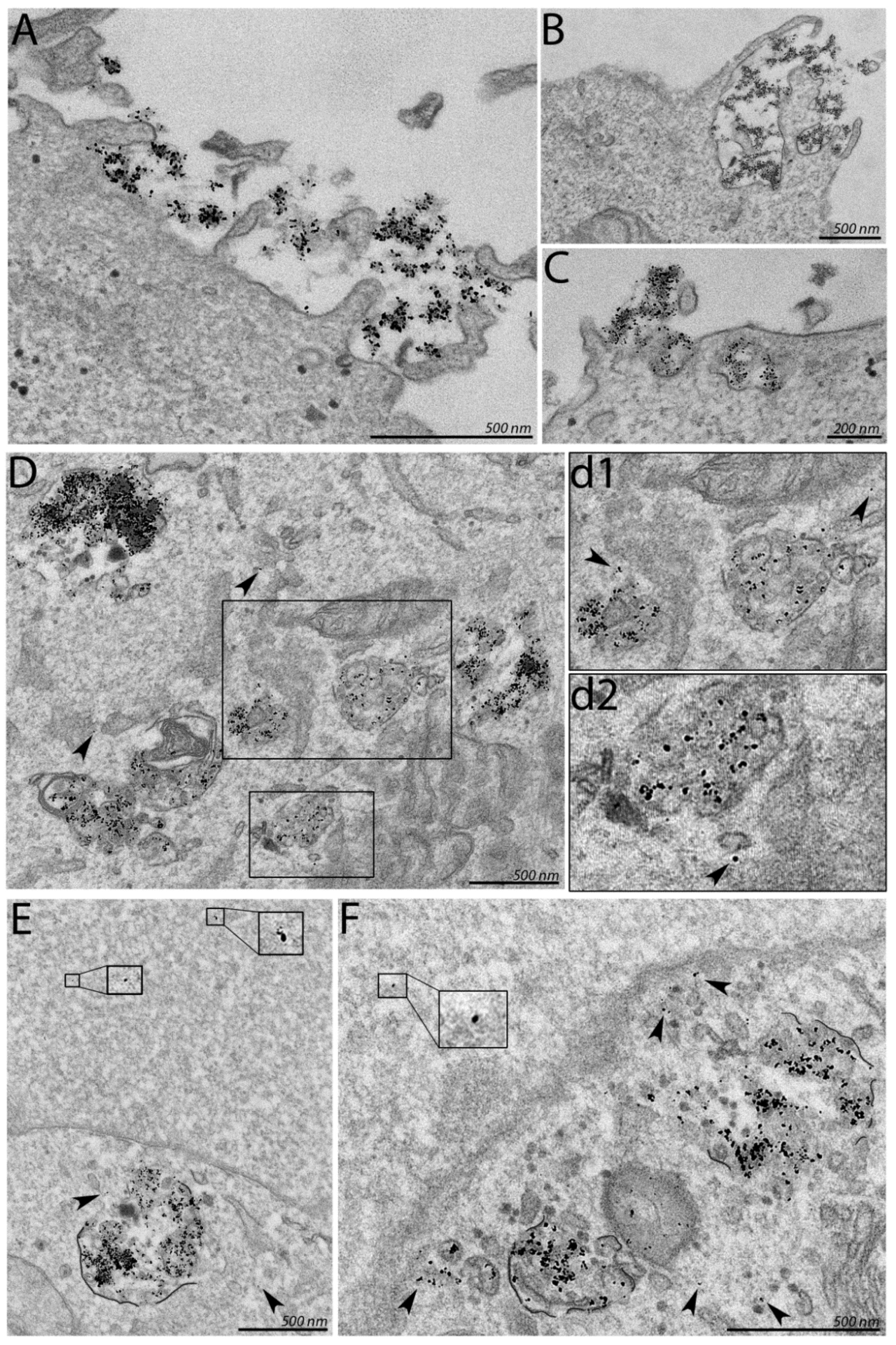
2.5. Electron Microscopy of Nanoparticles
2.6. Flow Cytometry
2.7. Luminescence Measurement
2.8. Assessment of the Role of Endosomal Escape and SR-As in Efficiency of PF14-SCO Nanoparticles
2.9. PCR Analysis
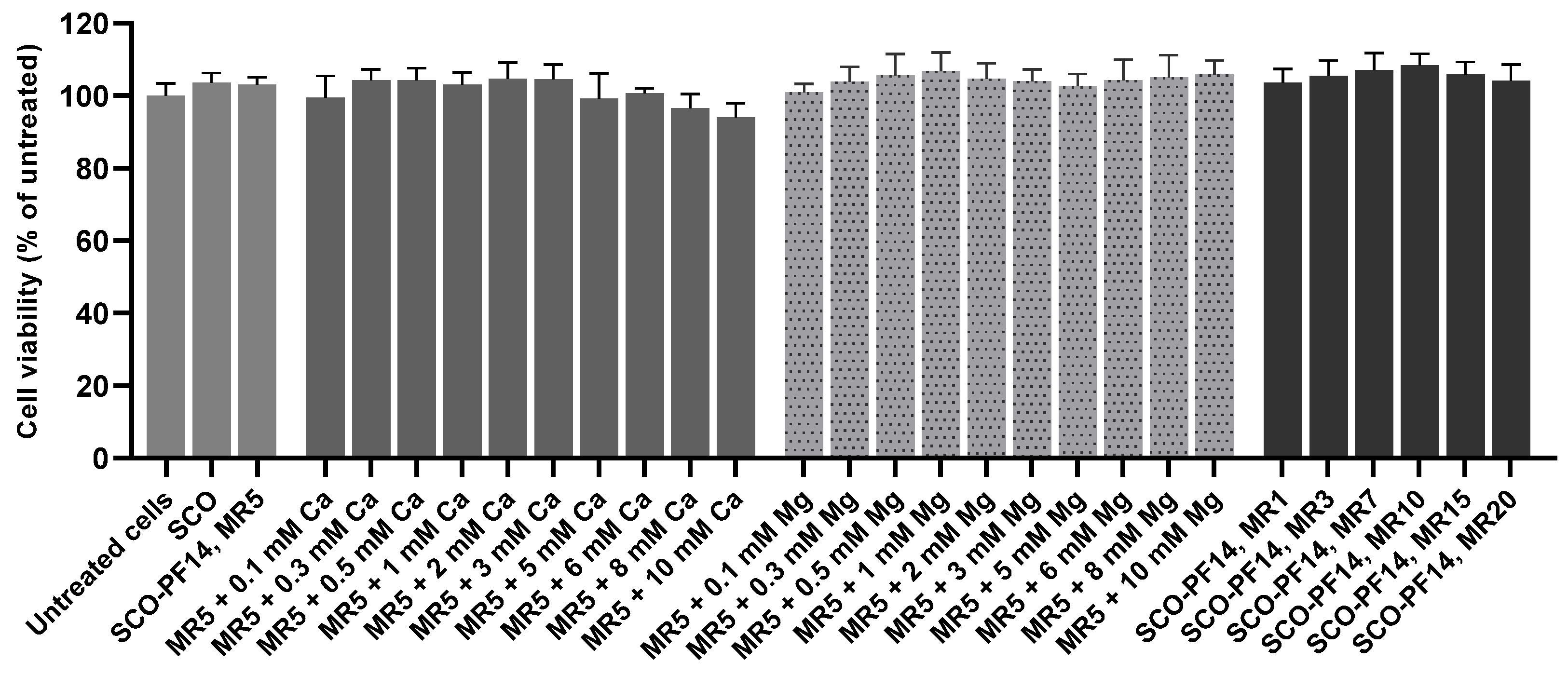
2.10. Nanoparticle Cytotoxicity Assessment
2.11. Measurement of Nanoparticle Size and Zeta Potential
2.12. Statistical Analysis
3. Results
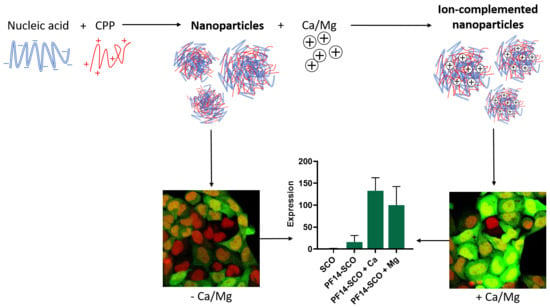
3.1. Addition of Divalent Metal Ions Enhances Splice-Correcting Activity of SCO in HeLa pLuc 705 Reporter Cells
3.2. Adding Calcium and Magnesium Ions to SCO-PF14 Nanoparticles Yields Nanoparticles with Similar Physical Properties and Does Not Impair Viability of the Cells
3.3. Association of PF14-SCO Complexes with Cells and Intracellular Distribution of SCO Changes upon Addition of Divalent Metal Ions into Nanoparticles
3.4. Calcium Ions Increase the Efficiency of Different CPPs
3.5. Calcium Ions Reduce the Effect of Endosome-Destabilizing Agent Chloroquine on Splicing Correction in Concentration-Dependent Manner
3.6. SR-A Inhibitors Reduce Biological Activity of Ion-Complemented Nanoparticles in a Concentration-Dependent Manner
3.7. Addition of Divalent Metal Ions Enhances RNA Interference Effect of siRNA-PF14 Nanoparticles in U87-luc2 Reporter Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Goncalves, G.A.R.; Paiva, R.D.A. Gene therapy: Advances, challenges and perspectives. Einstein-Sao Paulo 2017, 15, 369–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. Food and Drug Administration. What is Gene Therapy? Available online: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/what-gene-therapy#footnote1 (accessed on 29 September 2021).
- Baden, L.R.; El Sahly, H.M.; Essink, B.; Kotloff, K.; Frey, S.; Novak, R.; Diemert, D.; Spector, S.A.; Rouphael, N.; Creech, B.; et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021, 384, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Marc, G.P.; Moreira, E.D.; Zerbini, E.D.; et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Romero, N.B.; Braun, S.; Benveniste, O.; Leturcq, F.; Hogrel, J.-Y.; Morris, G.E.; Barois, A.; Eymard, B.; Payan, C.; Ortega, V.; et al. Phase I Study of Dystrophin Plasmid-Based Gene Therapy in Duchenne/Becker Muscular Dystrophy. Hum. Gene Ther. 2004, 15, 1065–1076. [Google Scholar] [CrossRef]
- Mendell, J.R.; Campbell, K.; Rodino-Klapac, L.; Sahenk, Z.; Shilling, C.; Lewis, S.; Bowles, D.; Gray, S.; Li, C.; Galloway, G.; et al. Dystrophin Immunity in Duchenne’s Muscular Dystrophy. N. Engl. J. Med. 2010, 363, 1429–1437. [Google Scholar] [CrossRef] [Green Version]
- Urgard, E.; Brjalin, A.; Langel, Ü.; Pooga, M.; Rebane, A.; Annilo, T. Comparison of Peptide- and Lipid-Based Delivery of miR-34a-5p Mimic into PPC-1 Cells. Nucleic Acid Ther. 2017, 27, 295–302. [Google Scholar] [CrossRef]
- Song, E.; Lee, S.-K.; Wang, J.; Ince, N.; Ouyang, N.; Min, J.; Chen, J.; Shankar, P.; Lieberman, J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003, 9, 347–351. [Google Scholar] [CrossRef]
- Bestas, B.; Moreno, P.; Blomberg, K.E.M.; Mohammad, D.K.; Saleh, A.F.; Sutlu, T.; Nordin, J.; Guterstam, P.; Gustafsson, M.O.; Kharazi, S.; et al. Splice-correcting oligonucleotides restore BTK function in X-linked agammaglobulinemia model. J. Clin. Investig. 2014, 124, 4067–4081. [Google Scholar] [CrossRef] [Green Version]
- Scoto, M.; Finkel, R.; Mercuri, E.; Muntoni, F. Genetic therapies for inherited neuromuscular disorders. Lancet Child Adolesc. Health 2018, 2, 600–609. [Google Scholar] [CrossRef]
- Hua, Y.; Sahashi, K.; Hung, G.; Rigo, F.; Passini, M.A.; Bennett, C.F.; Krainer, A.R. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010, 24, 1634–1644. [Google Scholar] [CrossRef] [Green Version]
- Hair, P.; Cameron, F.; McKeage, K. Mipomersen Sodium: First Global Approval. Drugs 2013, 73, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-H.; Cho, M.-J.; Kole, R. Up-Regulation of Luciferase Gene Expression with Antisense Oligonucleotides: Implications and Applications in Functional Assay Development. Biomaterials 1998, 37, 6235–6239. [Google Scholar] [CrossRef] [PubMed]
- Sazani, P.; Kang, S.-H.; Maier, M.A.; Wei, C.; Dillman, J.; Summerton, J.; Manoharan, M.; Kole, R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001, 29, 3965–3974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dagle, J.M.; Weeks, D.L.; Walder, J.A. Pathways of Degradation and Mechanism of Action of Antisense Oligonucleotides inXenopus laevisEmbryos. Antisense Res. Dev. 1991, 1, 11–20. [Google Scholar] [CrossRef]
- Boisguérin, P.; Deshayes, S.; Gait, M.J.; O’Donovan, L.; Godfrey, C.; Betts, C.A.; Wood, M.J.A.; Lebleu, B. Delivery of therapeutic oligonucleotides with cell penetrating peptides. Adv. Drug Deliv. Rev. 2015, 87, 52–67. [Google Scholar] [CrossRef]
- Monia, B.; Lesnik, E.; Gonzalez, C.; Lima, W.; McGee, D.; Guinosso, C.; Kawasaki, A.; Cook, P.; Freier, S. Evaluation of 2‘-modified oligonucleotides containing 2‘-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993, 268, 14514–14522. [Google Scholar] [CrossRef]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef] [Green Version]
- Mellet, C.O.; Fernández, J.M.G.; Benito, J.M. Cyclodextrin-based gene delivery systems. Chem. Soc. Rev. 2010, 40, 1586–1608. [Google Scholar] [CrossRef]
- Pooga, M.; Langel, U. Classes of Cell-Penetrating Peptides; Humana Press: New York, NY, USA, 2015; Volume 1324, pp. 3–28. [Google Scholar] [CrossRef]
- Filion, M.C.; Phillips, N.C. Toxicity and immunomodulatory activity of liposomal vectors formulated with cationic lipids toward immune effector cells. Biochim. Biophys. Acta-Biomembr. 1997, 1329, 345–356. [Google Scholar] [CrossRef] [Green Version]
- Kurrikoff, K.; Veiman, K.-L.; Künnapuu, K.; Peets, E.M.; Lehto, T.; Pärnaste, L.; Arukuusk, P.; Langel, Ü. Effective in vivo gene delivery with reduced toxicity, achieved by charge and fatty acid -modified cell penetrating peptide. Sci. Rep. 2017, 7, 17056. [Google Scholar] [CrossRef]
- Xie, J.; Bi, Y.; Zhang, H.; Dong, S.; Teng, L.; Lee, R.J.; Yang, Z. Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application. Front. Pharmacol. 2020, 11, 697. [Google Scholar] [CrossRef] [PubMed]
- Saar, K.; Lindgren, M.; Hansen, M.; Eiríksdóttir, E.; Jiang, Y.; Rosenthal-Aizman, K.; Sassian, M.; Langel, Ü. Cell-penetrating peptides: A comparative membrane toxicity study. Anal. Biochem. 2005, 345, 55–65. [Google Scholar] [CrossRef] [PubMed]
- EL Andaloussi, S.; Lehto, T.; Mäger, I.; Rosenthal-Aizman, K.; Oprea, I.I.; Simonson, O.E.; Sork, H.; Ezzat, K.; Copolovici, D.M.; Kurrikoff, K.; et al. Design of a peptide-based vector, PepFect6, for efficient delivery of siRNA in cell culture and systemically in vivo. Nucleic Acids Res. 2011, 39, 3972–3987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ezzat, K.; El Andaloussi, S.; Zaghloul, E.; Lehto, T.; Lindberg, S.; Moreno, P.; Viola, J.R.; Magdy, T.; Abdo, R.; Guterstam, P.; et al. PepFect 14, a novel cell-penetrating peptide for oligonucleotide delivery in solution and as solid formulation. Nucleic Acids Res. 2011, 39, 5284–5298. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Saltzman, W.M. Synthetic DNA delivery systems. Nat. Biotechnol. 2000, 18, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Juks, C.; Padari, K.; Margus, H.; Kriiska, A.; Etverk, I.; Arukuusk, P.; Koppel, K.; Ezzat, K.; Langel, Ü.; Pooga, M. The role of endocytosis in the uptake and intracellular trafficking of PepFect14–nucleic acid nanocomplexes via class A scavenger receptors. Biochim. Biophys. Acta-Biomembr. 2015, 1848, 3205–3216. [Google Scholar] [CrossRef] [Green Version]
- Ezzat, K.; Helmfors, H.; Tudoran, O.; Juks, C.; Lindberg, S.; Padari, K.; El-Andaloussi, S.; Pooga, M.; Langel, Ü. Scavenger receptor-mediated uptake of cell-penetrating peptide nanocomplexes with oligonucleotides. FASEB J. 2011, 26, 1172–1180. [Google Scholar] [CrossRef]
- Van Asbeck, A.H.; Beyerle, A.; McNeill, H.; Bovee-Geurts, P.H.; Lindberg, S.; Verdurmen, W.P.R.; Hällbrink, M.; Langel, Ü.; Heidenreich, O.; Brock, R. Molecular Parameters of siRNA–Cell Penetrating Peptide Nanocomplexes for Efficient Cellular Delivery. ACS Nano 2013, 7, 3797–3807. [Google Scholar] [CrossRef]
- Srimanee, A.; Arvanitidou, M.; Kim, K.; Hällbrink, M.; Langel, Ü. Cell-penetrating peptides for siRNA delivery to glioblastomas. Peptides 2018, 104, 62–69. [Google Scholar] [CrossRef]
- Carreras-Badosa, G.; Maslovskaja, J.; Periyasamy, K.; Urgard, E.; Padari, K.; Vaher, H.; Tserel, L.; Gestin, M.; Kisand, K.; Arukuusk, P.; et al. NickFect type of cell-penetrating peptides present enhanced efficiency for microRNA-146a delivery into dendritic cells and during skin inflammation. Biomaterials 2020, 262, 120316. [Google Scholar] [CrossRef]
- Lorents, A.; Urgard, E.; Runnel, T.; Wawrzyniak, P.; Langel, Ü.; Pooga, M.; Rebane, A. Functional delivery of miR-146a to human primary keratinocytes and skin equivalents with cell-penetrating peptides. Allergy 2014, 69, 618. [Google Scholar]
- Urgard, E.; Lorents, A.; Klaas, M.; Padari, K.; Viil, J.; Runnel, T.; Langel, K.; Kingo, K.; Tkaczyk, E.; Langel, Ü.; et al. Pre-administration of PepFect6-microRNA-146a nanocomplexes inhibits inflammatory responses in keratinocytes and in a mouse model of irritant contact dermatitis. J. Control. Release 2016, 235, 195–204. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Lehto, T.; Lundin, P.; Langetl, U. Application of PepFect Peptides for the Delivery of Splice-Correcting Oligonucleotides. Methods Mol. Biol. 2011, 683, 361–373. [Google Scholar] [CrossRef]
- Juks, C.; Lorents, A.; Arukuusk, P.; Langel, U.; Pooga, M. Cell-penetrating peptides recruit type A scavenger receptors to the plasma membrane for cellular delivery of nucleic acids. FASEB J. 2016, 31, 975–988. [Google Scholar] [CrossRef] [Green Version]
- Lorents, A.; Kodavali, P.K.; Oskolkov, N.; Langel, U.; Hällbrink, M.; Pooga, M. Cell-penetrating Peptides Split into Two Groups Based on Modulation of Intracellular Calcium Concentration. J. Biol. Chem. 2012, 287, 16880–16889. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Moyle, P.; Toth, I. Endosome Escape Strategies for Improving the Efficacy of Oligonucleotide Delivery Systems. Curr. Med. Chem. 2015, 22, 3326–3346. [Google Scholar] [CrossRef] [PubMed]
- Porosk, L.; Arukuusk, P.; Põhako, K.; Kurrikoff, K.; Kiisholts, K.; Padari, K.; Pooga, M.; Langel, U. Enhancement of siRNA transfection by the optimization of fatty acid length and histidine content in the CPP. Biomater. Sci. 2019, 7, 4363–4374. [Google Scholar] [CrossRef] [Green Version]
- Graham, F.L.; Van Der Eb, A.J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 1973, 52, 456–467. [Google Scholar] [CrossRef]
- Baoum, A.; Xie, S.-X.; Fakhari, A.; Berkland, C. “Soft” calcium crosslinks enable highly efficient gene transfection using TAT peptide. Pharm. Res. 2009, 26, 2619–2629. [Google Scholar] [CrossRef] [Green Version]
- Baoum, A.; Ovcharenko, D.; Berkland, C. Calcium condensed cell penetrating peptide complexes offer highly efficient, low toxicity gene silencing. Int. J. Pharm. 2012, 427, 134–142. [Google Scholar] [CrossRef]
- Baoum, A.A.; Berkland, C. Calcium Condensation of DNA Complexed with Cell-Penetrating Peptides Offers Efficient, Noncytotoxic Gene Delivery. J. Pharm. Sci. 2011, 100, 1637–1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melikov, K.; Hara, A.; Yamoah, K.; Zaitseva, E.; Zaitsev, E.; Chernomordik, L.V. Efficient entry of cell-penetrating peptide nona-arginine into adherent cells involves a transient increase in intracellular calcium. Biochem. J. 2015, 471, 221–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldshtein, M.; Forti, E.; Ruvinov, E.; Cohen, S. Mechanisms of cellular uptake and endosomal escape of calcium-siRNA nanocomplexes. Int. J. Pharm. 2016, 515, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.-I.; Yamamoto, T.; Waki, R.; Wada, S.; Wada, F.; Noda, M.; Obika, S. Ca2+enrichment in culture medium potentiates effect of oligonucleotides. Nucleic Acids Res. 2015, 43, e128. [Google Scholar] [CrossRef] [Green Version]
- Santos, R.C.D.; Bautista, S.; Lucarelli, S.; Bone, L.N.; Dayam, R.M.; Abousawan, J.; Botelho, R.J.; Antonescu, C.N. Selective regulation of clathrin-mediated epidermal growth factor receptor signaling and endocytosis by phospholipase C and calcium. Mol. Biol. Cell 2017, 28, 2802–2818. [Google Scholar] [CrossRef]
- Helmfors, H.; Eriksson, J.; Langel, Ü. Optimized luciferase assay for cell-penetrating peptide-mediated delivery of short oligonucleotides. Anal. Biochem. 2015, 484, 136–142. [Google Scholar] [CrossRef]
- Rocha, C.S.; Lundin, K.E.; Behlke, M.A.; Zain, R.; EL Andaloussi, S.; Smith, C.E. Four Novel Splice-Switch Reporter Cell Lines: Distinct Impact of Oligonucleotide Chemistry and Delivery Vector on Biological Activity. Nucleic Acid Ther. 2016, 26, 381–391. [Google Scholar] [CrossRef]
- Saher, O.; Rocha, C.S.; Zaghloul, E.M.; Wiklander, O.P.; Zamolo, S.; Heitz, M.; Ezzat, K.; Gupta, D.; Reymond, J.-L.; Zain, R.; et al. Novel peptide-dendrimer/lipid/oligonucleotide ternary complexes for efficient cellular uptake and improved splice-switching activity. Eur. J. Pharm. Biopharm. 2018, 132, 29–40. [Google Scholar] [CrossRef]
- Expasy. Cellosaurus HeLa EGFP-654 (CVCL_VS42). Available online: https://web.expasy.org/cellosaurus/CVCL_VS42 (accessed on 29 September 2021).
- Expasy. Cellosaurus U-87 MG-luc2 (CVCL_5J15). Available online: https://web.expasy.org/cellosaurus/CVCL_5J15 (accessed on 4 October 2021).
- Lehto, T.; Vasconcelos, L.; Margus, H.; Figueroa, R.; Pooga, M.; Hällbrink, M.; Langel, U. Saturated Fatty Acid Analogues of Cell-Penetrating Peptide PepFect14: Role of Fatty Acid Modification in Complexation and Delivery of Splice-Correcting Oligonucleotides. Bioconjugate Chem. 2017, 28, 782–792. [Google Scholar] [CrossRef]
- Cardoso, A.M.; Trabulo, S.; Cardoso, A.L.; Lorents, A.; Morais, C.M.; Gomes, P.; Nunes, C.; Lúcio, M.; Reis, S.; Padari, K.; et al. S4(13)-PV cell-penetrating peptide induces physical and morphological changes in membrane-mimetic lipid systems and cell membranes: Implications for cell internalization. Biochim. Biophys. Acta 2012, 1818, 877–888. [Google Scholar] [CrossRef]
- Luthman, H.; Magnusson, G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983, 11, 1295–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kneuer, C.; Sameti, M.; Bakowsky, U.; Schiestel, T.; Schirra, H.; Schmidt, H.; Lehr, C.-M. A Nonviral DNA Delivery System Based on Surface Modified Silica-Nanoparticles Can Efficiently Transfect Cells in Vitro. Bioconjugate Chem. 2000, 11, 926–932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al-Bari, M.A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017, 5, e0029. [Google Scholar] [CrossRef] [PubMed]
- Pearson, A.M.; Rich, A.; Krieger, M. Polynucleotide Binding to Macrophage Scavenger Receptors Depends on the Formation of Base-Quartet-Stabilized 4-Stranded Helices. J. Biol. Chem. 1993, 268, 3546–3554. [Google Scholar] [CrossRef]
- Terpstra, V.; Van Berkel, T.J.C. Scavenger receptors on liver Kupffer cells mediate the in vivo uptake of oxidatively damaged red blood cells in mice. Blood 2000, 95, 2157–2163. [Google Scholar] [CrossRef]
- Kawabata, K.; Takakura, Y.; Hashida, M. The Fate of Plasmid DNA After Intravenous Injection in Mice: Involvement of Scavenger Receptors in Its Hepatic Uptake. Pharm. Res. 1995, 12, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Beppu, M.; Hayashi, T.; Hasegawa, T.; Kikugawa, K. Recognition of sialosaccharide chains of glycophorin on damaged erythrocytes by macrophage scavenger receptors. Biochim. Biophys. Acta 1995, 1268, 9–19. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.S.; Basu, S.K.; Falck, J.R.; Ho, Y.K.; Goldstein, J.L. The scavenger cell pathway for lipoprotein degradation: Specificity of the binding site that mediates the uptake of negatively-charged LDL by macrophages. J. Supramol. Struct. 1980, 13, 67–81. [Google Scholar] [CrossRef]
- Thelen, T.; Hao, Y.; Medeiros, A.I.; Curtis, J.; Serezani, C.H.; Kobzik, L.; Harris, L.H.; Aronoff, D.M. The Class A Scavenger Receptor, Macrophage Receptor with Collagenous Structure, Is the Major Phagocytic Receptor forClostridium sordelliiExpressed by Human Decidual Macrophages. J. Immunol. 2010, 185, 4328–4335. [Google Scholar] [CrossRef] [Green Version]
- Lorents, A.; Maloverjan, M.; Padari, K.; Pooga, M. Internalisation and Biological Activity of Nucleic Acids Delivering Cell-Penetrating Peptide Nanoparticles Is Controlled by the Biomolecular Corona. Pharmaceuticals 2021, 14, 667. [Google Scholar] [CrossRef]
- Freimann, K.; Arukuusk, P.; Kurrikoff, K.; Vasconcelos, L.; Veiman, K.-L.; Uusna, J.; Margus, H.; Garcia-Sosa, A.T.; Pooga, M.; Langel, Ü. Optimization of in vivo DNA delivery with NickFect peptide vectors. J. Control. Release 2016, 241, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Dubikovskaya, E.A.; Thorne, S.H.; Pillow, T.H.; Contag, C.H.; Wender, P.A. Overcoming multidrug resistance of small-molecule therapeutics through conjugation with releasable octaarginine transporters. Proc. Natl. Acad. Sci. USA 2008, 105, 12128–12133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.-Y.; Choi, Y.-S.; Suh, J.-S.; Kwon, Y.-M.; Yang, V.C.; Lee, S.-J.; Chung, C.-P.; Park, Y.-J. Cell-penetrating chitosan/doxorubicin/TAT conjugates for efficient cancer therapy. Int. J. Cancer 2010, 128, 2470–2480. [Google Scholar] [CrossRef] [PubMed]
- Myrberg, H.; Zhang, L.; Mäe, M.; Langel, U. Design of a Tumor-Homing Cell-Penetrating Peptide. Bioconjugate Chem. 2007, 19, 70–75. [Google Scholar] [CrossRef]
- Ezzat, K.; Zaghloul, E.M.; EL Andaloussi, S.; Lehto, T.; El-Sayed, R.; Magdy, T.; Smith, C.E.; Langel, Ü. Solid formulation of cell-penetrating peptide nanocomplexes with siRNA and their stability in simulated gastric conditions. J. Control. Release 2012, 162, 1–8. [Google Scholar] [CrossRef]
- Veiman, K.-L.; Mäger, I.; Ezzat, K.; Margus, H.; Lehto, T.; Langel, K.; Kurrikoff, K.; Arukuusk, P.; Suhorutšenko, J.; Padari, K.; et al. PepFect14 Peptide Vector for Efficient Gene Delivery in Cell Cultures. Mol. Pharm. 2012, 10, 199–210. [Google Scholar] [CrossRef]
- Helmfors, H.; Lindberg, S.; Langel, U. SCARA Involvement in the Uptake of Nanoparticles Formed by Cell-Penetrating Peptides. Cell-Penetrating Pepti. 2015, 1324, 163–174. [Google Scholar] [CrossRef]


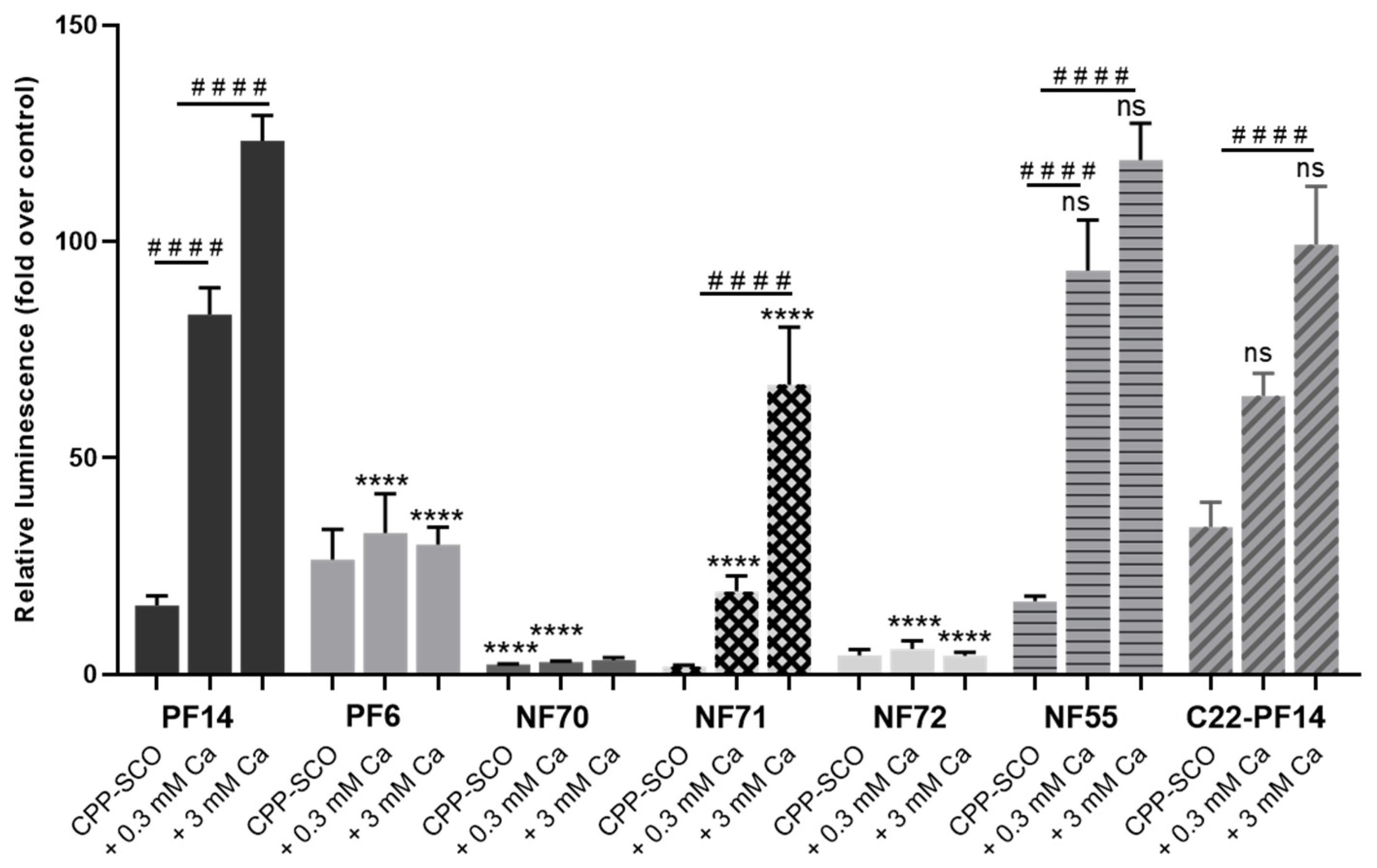

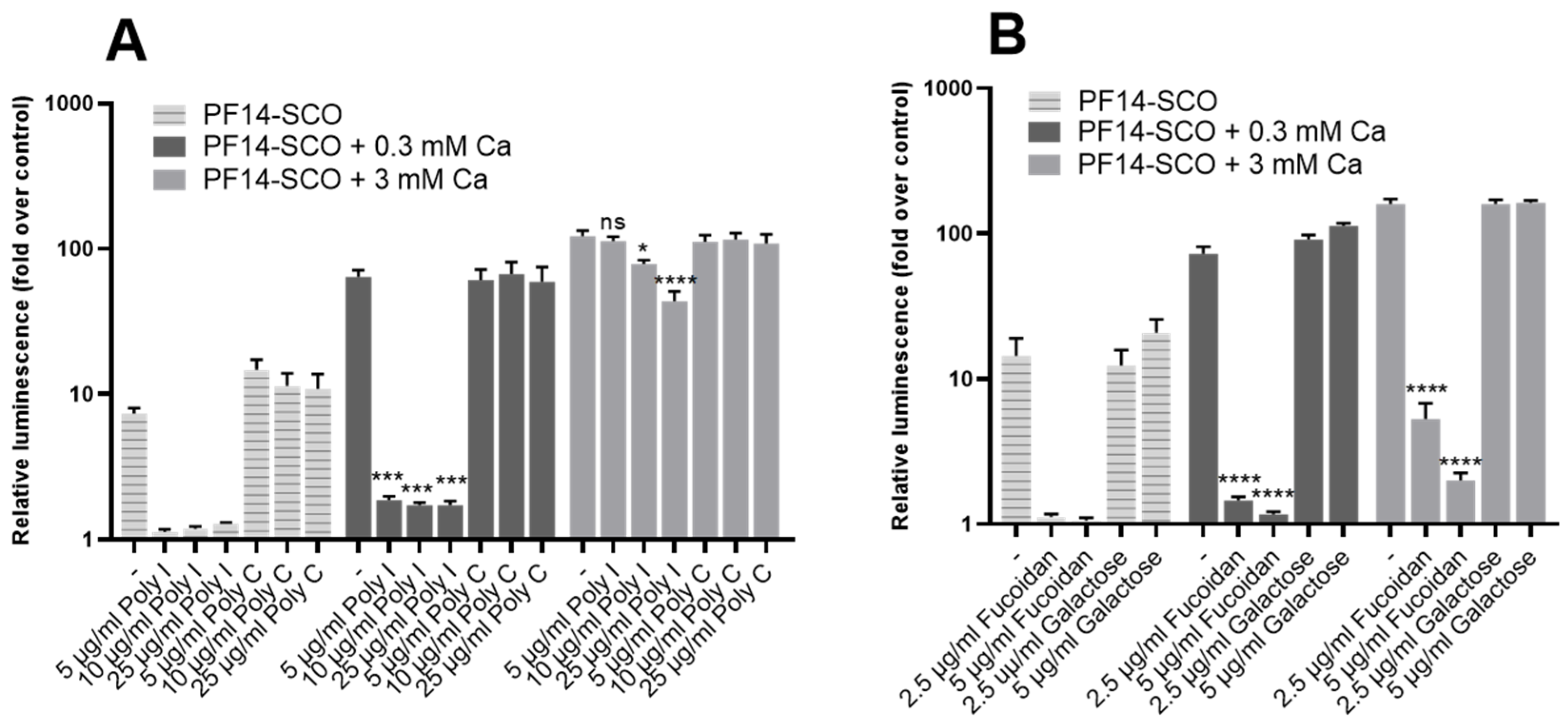
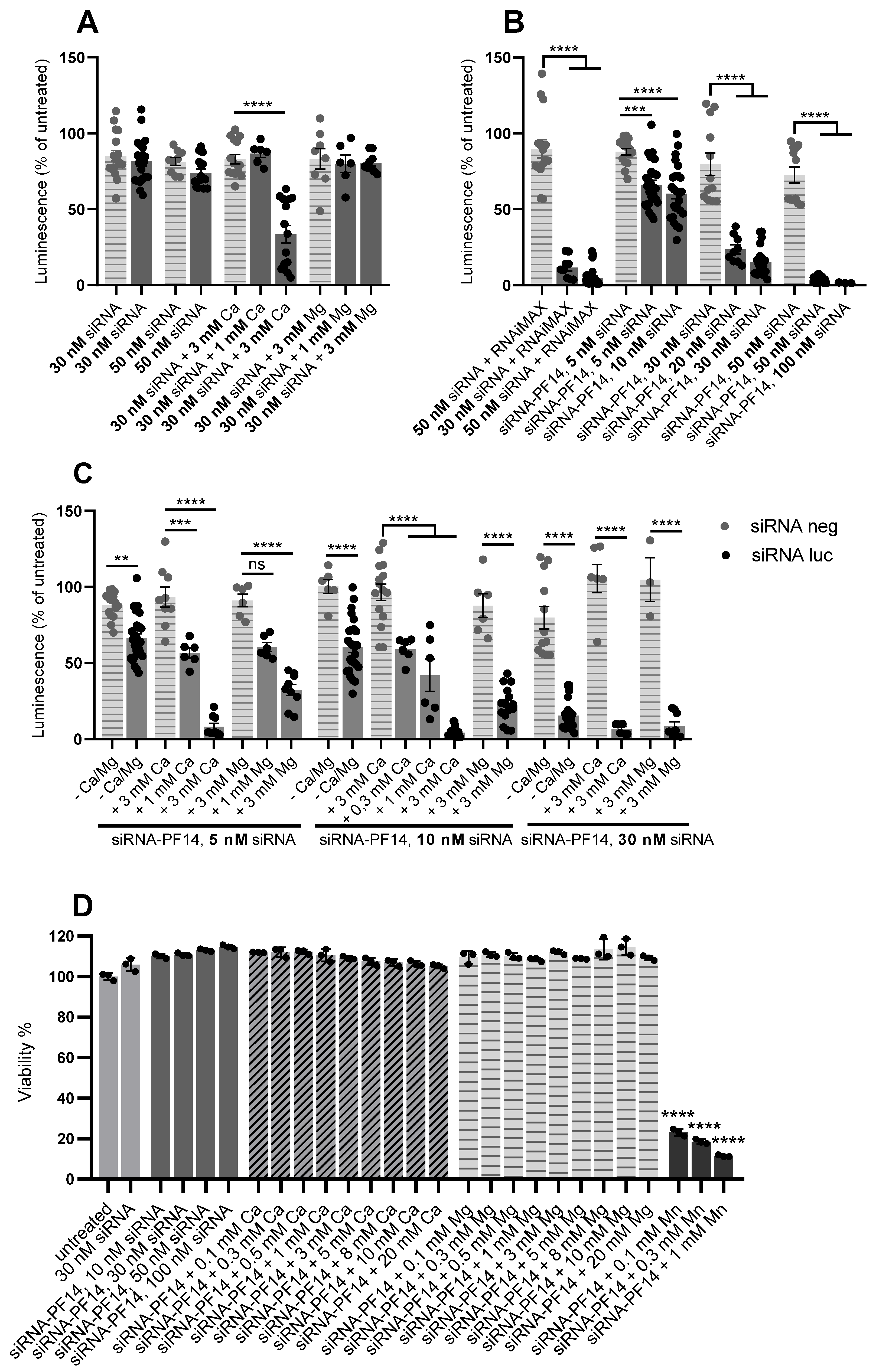
| Nanoparticle | Size (nm) | Zeta Potential (mV) |
|---|---|---|
| PF14-SCO | 231 ± 22 | 9.3 ± 1.1 |
| PF14-SCO + 0.3 mM CaCl2 | 327 ± 60 | 10.1 ± 1.8 |
| PF14-SCO + 3 mM CaCl2 | 305 ± 51 | 13.1 ± 1.0 |
| PF14-SCO + 0.3 mM MgCl2 | 352 ± 65 | 10.6 ± 1.7 |
| PF14-SCO + 3 mM MgCl2 | 393 ± 60 | 15.0 ± 2.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maloverjan, M.; Padari, K.; Abroi, A.; Rebane, A.; Pooga, M. Divalent Metal Ions Boost Effect of Nucleic Acids Delivered by Cell-Penetrating Peptides. Cells 2022, 11, 756. https://doi.org/10.3390/cells11040756
Maloverjan M, Padari K, Abroi A, Rebane A, Pooga M. Divalent Metal Ions Boost Effect of Nucleic Acids Delivered by Cell-Penetrating Peptides. Cells. 2022; 11(4):756. https://doi.org/10.3390/cells11040756
Chicago/Turabian StyleMaloverjan, Maria, Kärt Padari, Aare Abroi, Ana Rebane, and Margus Pooga. 2022. "Divalent Metal Ions Boost Effect of Nucleic Acids Delivered by Cell-Penetrating Peptides" Cells 11, no. 4: 756. https://doi.org/10.3390/cells11040756
APA StyleMaloverjan, M., Padari, K., Abroi, A., Rebane, A., & Pooga, M. (2022). Divalent Metal Ions Boost Effect of Nucleic Acids Delivered by Cell-Penetrating Peptides. Cells, 11(4), 756. https://doi.org/10.3390/cells11040756