Silencing of Ago-2 Interacting Protein SERBP1 Relieves KCC2 Repression by miR-92 in Neurons
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmid SPORT6-MycSERBP1
2.2. Cell Cultures
2.3. Immunoprecipitation
2.4. Protein Identification by LC-MS/MS
2.5. Western Blot Analysis
2.6. RT-PCR Assay and RNA Isolation
2.7. RT- Quantitative PCR for miR-92
2.8. Luciferase Assay
2.9. Ribonucleoprotein Immunoprecipitation
2.9.1. Preparation of mRNP Lysate
2.9.2. Preparation of Beads
2.9.3. Immunoprecipitation Reaction and RNA Precipitation
2.10. RiboTrap Kit
2.11. Statistics
3. Results
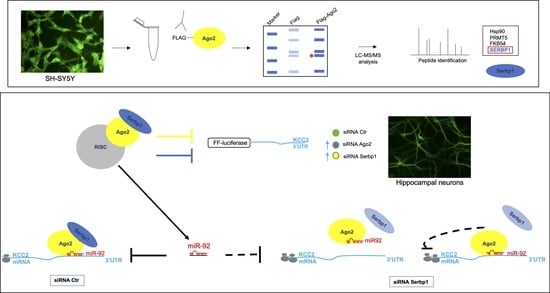
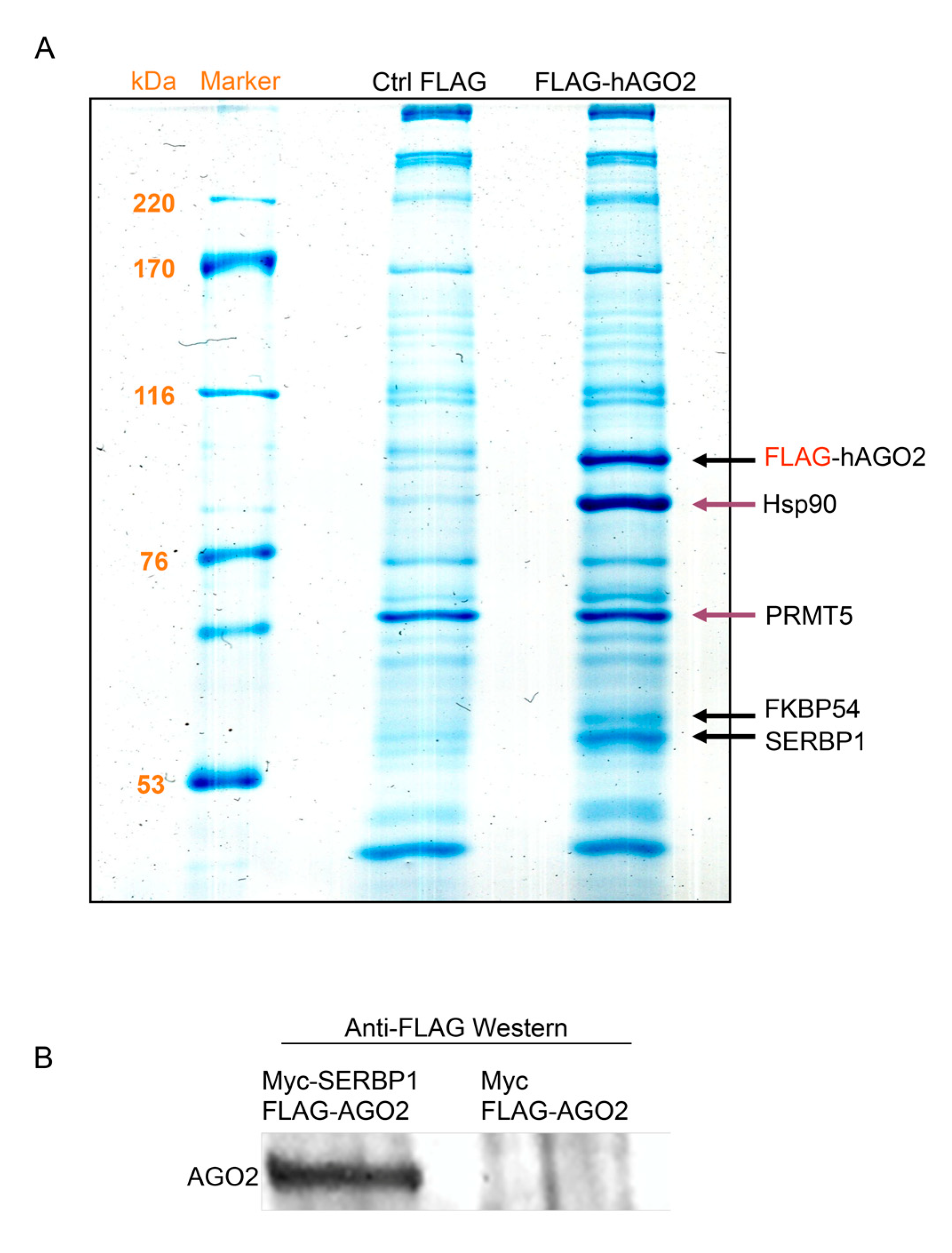
3.1. Ago2 Interacts with SERBP1 in SH-SY5Y Neuroblastoma Cells
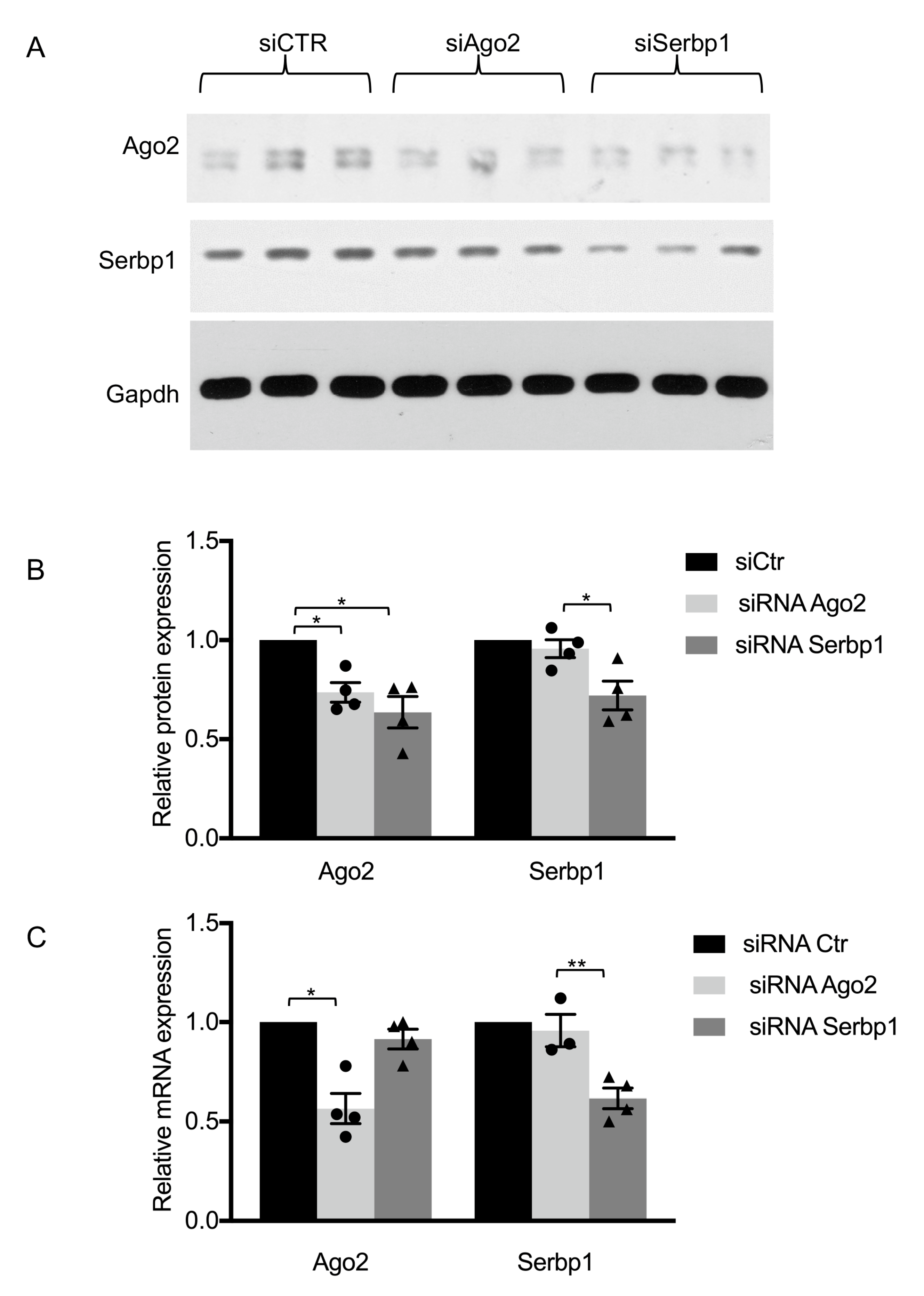
3.2. RNAi of SERBP1 Reduces Ago2 Protein
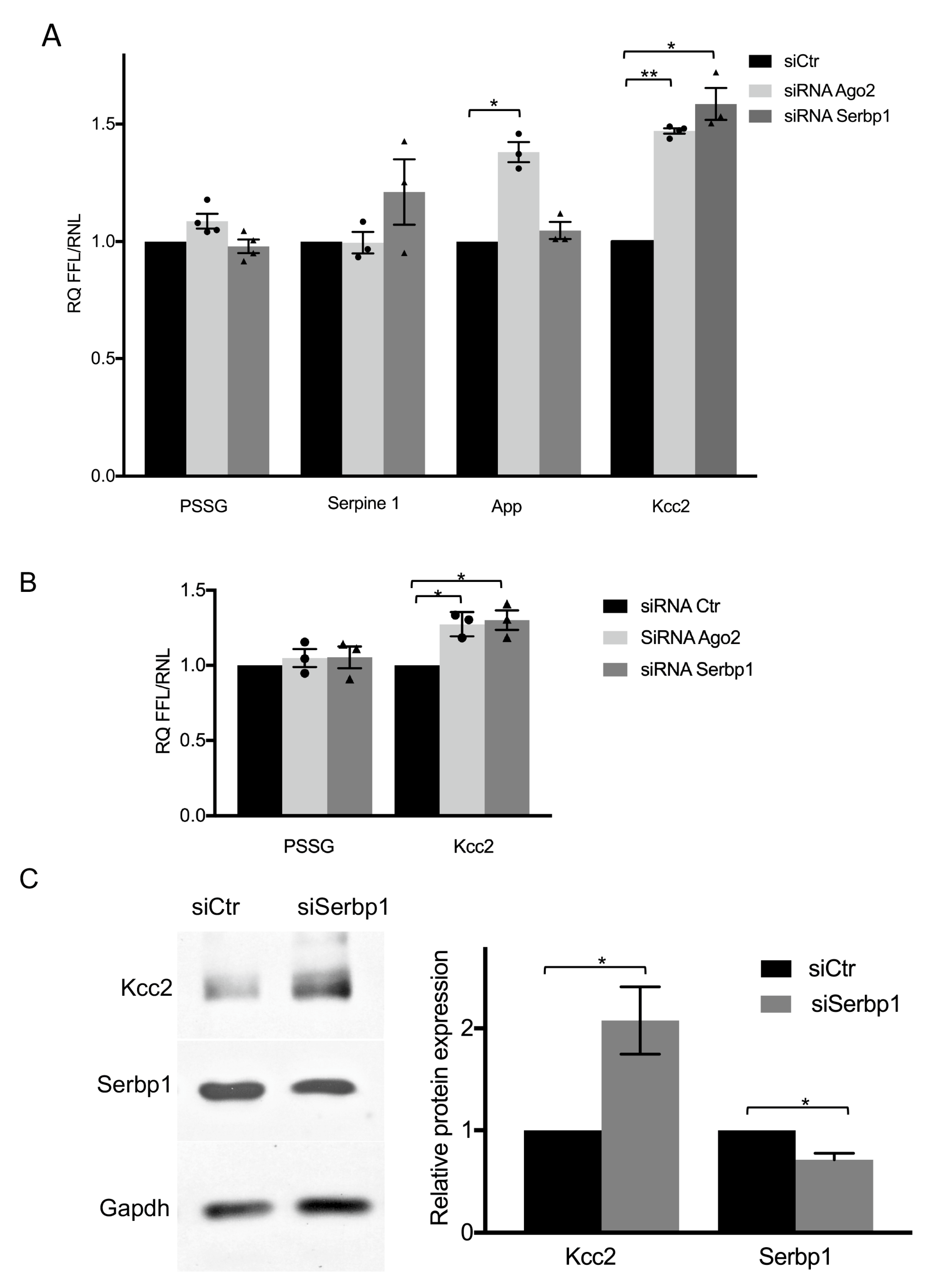
3.3. SERBP1 Regulates KCC2 Expression through the 3′UTR in Neurons
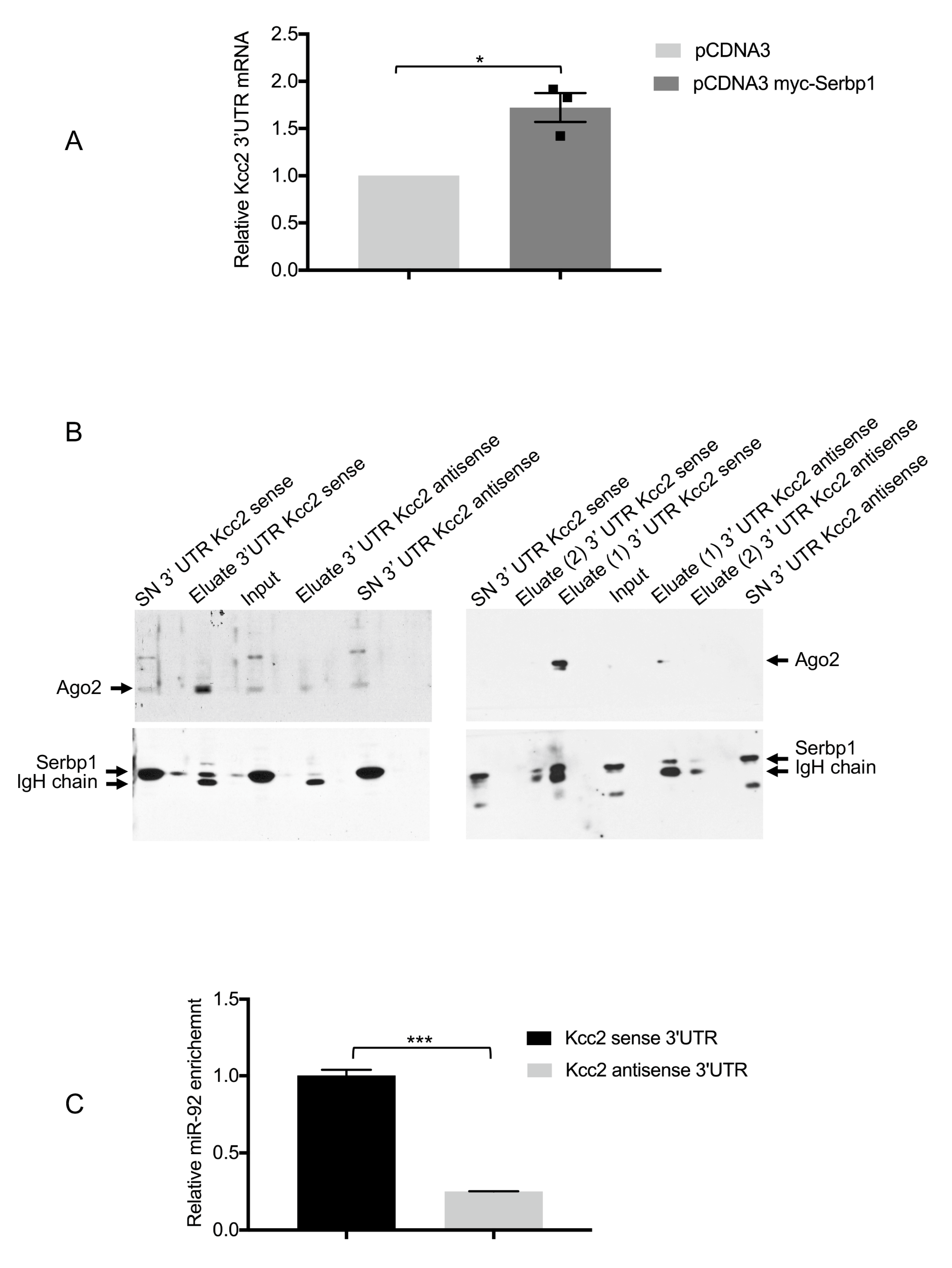
3.4. SERBP1, Ago2 and miR-92 Are Associated to the KCC2 3′UTR
3.5. miR-92 Post-Transcriptional Regulation of KCC2 3′UTR Requires SERBP1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dexheimer, P.J.; Cochella, L. MicroRNAs: From Mechanism to Organism. Front. Cell. Dev. Biol. 2020, 8, 409. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell. Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, X.; Cheng, Z.; Zhu, Q. AGO2 and its partners: A silencing complex, a chromatin modulator, and new features. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 33–53. [Google Scholar] [CrossRef] [PubMed]
- Nawalpuri, B.; Ravindran, S.; Muddashetty, R.S. The Role of Dynamic miRISC During Neuronal Development. Front. Mol. Biosci. 2020, 7, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Störchel, P.H.; Thümmler, J.; Siegel, G.; Aksoy-Aksel, A.; Zampa, F.; Sumer, S.; Schratt, G. A large-scale functional screen identifies Nova1 and Ncoa3 as regulators of neuronal miRNA function. EMBO J. 2015, 34, 2237–2254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Antoniou, A.; Baptista, M.; Carney, N.; Hanley, J.G. PICK1 links Argonaute 2 to endosomes in neuronal dendrites and regulates miRNA activity. EMBO Rep. 2014, 15, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Rajgor, D.; Sanderson, T.M.; Amici, M.; Collingridge, G.L.; Hanley, J.G. NMDAR-dependent Argonaute 2 phosphorylation regulates miRNA activity and dendritic spine plasticity. EMBO J. 2018, 37, e97943. [Google Scholar] [CrossRef]
- Parisi, C.; Giorgi, C.; Batassa, E.M.; Braccini, L.; Maresca, G.; D’agnano, I.; Caputo, V.; Salvatore, A.; Pietrolati, F.; Cogoni, C. Ago1 and Ago2 differentially affect cell proliferation, motility and apoptosis when overexpressed in SH-SY5Y neuroblastoma cells. FEBS Lett. 2011, 585, 2965–2971. [Google Scholar] [CrossRef]
- Heaton, J.H.; Dlakic, W.M.; Dlakic, M.; Gelehrter, T.D. Identification and cDNA Cloning of a Novel RNA-binding Protein That Interacts with the Cyclic Nucleotide-responsive Sequence in the Type-1 Plasminogen Activator Inhibitor mRNA. J. Biol. Chem. 2011, 276, 3341–3347. [Google Scholar] [CrossRef] [Green Version]
- Mondal, S.; Begum, N.A.; Hu, W.; Honjo, T. Functional requirements of AID’s higher order structures and their interaction with RNA-binding proteins. Proc. Natl. Acad. Sci. USA 2016, 113, E1545–E1554. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.W.; Kim, S.; Na, W.; Baek, S.J.; Kim, J.H.; Min, K.; Yeom, J.; Kwak, H.; Jeong, S.; Lee, C. SERBP1 affects homologous recombination-mediated DNA repair by regulation of CtIP translation during S phase. Nucleic Acids Res. 2015, 43, 6321–6333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemos, T.A.; Passos, D.O.; Nery, F.C.; Kobarg, J. Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 2003, 533, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Mari, Y.; West, G.M.; Scharager-Tapia, C.; Pascal, B.D.; Garcia-Ordonez, R.D.; Griffin, P.R. SERBP1 Is a Component of the Liver Receptor Homologue-1 Transcriptional Complex. J. Proteome Res. 2015, 14, 4571–4580. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Hsieh, W.Y.; Chen, L.Y.; Li, C. Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution. J. Cell. Biochem. 2012, 113, 2721–2728. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Wei, H.M.; Chen, L.Y.; Li, C. Localization of SERBP1 in stress granules and nucleoli. FEBS J. 2012, 281, 352–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, T.G.; Peaston, A.; Lim, A.K.; Lorthongpanich, C.; Knowles, B.B.; Solter, D. A tudor domain protein SPINDLIN1 interacts with the mRNA-binding protein SERBP1 and is involved in mouse oocyte meiotic resumption. PLoS ONE 2013, 8, e69764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muto, A.; Sugihara, Y.; Shibakawa, M.; Oshima, K.; Matsuda, T.; Nadano, D. The mRNA-binding protein Serbp1 as an auxiliary protein associated with mammalian cytoplasmic ribosomes. Cell. Biochem. Funct. 2018, 36, 312–322. [Google Scholar] [CrossRef] [PubMed]
- Kosti, A.; de Araujo, P.R.; Li, W.Q.; Guardia, G.D.A.; Chiou, J.; Yi, C.; Ray, D.; Meliso, F.; Li, Y.M.; Delambre, T.; et al. The RNA-binding protein SERBP1 functions as a novel oncogenic factor in glioblastoma by bridging cancer metabolism and epigenetic regulation. Genome Biol. 2020, 21, 195. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ari, Y. Excitatory actions of gaba during development: The nature of the nurture. Nature Rev. Neurosci. 2002, 3, 728–739. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C.; Voipio, J.; Payne, J.A.; Ruusuvuori, E.; Lahtinen, H.; Lamsa, K.; Pirvola, U.; Saarma, M.; Kaila, K. The K+/Cl- cotransporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature 1999, 397, 251–255. [Google Scholar] [CrossRef]
- Virtanen, M.A.; Uvarov, P.; Mavrovic, M.; Poncer, J.C.; Kaila, K. The Multifaceted Roles of KCC2 in Cortical Development. Trends Neurosci. 2021, 44, 378–392. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006, 1, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Pare, J.M.; Tahbaz, N.; Lopez-Orozco, J.; LaPointe, P.; Lasko, P.; Hobman, T.C. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol. Biol. Cell. 2009, 20, 3273–3284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Criado-Marrero, M.; Rein, T.; Binder, E.B.; Porter, J.T.; Koren, J., III; Blair, L.J. Hsp90 and FKBP51: Complex regulators of psychiatric diseases. Philos. Trans. Biol. Sci. 2018, 373, 20160532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, N.J.; Chang, H.M.; Borrajo, J.R.; Gregory, R.I. The co-chaperones Fkbp4/5 control Argonaute2 expression and facilitate RISC assembly. RNA 2013, 19, 1583–1593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frohn, A.; Eberl, C.; Stöhr, J.; Glasmacher, E.; Rüdel, S.; Heissmeyr, V.; Mann, M.; Meister, G. Dicer-dependent and independent Argonaute2 Protein Interaction Networks in Mammalian Cells. Mol. Cell. Proteom. 2012, 11, 1442–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schopp, I.M.; Amaya Ramirez, C.C.; Debeljak, J.; Kreibich, E.; Skribbe, M.; Wild, K.; Bethune, J. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 2017, 8, 15690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meister, G.; Landthaler, M.; Peters, L.; Chen, P.Y.; Urlaub, H.; Lührmann, R.; Tuschl, T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005, 15, 2149–2155. [Google Scholar] [CrossRef] [Green Version]
- Hu, P.; Zhao, H.; Zhu, P.; Xiao, Y.; Miao, W.; Wang, Y.; Jin, H. Dual regulation of Arabidopsis AGO2 by arginine methylation. Nat. Commun. 2019, 10, 844. [Google Scholar] [CrossRef] [Green Version]
- Rybak, A.; Fuchs, H.; Hadian, K.; Smirnova, L.; Wulczyn, E.A.; Michel, G.; Nitsch, R.; Krappmann, D.; Wulczyn, F.G. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat. Cell. Biol. 2009, 11, 1411–1420. [Google Scholar] [CrossRef]
- Johnston, M.; Geoffroy, M.C.; Sobala, A.; Hay, R.; Hutvagner, G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol. Biol. Cell. 2010, 21, 1462–1469. [Google Scholar] [CrossRef] [PubMed]
- Colleti, C.; Melo-Hanchuk, T.D.; da Silva, F.R.M.; Saito, Â.; Kobarg, J. Complex interactomes and post-translational modifications of the regulatory proteins HABP4 and SERBP1 suggest pleiotropic cellular functions. World J. Biol. Chem. 2019, 10, 44–64. [Google Scholar] [CrossRef]
- Asakura, T.; Iwaki, S.; Okada, H.; Sobel, B.E.; Fujii, S. Posttranscriptional regulation of expression of plasminogen activator inhibitor type-1 by cAMP in HepG2 liver cells. J. Biochem. 2011, 150, 687–694. [Google Scholar] [CrossRef]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villadsen, S.B.; Bramsen, J.B.; Ostenfeld, M.S.; Wiklund, E.D.; Fristrup, N.; Gao, S.; Hansen, T.B.; Jensen, T.I.; Borre, M.; Ørntoft, T.F.; et al. The miR-143/-145 cluster regulates plasminogen activator inhibitor-1 in bladder cancer. Br. J. Cancer. 2012, 106, 366–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broytman, O.; Westmark, P.R.; Gurel, Z.; Malter, J.S. Rck/p54 interacts with APP mRNA as part of a multi-protein complex and enhances APP mRNA and protein expression in neuronal cell lines. Neurobiol. Aging 2009, 30, 1962–1974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruberti, F.; Barbato, C.; Cogoni, C. Post-transcriptional regulation of amyloid precursor protein by microRNAs and RNA binding proteins. Commun. Integr. Biol. 2010, 3, 499–503. [Google Scholar] [CrossRef] [PubMed]
- Barbato, C.; Ruberti, F.; Pieri, M.; Vilardo, E.; Costanzo, M.; Ciotti, M.T.; Zona, C.; Cogoni, C. MicroRNA-92 modulates K (+) Cl (-) co-transporter KCC2 expression in cerebellar granule neurons. J. Neurochem. 2010, 113, 591–600. [Google Scholar] [CrossRef]
- Korecka, J.A.; van Kesteren, R.E.; Blaas, E.; Spitzer, S.O.; Kamstra, J.H.; Smit, A.B.; Swaab, D.F.; Verhaagen, J.; Bossers, K. Phenotypic characterization of retinoic acid differentiated SH-SY5Y cells by transcriptional profiling. PLoS ONE 2013, 28, e63862. [Google Scholar] [CrossRef] [Green Version]
- Fukao, A.; Aoyama, T.; Fujiwara, T. The molecular mechanism of translational control via the communication between the microRNA pathway and RNA-binding proteins. RNA Biol. 2015, 12, 922–926. [Google Scholar] [CrossRef] [Green Version]
- Muddashetty, R.S.; Nalavadi, V.C.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.T.; Bassell, G.J. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011, 42, 673–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim, S.; Chang, H.R.; Kim, D.; Park, J.; Son, N.; Park, J.; Yoon, M.; Chae, G.; Kim, Y.K.; et al. The regulatory impact of RNA-binding proteins on microRNA targeting. Nat. Commun. 2021, 12, 5057. [Google Scholar] [CrossRef]
- Degrauwe, N.; Schlumpf, T.B.; Janiszewska, M.; Martin, P.; Cauderay, A.; Provero, P.; Riggi, N.; Suvà, M.L.; Paro, R.; Stamenkovic, I. The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell. Rep. 2016, 15, 1634–1647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kedde, M.; Strasser, M.J.; Boldajipour, B.; Oude Vrielink, J.A.; Slanchev, K.; le Sage, C.; Nagel, R.; Voorhoeve, P.M.; van Duijse, J.; Ørom, U.A.; et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell 2007, 131, 1273–1286. [Google Scholar] [CrossRef] [Green Version]
- Vetere, G.; Barbato, C.; Pezzola, S.; Frisone, P.; Aceti, M.; Ciotti, M.; Cogoni, C.; Ammassari-Teule, M.; Ruberti, F. Selective inhibition of miR-92 in hippocampal neurons alters contextual fear memory. Hippocampus 2014, 24, 1458–1465. [Google Scholar] [CrossRef]
- Mi, T.; Sun, X.; Wang, Z.; Wang, Y.; He, X.; Liu, C.; Zhang, S.; Du, H.; Liu, C.; Teng, Z. Loss of MicroRNA-137 Impairs the Homeostasis of Potassium in Neurons via KCC2. Exp. Neurobiol. 2020, 29, 138–149. [Google Scholar] [CrossRef]
- Hentze, M.W.; Castello, A.; Schwarzl, T.; Preiss, T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell. Biol. 2018, 19, 327–341. [Google Scholar] [CrossRef]
- Youn, J.-Y.; Dunham, W.H.; Hong, S.J.; Knight, J.D.R.; Bashkurov, M.; Chen, G.I.; Bagci, H.; Rathod, B.; MacLeod, G.; Eng, S.W.M.; et al. High-Density Proximity Mapping Reveals the Subcellular Organization of mRNA-Associated Granules and Bodies. Mol. Cell 2018, 69, 517–532. [Google Scholar] [CrossRef]
- Go, C.D.; Knight, J.D.R.; Rajasekharan, A.; Rathod, B.; Hesketh, G.G.; Abe, K.T.; Youn, J.-Y.; Samavarchi-Tehrani, P.; Zhang, H.; Zhu, L.Y.; et al. Proximity-Dependent Biotinylation Map of a Human Cell. Nature 2021, 595, 120–124. [Google Scholar] [CrossRef]
- Khudayberdiev, S.; Soutschek, M.; Ammann, I.; Heinze, A.; Rust, M.B.; Baumeister, S.; Schratt, G. The cytoplasmic SYNCRIP mRNA interactome of mammalian neurons. RNA Biol. 2021, 18, 1252–1264. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.L. The Expanding Therapeutic Potential of Neuronal KCC2. Cells 2020, 9, 240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oyarzabal, A.; Xiol, C.; Castells, A.A.; Grau, C.; O’Callaghan, M.; Fernández, G.; Alcantara, S.; Pineda, M.; Armstrong, J.; Altafaj, X.; et al. Comprehensive Analysis of GABAA-A1R developmental alterations in Rett Syndrome: Setting the focus for therapeutic targets in the time frame of the disease. Int. J. Mol. Sci. 2020, 21, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinz, L.; Torrella Barrufet, J.; Heine, V.M. KCC2 expression levels are reduced in post mortem brain tissue of Rett syndrome patients. Acta Neuropathol. Commun. 2019, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.; Park, S.K.; Xu, T.; Vanderklish, P.; Yates, J.R., III. Quantitative proteomic analysis of primary neurons reveals diverse changes in synaptic protein content in fmr1 knockout mice. Proc. Natl. Acad. Sci. USA 2008, 105, 15281–15286. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbato, C.; Frisone, P.; Braccini, L.; D’Aguanno, S.; Pieroni, L.; Ciotti, M.T.; Catalanotto, C.; Cogoni, C.; Ruberti, F. Silencing of Ago-2 Interacting Protein SERBP1 Relieves KCC2 Repression by miR-92 in Neurons. Cells 2022, 11, 1052. https://doi.org/10.3390/cells11061052
Barbato C, Frisone P, Braccini L, D’Aguanno S, Pieroni L, Ciotti MT, Catalanotto C, Cogoni C, Ruberti F. Silencing of Ago-2 Interacting Protein SERBP1 Relieves KCC2 Repression by miR-92 in Neurons. Cells. 2022; 11(6):1052. https://doi.org/10.3390/cells11061052
Chicago/Turabian StyleBarbato, Christian, Paola Frisone, Laura Braccini, Simona D’Aguanno, Luisa Pieroni, Maria Teresa Ciotti, Caterina Catalanotto, Carlo Cogoni, and Francesca Ruberti. 2022. "Silencing of Ago-2 Interacting Protein SERBP1 Relieves KCC2 Repression by miR-92 in Neurons" Cells 11, no. 6: 1052. https://doi.org/10.3390/cells11061052
APA StyleBarbato, C., Frisone, P., Braccini, L., D’Aguanno, S., Pieroni, L., Ciotti, M. T., Catalanotto, C., Cogoni, C., & Ruberti, F. (2022). Silencing of Ago-2 Interacting Protein SERBP1 Relieves KCC2 Repression by miR-92 in Neurons. Cells, 11(6), 1052. https://doi.org/10.3390/cells11061052