Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Antibodies and Reagents
2.3. Transfection of 2xFYVE and p40PX PI3P-Binding Domains
2.4. Labeling of Transferrin-Loaded Compartments and Internalization of Transferrin Receptors
2.5. Flow Cytometric Quantification of Recycling
2.6. Immunofluorescence and Confocal Analysis
2.7. Image Analysis
2.8. Western Blot
2.9. Data Presentation and Statistics
3. Results
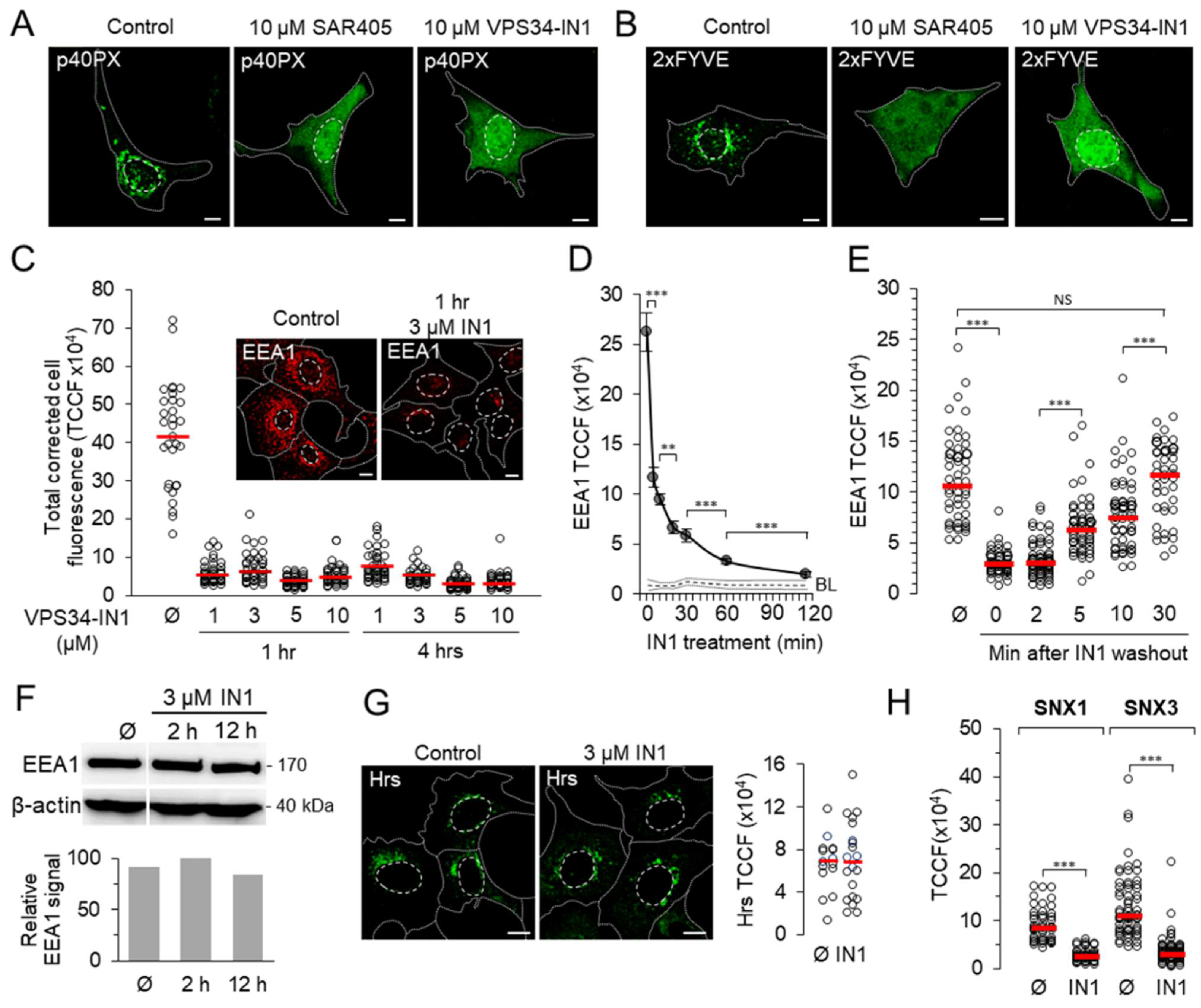
3.1. Pharmacological Inhibition of Vps34 Rapidly and Reversibly Depletes Endosomal PI3P Pool and Alters PI3P-Associated Functions
3.2. Depletion of Vps34-Derived PI3P Arrests Internalized Tf in Perinuclear Endosomes
3.3. PI3P Depletion Does Not Inhibit Tf Recycling
3.4. PI3P Depletion Traps Internalized Tf in Rab5a/Rab4-Positive Endosomes and Prevents the Loading of Rab11a-Positive Endosomes
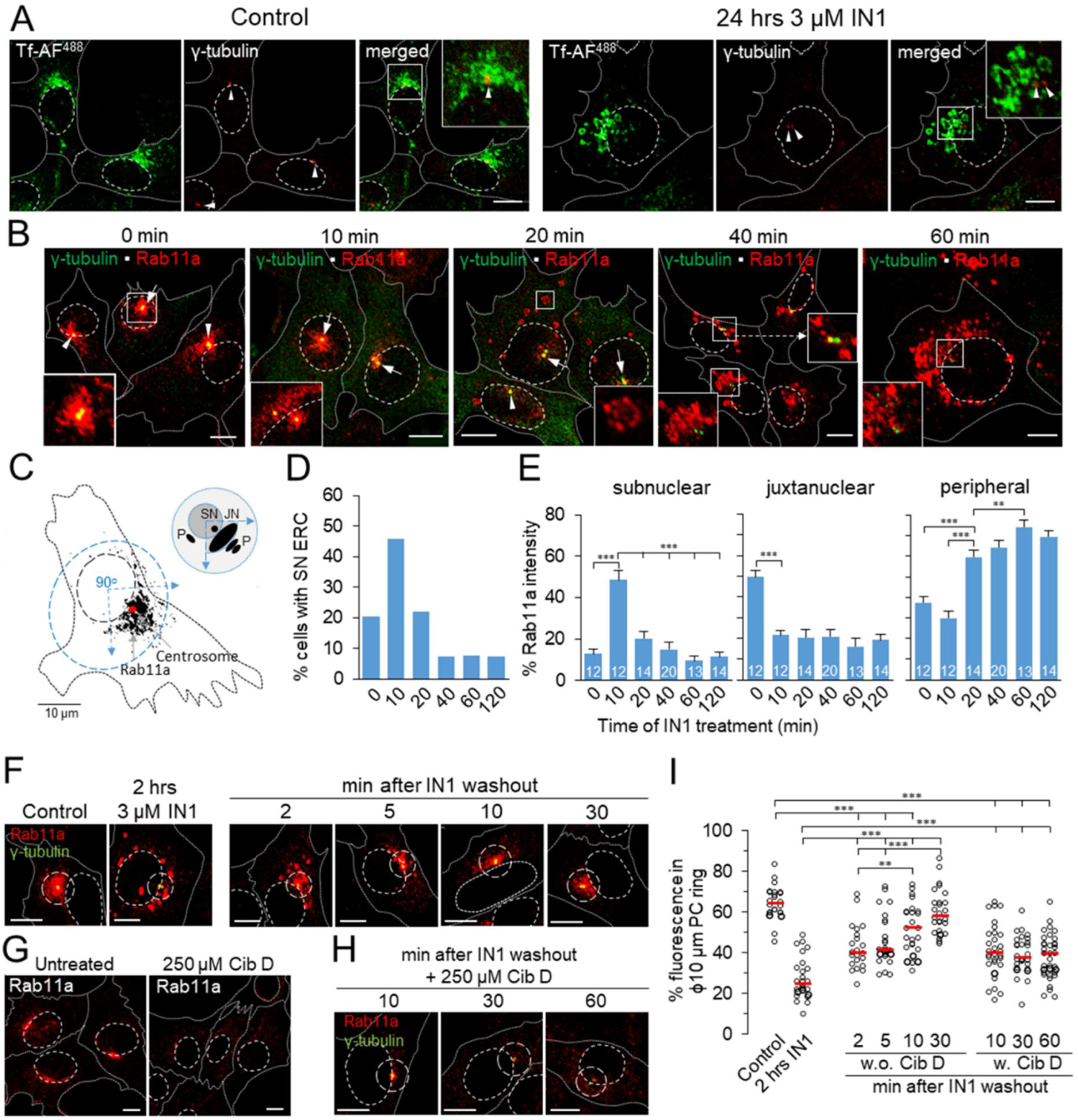
3.5. PI3P Depletion Reorganizes the Pericentriolar Recycling System
3.6. IN1 Treatment DEPLETES the Rab8a-Positive Subset of ERC
3.7. Rab11a-Positive Endosomes Acquire Rab11-FIP5 However, Not Rab11-FIP3
3.8. Segregation of Endosomal PI3P by Expression of PI3P-Binding Modules Alters Endosomal Trafficking of TfR
4. Discussion
4.1. PI3P-Dependent Maturation of Vacuolar EE Domain
4.2. PI3P-Dependent Cargo Sorting
4.3. PI3P-Independent Biogenesis of Rab11 Endosomes
4.4. PI3P-Dependent Maturation of the Rab11-Dependent Pathway
4.5. PI3P-Dependent Biogenesis of the ERC
4.6. Possible off-Target Effects of VPS34-IN1
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bitsikas, V.; Corrêa, I.R.; Nichols, B.J. Clathrin-independent pathways do not contribute significantly to endocytic flux. eLife 2014, 3, e03970. [Google Scholar] [CrossRef] [PubMed]
- Wandinger-Ness, A.; Zerial, M. Rab Proteins and the Compartmentalization of the Endosomal System. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny Lipids With Giant Impact on Cell Regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Schink, K.O.; Tan, K.-W.; Stenmark, H. Phosphoinositides in Control of Membrane Dynamics. Annu. Rev. Cell Dev. Biol. 2016, 32, 143–171. [Google Scholar] [CrossRef] [PubMed]
- Leonard, D.; Hayakawa, A.; Lawe, D.; Lambright, D.; Bellve, K.D.; Standley, C.; Lifshitz, L.M.; Fogarty, K.E.; Corvera, S. Sorting of EGF and transferrin at the plasma membrane and by cargo-specific signaling to EEA1-enriched endosomes. J. Cell Sci. 2008, 121, 3445–3458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marat, A.L.; Haucke, V. Phosphatidylinositol 3-phosphates—At the interface between cell signalling and membrane traffic. EMBO J. 2016, 35, 561–579. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lo, W.-T.; Haucke, V. Phosphoinositide switches in endocytosis and in the endolysosomal system. Curr. Opin. Cell Biol. 2019, 59, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Redpath, G.M.I.; Betzler, V.M.; Rossatti, P.; Rossy, J. Membrane Heterogeneity Controls Cellular Endocytic Trafficking. Front. Cell Dev. Biol. 2020, 8, 757. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Etoh, K.; Ohbayashi, N.; Fukuda, M. Rab35 promotes the recruitment of Rab8, Rab13 and Rab36 to recycling endosomes through MICAL-L1 during neurite outgrowth. Biol. Open 2014, 3, 803–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinkert, K.; Echard, A. Rab35 GTPase: A Central Regulator of Phosphoinositides and F-actin in Endocytic Recycling and Beyond. Traffic 2016, 17, 1063–1077. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Liu, H.; Wang, Y.; Liu, O.; Zhang, J.; Gleason, A.; Yang, Z.; Wang, H.; Shi, A.; Grant, B.D. RAB-10 Promotes EHBP-1 Bridging of Filamentous Actin and Tubular Recycling Endosomes. PLoS Genet. 2016, 12, e1006093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linford, A.; Yoshimura, S.-I.; Bastos, R.N.; Langemeyer, L.; Gerondopoulos, A.; Rigden, D.J.; Barr, F.A. Rab14 and Its Exchange Factor FAM116 Link Endocytic Recycling and Adherens Junction Stability in Migrating Cells. Dev. Cell 2012, 22, 952–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strick, D.J.; Elferink, L.A. Rab15 Effector Protein: A Novel Protein for Receptor Recycling from the Endocytic Recycling Compartment. Mol. Biol. Cell 2005, 16, 5699–5709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wallroth, A.; Haucke, V. Phosphoinositide conversion in endocytosis and the endolysosomal system. J. Biol. Chem. 2018, 293, 1526–1535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gharbi, S.I.; Zvelebil, M.J.; Shuttleworth, S.J.; Hancox, T.; Saghir, N.; Timms, J.F.; Waterfield, M.D. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem. J. 2007, 404, 15–21. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.E.; Overmeyer, J.H.; Gunning, W.T.; Maltese, W.A. Gene silencing reveals a specific function of hVps34 phosphatidylinositol 3-kinase in late versus early endosomes. J. Cell Sci. 2006, 119, 1219–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Law, F.; Seo, J.H.; Wang, Z.; DeLeon, J.L.; Bolis, Y.; Brown, A.; Zong, W.-X.; Du, G.; Rocheleau, C.E. The VPS-34 PI3 kinase negatively regulates RAB-5 during endosome maturation. J. Cell Sci. 2017, 130, 2007–2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaber, N.; Dou, Z.; Chen, J.-S.; Catanzaro, J.; Jiang, Y.-P.; Ballou, L.M.; Selinger, E.; Ouyang, X.; Lin, R.Z.; Zhang, J.; et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc. Natl. Acad. Sci. USA 2012, 109, 2003–2008. [Google Scholar] [CrossRef] [Green Version]
- Jaber, N.; Mohd-Naim, N.F.; Wang, Z.; DeLeon, J.L.; Kim, S.; Zhong, H.; Sheshadri, N.; Dou, Z.; Edinger, A.L.; Du, G.; et al. Vps34 regulates Rab7 and late endocytic trafficking through recruitment of the GTPase activating protein Armus. J. Cell Sci. 2016, 129, 4424–4435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siddhanta, U.; McIlroy, J.; Shah, A.; Zhang, Y.; Backer, J.M. Distinct Roles for the p110α and hVPS34 Phosphatidylinositol 3′-Kinases in Vesicular Trafficking, Regulation of the Actin Cytoskeleton, and Mitogenesis. J. Cell Biol. 1998, 143, 1647–1659. [Google Scholar] [CrossRef] [Green Version]
- Zoncu, R.; Perera, R.M.; Balkin, D.M.; Pirruccello-Straub, M.; Toomre, D.; De Camilli, P. A Phosphoinositide Switch Controls the Maturation and Signaling Properties of APPL Endosomes. Cell 2009, 136, 1110–1121. [Google Scholar] [CrossRef] [Green Version]
- Bago, R.; Malik, N.; Munson, M.J.; Prescott, A.R.; Davies, P.; Sommer, E.; Shpiro, N.; Ward, R.; Cross, D.; Ganley, I.G.; et al. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem. J. 2014, 463, 413–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowdle, W.E.; Nyfeler, B.; Nagel, J.; Elling, R.A.; Liu, S.; Triantafellow, E.; Menon, S.; Wang, Z.; Honda, A.; Pardee, G.; et al. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat. Cell Biol. 2014, 16, 1069–1079. [Google Scholar] [CrossRef]
- Ronan, B.; Flamand, O.; Vescovi, L.; Dureuil, C.; Durand, L.; Fassy, F.; Bachelot, M.-F.; Lamberton, A.; Mathieu, M.; Bertrand, T.; et al. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat. Chem. Biol. 2014, 10, 1013–1019. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, E.M.; Schultz, S.W.; Schink, K.O.; Pedersen, N.M.; Nähse, V.; Carlson, A.; Brech, A.; Stenmark, H.; Raiborg, C. Concerted ESCRT and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat. Commun. 2018, 9, 2932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kano, F.; Murata, M. Phosphatidylinositol-3-phosphate-mediated actin domain formation linked to DNA synthesis upon insulin treatment in rat hepatoma-derived H4IIEC3 cells. Biochim. Biophys. Acta 2019, 1866, 793–805. [Google Scholar] [CrossRef]
- Campa, C.C.; Margaria, J.P.; Derle, A.; Del Giudice, M.; De Santis, M.C.; Gozzelino, L.; Copperi, F.; Bosia, C.; Hirsch, E. Rab11 activity and PtdIns(3)P turnover removes recycling cargo from endosomes. Nat. Chem. Biol. 2018, 14, 801–810. [Google Scholar] [CrossRef]
- Bertović, I.; Kurelić, R.; Milosevic, I.; Bender, M.; Krauss, M.; Haucke, V.; Begonja, A.J. Vps34 derived phosphatidylinositol 3-monophosphate modulates megakaryocyte maturation and proplatelet production through late endosomes/lysosomes. J. Thromb. Haemost. 2020, 18, 1756–1772. [Google Scholar] [CrossRef] [PubMed]
- Zagorac, G.B.; Mahmutefendić, H.; Maćešić, S.; Karleuša, L.; Lučin, P. Quantitative Analysis of Endocytic Recycling of Membrane Proteins by Monoclonal Antibody-Based Recycling Assays. J. Cell. Physiol. 2017, 232, 463–476. [Google Scholar] [CrossRef]
- Mahmutefendić, H.; Zagorac, G.B.; Grabušić, K.; Karleuša, L.; Maćešić, S.; Momburg, F.; Lučin, P. Late Endosomal Recycling of Open MHC-I Conformers. J. Cell. Physiol. 2017, 232, 872–887. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelières, F.P. A Guided Tour into Subcellular Colocalization Analysis in Light Microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- McCloy, R.; Rogers, S.; Caldon, C.E.; Lorca, T.; Castro, A.; Burgess, A. Partial inhibition of Cdk1 in G2phase overrides the SAC and decouples mitotic events. Cell Cycle 2014, 13, 1400–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanai, F.; Liu, H.; Field, S.; Akbary, H.; Matsuo, T.; Brown, G.E.; Cantley, L.; Yaffe, M.B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001, 3, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Le Blanc, I.; Luyet, P.-P.; Pons, V.; Ferguson, C.; Emans, N.; Petiot, A.; Mayran, N.; Demaurex, N.; Fauré, J.; Sadoul, R.; et al. Endosome-to-cytosol transport of viral nucleocapsids. Nat. Cell Biol. 2005, 7, 653–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christoforidis, S.; Miaczynska, M.; Ashman, K.; Wilm, M.; Zhao, L.; Yip, S.-C.; Waterfield, M.D.; Backer, J.M.; Zerial, M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell Biol. 1999, 1, 249–252. [Google Scholar] [CrossRef]
- Simonsen, A.; Lippe, R.; Christoforidis, S.; Gaullier, J.-M.; Brech, A.; Callaghan, J.M.; Toh, B.-H.; Murphy, C.; Zerial, M.; Stenmark, H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 1998, 394, 494–498. [Google Scholar] [CrossRef]
- Gillooly, D.J.; Raiborg, C.; Stenmark, H. Phosphatidylinositol 3-phosphate is found in microdomains of early endosomes. Histochem. Cell Biol. 2003, 120, 445–453. [Google Scholar] [CrossRef]
- Sachse, M.; Urbé, S.; Oorschot, V.; Strous, G.J.; Klumperman, J. Bilayered Clathrin Coats on Endosomal Vacuoles Are Involved in Protein Sorting toward Lysosomes. Mol. Biol. Cell 2002, 13, 1313–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cullen, P.J.; Steinberg, F. To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2018, 19, 679–696. [Google Scholar] [CrossRef]
- van Weering, J.R.; Verkade, P.; Cullen, P.J. SNX-BAR-Mediated Endosome Tubulation is Co-ordinated with Endosome Maturation. Traffic 2011, 13, 94–107. [Google Scholar] [CrossRef] [PubMed]
- Solinger, J.A.; Rashid, H.-O.; Prescianotto-Baschong, C.; Spang, A. FERARI is required for Rab11-dependent endocytic recycling. Nat. Cell Biol. 2020, 22, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Garcia-Santos, D.; Ishikawa, Y.; Seguin, A.; Li, L.; Fegan, K.H.; Hildick-Smith, G.J.; Shah, D.I.; Cooney, J.D.; Chen, W.; et al. Snx3 Regulates Recycling of the Transferrin Receptor and Iron Assimilation. Cell Metab. 2013, 17, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Kang, Q.; Shi, X.; Wang, Y.; Zhang, N.; Ye, H.; Xu, Q.; Xu, T.; Zhang, R. SNX-3 mediates retromer-independent tubular endosomal recycling by opposing EEA-1-facilitated trafficking. PLoS Genet. 2021, 17, e1009607. [Google Scholar] [CrossRef] [PubMed]
- Maxfield, F.R.; McGraw, T.E. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 2004, 5, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Mahmutefendić, H.; Zagorac, G.B.; Maćešić, S.; Lučin, P. Rapid Endosomal Recycling. In Peripheral Membrane Proteins; Tanabe, S., Ed.; IntechOpen: London, UK, 2018; pp. 83–104. [Google Scholar]
- Kalaidzidis, I.; Miaczynska, M.; Brewinska-Olchowik, M.; Hupalowska, A.; Ferguson, C.; Parton, R.; Kalaidzidis, Y.; Zerial, M. APPL endosomes are not obligatory endocytic intermediates but act as stable cargo-sorting compartments. J. Cell Biol. 2015, 211, 123–144. [Google Scholar] [CrossRef] [PubMed]
- Sposini, S.; Jean-Alphonse, F.; Ayoub, M.A.; Oqua, A.; West, C.; Lavery, S.; Brosens, J.; Reiter, E.; Hanyaloglu, A.C. Integration of GPCR Signaling and Sorting from Very Early Endosomes via Opposing APPL1 Mechanisms. Cell Rep. 2017, 21, 2855–2867. [Google Scholar] [CrossRef] [Green Version]
- Progida, C.; Cogli, L.; Piro, F.; De Luca, A.; Bakke, O.; Bucci, C. Rab7b controls trafficking from endosomes to the TGN. J. Cell Sci. 2010, 123, 1480–1491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucera, A.; Distefano, M.B.; Berg-Larsen, A.; Skjeldal, F.; Repnik, U.; Bakke, O.; Progida, C. Spatiotemporal Resolution of Rab9 and CI-MPR Dynamics in the Endocytic Pathway. Traffic 2016, 17, 211–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbero, P.; Bittova, L.; Pfeffer, S.R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002, 156, 511–518. [Google Scholar] [CrossRef]
- Poteryaev, D.; Datta, S.; Ackema, K.; Zerial, M.; Spang, A. Identification of the Switch in Early-to-Late Endosome Transition. Cell 2010, 141, 497–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucera, A.; Bakke, O.; Progida, C. The multiple roles of Rab9 in the endolysosomal system. Commun. Integr. Biol. 2016, 9, e1204498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, S.; Bahl, K.; Reinecke, J.B.; Hammond, G.; Naslavsky, N.; Caplan, S. The endocytic recycling compartment maintains cargo segregation acquired upon exit from the sorting endosome. Mol. Biol. Cell 2016, 27, 108–126. [Google Scholar] [CrossRef] [PubMed]
- Naslavsky, N.; Caplan, S. The enigmatic endosome—Sorting the ins and outs of endocytic trafficking. J. Cell Sci. 2018, 131, jcs216499. [Google Scholar] [CrossRef] [Green Version]
- Homma, Y.; Fukuda, M. Rabin8 regulates neurite outgrowth in both GEF activity–dependent and –independent manners. Mol. Biol. Cell 2016, 27, 2107–2118. [Google Scholar] [CrossRef]
- Hehnly, H.; Chen, C.-T.; Powers, C.M.; Liu, H.-L.; Doxsey, S. The Centrosome Regulates the Rab11- Dependent Recycling Endosome Pathway at Appendages of the Mother Centriole. Curr. Biol. 2012, 22, 1944–1950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marie, M.; Dale, H.A.; Sannerud, R.; Saraste, J. The Function of the Intermediate Compartment in Pre-Golgi Trafficking Involves its Stable Connection with the Centrosome. Mol. Biol. Cell 2009, 20, 4458–4470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firestone, A.J.; Weinger, J.S.; Maldonado, M.T.; Barlan, K.; Langston, L.D.; O’Donnell, M.; Gelfand, V.I.; Kapoor, T.M.; Chen, J.K. Small-molecule inhibitors of the AAA+ ATPase motor cytoplasmic dynein. Nature 2012, 484, 125–129. [Google Scholar] [CrossRef] [Green Version]
- Welz, T.; Wellbourne-Wood, J.; Kerkhoff, E. Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 2014, 24, 407–415. [Google Scholar] [CrossRef] [PubMed]
- Parmar, H.B.; Duncan, R. A novel tribasic Golgi export signal directs cargo protein interaction with activated Rab11 and AP-1–dependent Golgi–plasma membrane trafficking. Mol. Biol. Cell 2016, 27, 1320–1331. [Google Scholar] [CrossRef] [PubMed]
- Goldenring, J.R. Recycling endosomes. Curr. Opin. Cell Biol. 2015, 35, 117–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baetz, N.W.; Goldenring, J.R. Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol. Biol. Cell 2013, 24, 643–658. [Google Scholar] [CrossRef] [PubMed]
- Meyers, J.M.; Prekeris, R. Formation of Mutually Exclusive Rab11 Complexes with Members of the Family of Rab11-interacting Proteins Regulates Rab11 Endocytic Targeting and Function. J. Biol. Chem. 2002, 277, 49003–49010. [Google Scholar] [CrossRef] [Green Version]
- Horgan, C.P.; Hanscom, S.R.; Jolly, R.S.; Futter, C.; McCaffrey, M.W. Rab11-FIP3 links the Rab11 GTPase and cytoplasmic dynein to mediate transport to the endosomal-recycling compartment. J. Cell Sci. 2010, 123, 181–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petiot, A.; Fauré, J.; Stenmark, H.; Gruenberg, J. PI3P signaling regulates receptor sorting but not transport in the endosomal pathway. J. Cell Biol. 2003, 162, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, E.M.; Stoorvogel, W. Dynamin-dependent Transferrin Receptor Recycling by Endosome-derived Clathrin-coated Vesicles. Mol. Biol. Cell 2002, 13, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Hayakawa, A.; Hayes, S.J.; Lawe, D.C.; Sudharshan, E.; Tuft, R.; Fogarty, K.; Lambright, D.; Corvera, S. Structural Basis for Endosomal Targeting by FYVE Domains. J. Biol. Chem. 2004, 279, 5958–5966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, G.R.; Burke, J.E. Novel roles of phosphoinositides in signaling, lipid transport, and disease. Curr. Opin. Cell Biol. 2020, 63, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Flores-Rodriguez, N.; Kenwright, D.A.; Chung, P.-H.; Harrison, A.W.; Stefani, F.; Waigh, T.A.; Allan, V.J.; Woodman, P.G. ESCRT-0 marks an APPL1-independent transit route for EGFR between the cell surface and the EEA1-positive early endosome. J. Cell Sci. 2015, 128, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Law, F.; Rocheleau, C.E. Vps34 and the Armus/TBC-2 Rab GAPs: Putting the brakes on the endosomal Rab5 and Rab7 GTPases. Cell. Logist. 2017, 7, e1403530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stenmark, H.; Parton, R.; Steele-Mortimer, O.; Lütcke, A.; Gruenberg, J.; Zerial, M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994, 13, 1287–1296. [Google Scholar] [CrossRef] [PubMed]
- Spang, A. Membrane Tethering Complexes in the Endosomal System. Front. Cell Dev. Biol. 2016, 4, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gillingham, A.K.; Munro, S. Transport carrier tethering—How vesicles are captured by organelles. Curr. Opin. Cell Biol. 2019, 59, 140–146. [Google Scholar] [CrossRef]
- Gillingham, A.K.; Sinka, R.; Torres, I.L.; Lilley, K.S.; Munro, S. Toward a Comprehensive Map of the Effectors of Rab GTPases. Dev. Cell 2014, 31, 358–373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, B.D.; Donaldson, J.G. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2009, 10, 597–608. [Google Scholar] [CrossRef] [Green Version]
- Chandra, M.; Chin, Y.K.-Y.; Mas, C.; Feathers, J.R.; Paul, B.; Datta, S.; Chen, K.-E.; Jia, X.; Yang, Z.; Norwood, S.J.; et al. Classification of the human phox homology (PX) domains based on their phosphoinositide binding specificities. Nat. Commun. 2019, 10, 1528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wu, Y.; Belenkaya, T.Y.; Lin, X. SNX3 controls Wingless/Wnt secretion through regulating retromer-dependent recycling of Wntless. Cell Res. 2011, 21, 1677–1690. [Google Scholar] [CrossRef] [Green Version]
- Sönnichsen, B.; De Renzis, S.; Nielsen, E.; Rietdorf, J.; Zerial, M. Distinct Membrane Domains on Endosomes in the Recycling Pathway Visualized by Multicolor Imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 2000, 149, 901–914. [Google Scholar] [CrossRef]
- Horgan, C.P.; McCaffrey, M.W. The dynamic Rab11-FIPs. Biochem. Soc. Trans. 2009, 37, 1032–1036. [Google Scholar] [CrossRef]
- Jonker, C.T.H.; Galmes, R.; Veenendaal, T.; Brink, C.T.; van der Welle, R.E.N.; Liv, N.; de Rooij, J.; Peden, A.A.; van der Sluijs, P.; Margadant, C.; et al. Vps3 and Vps8 control integrin trafficking from early to recycling endosomes and regulate integrin-dependent functions. Nat. Commun. 2018, 9, 792. [Google Scholar] [CrossRef]
- Banerjee, S.; Kane, P.M. Regulation of V-ATPase Activity and Organelle pH by Phosphatidylinositol Phosphate Lipids. Front. Cell Dev. Biol. 2020, 8, 510. [Google Scholar] [CrossRef]
- Schöneberg, J.; Lee, I.-H.; Iwasa, J.H.; Hurley, J.H. Reverse-topology membrane scission by the ESCRT proteins. Nat. Rev. Mol. Cell Biol. 2017, 18, 5–17. [Google Scholar] [CrossRef]
- Redpath, G.; Ecker, M.; Kapoor-Kaushik, N.; Vartoukian, H.; Carnell, M.; Kempe, D.; Biro, M.; Ariotti, N.; Rossy, J. Flotillins promote T cell receptor sorting through a fast Rab5–Rab11 endocytic recycling axis. Nat. Commun. 2019, 10, 4392. [Google Scholar] [CrossRef] [Green Version]
- Horgan, C.P.; Oleksy, A.; Zhdanov, A.V.; Lall, P.Y.; White, I.J.; Khan, A.R.; Futter, C.E.; McCaffrey, J.G.; McCaffrey, M.W. Rab11-FIP3 Is Critical for the Structural Integrity of the Endosomal Recycling Compartment. Traffic 2007, 8, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Inoue, H.; Ha, V.L.; Prekeris, R.; Randazzo, P.A. Arf GTPase-activating Protein ASAP1 Interacts with Rab11 Effector FIP3 and Regulates Pericentrosomal Localization of Transferrin Receptor–positive Recycling Endosome. Mol. Biol. Cell 2008, 19, 4224–4237. [Google Scholar] [CrossRef] [Green Version]
- Traer, C.J.; Rutherford, A.C.; Palmer, K.J.; Wassmer, T.; Oakley, J.; Attar, N.; Carlton, J.; Kremerskothen, J.; Stephens, D.; Cullen, P. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat. Cell Biol. 2007, 9, 1370–1380. [Google Scholar] [CrossRef]
- Ullrich, O.; Reinsch, S.; Urbe, S.; Zerial, M.; Parton, R. Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 1996, 135, 913–924. [Google Scholar] [CrossRef]
- Ren, M.; Xu, G.; Zeng, J.; De Lemos-Chiarandini, C.; Adesnik, M.; Sabatini, D.D. Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 6187–6192. [Google Scholar] [CrossRef] [Green Version]
- Wilcke, M.; Johannes, L.; Galli, T.; Mayau, V.; Goud, B.; Salamero, J. Rab11 Regulates the Compartmentalization of Early Endosomes Required for Efficient Transport from Early Endosomes to the Trans-Golgi Network. J. Cell Biol. 2000, 151, 1207–1220. [Google Scholar] [CrossRef] [Green Version]
- Jean, S.; Kiger, A.A. Coordination between RAB GTPase Functions. Nature reviews. Mol. Cell Biol. 2012, 13, 463. [Google Scholar]
- Knödler, A.; Feng, S.; Zhang, J.; Zhang, X.; Das, A.; Peränen, J.; Guo, W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 6346–6351. [Google Scholar] [CrossRef] [Green Version]
- Vetter, M.; Wang, J.; Lorentzen, E.; Deretic, D. Novel topography of the Rab11-effector interaction network within a ciliary membrane targeting complex. Small GTPases 2015, 6, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheff, D.; Pelletier, L.; O’Connell, C.B.; Warren, G.; Mellman, I. Transferrin receptor recycling in the absence of perinuclear recycling endosomes. J. Cell Biol. 2002, 156, 797–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Ghosh, C.; Xing, Y.; Sun, Y. Phosphatidylinositol 4,5-bisphosphate in the Control of Membrane Trafficking. Int. J. Biol. Sci. 2020, 16, 2761–2774. [Google Scholar] [CrossRef] [PubMed]
- Howe, E.N.; Burnette, M.D.; Justice, M.E.; Schnepp, P.M.; Hedrick, V.; Clancy, J.W.; Guldner, I.H.; Lamere, A.T.; Li, J.; Aryal, U.K.; et al. Rab11b-mediated integrin recycling promotes brain metastatic adaptation and outgrowth. Nat. Commun. 2020, 11, 3017. [Google Scholar] [CrossRef]
- Marcelić, M.; Lučin, H.M.; Begonja, A.J.; Zagorac, G.B.; Lisnić, V.J.; Lučin, P. Endosomal Phosphatidylinositol-3-Phosphate-Associated Functions Are Dispensable for Establishment of the Cytomegalovirus Pre-Assembly Compartment but Essential for the Virus Growth. Life 2021, 11, 859. [Google Scholar] [CrossRef]
- Lučin, P.; Kareluša, L.; Zagorac, G.B.; Lučin, H.M.; Pavišić, V.; Vučko, N.J.; Jurić, S.L.; Marcelić, M.; Lisnić, B.; Jonjić, S. Cytomegaloviruses Exploit Recycling Rab Proteins in the Sequential Establishment of the Assembly Compartment. Front. Cell Dev. Biol. 2018, 6, 165. [Google Scholar] [CrossRef] [Green Version]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcelić, M.; Mahmutefendić Lučin, H.; Jurak Begonja, A.; Blagojević Zagorac, G.; Lučin, P. Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment. Cells 2022, 11, 962. https://doi.org/10.3390/cells11060962
Marcelić M, Mahmutefendić Lučin H, Jurak Begonja A, Blagojević Zagorac G, Lučin P. Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment. Cells. 2022; 11(6):962. https://doi.org/10.3390/cells11060962
Chicago/Turabian StyleMarcelić, Marina, Hana Mahmutefendić Lučin, Antonija Jurak Begonja, Gordana Blagojević Zagorac, and Pero Lučin. 2022. "Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment" Cells 11, no. 6: 962. https://doi.org/10.3390/cells11060962
APA StyleMarcelić, M., Mahmutefendić Lučin, H., Jurak Begonja, A., Blagojević Zagorac, G., & Lučin, P. (2022). Early Endosomal Vps34-Derived Phosphatidylinositol-3-Phosphate Is Indispensable for the Biogenesis of the Endosomal Recycling Compartment. Cells, 11(6), 962. https://doi.org/10.3390/cells11060962







