Proteomics Approach Highlights Early Changes in Human Fibroblasts-Pancreatic Ductal Adenocarcinoma Cells Crosstalk
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Production of MIA-PaCa2 Cells Conditioned Medium (CM)
2.3. Collection of HDFs after MIAPaCa2-CM Treatment for Proteomic Analysis
2.4. Label-Free Proteomics Analysis
2.5. Bioinformatics and Proteomics Data Processing
2.6. Western Blot Analysis
2.7. In Vitro Scratch Assay
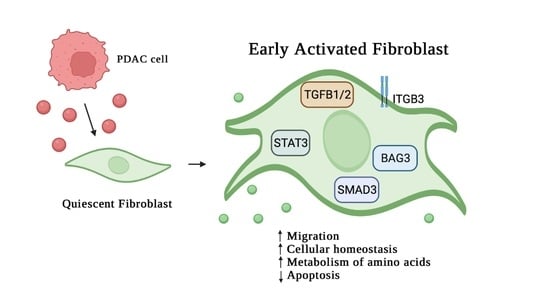
3. Results
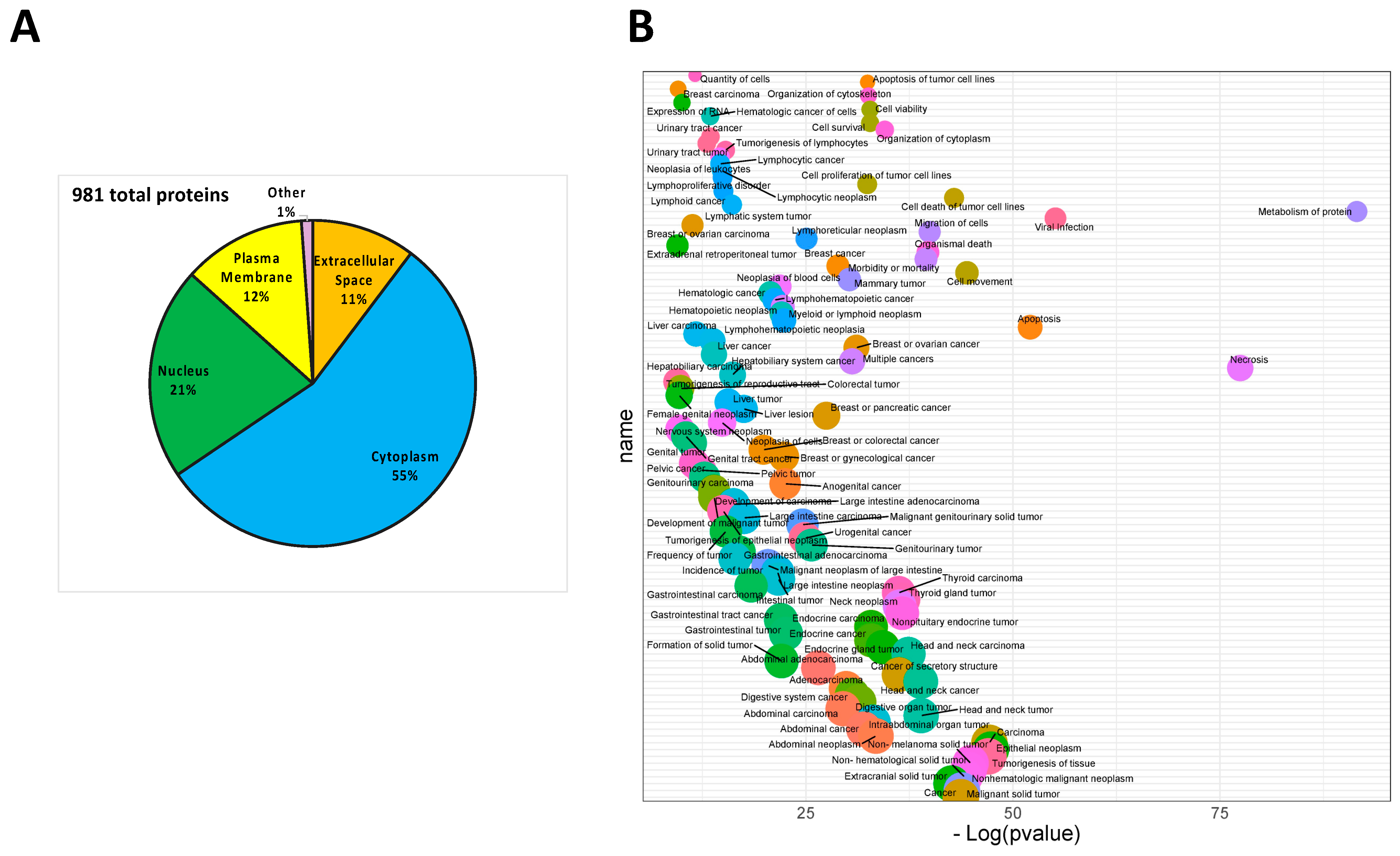
3.1. Analysis of Protein Secreted by PDAC Cells
3.2. Effect of MIA-PaCa2 Conditioned Media on HDFs
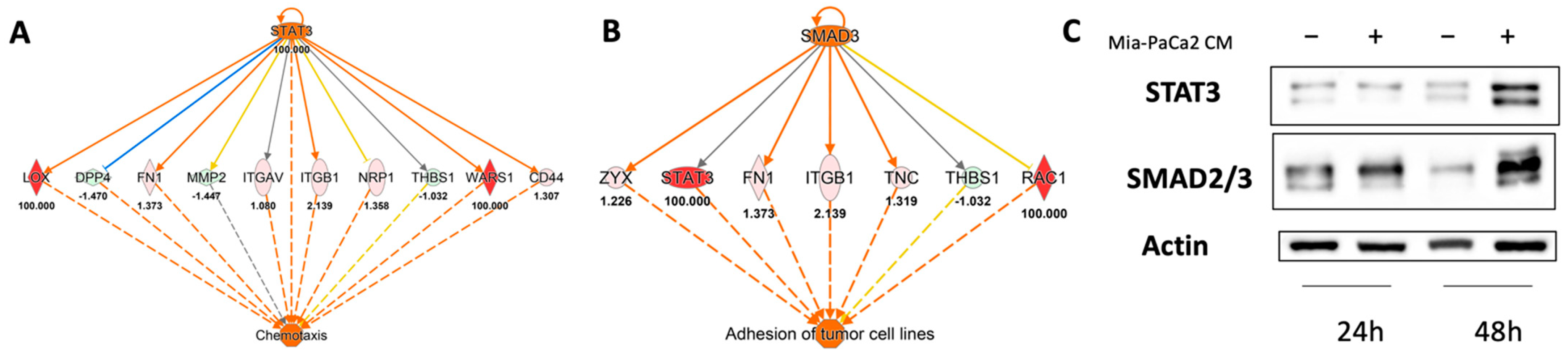
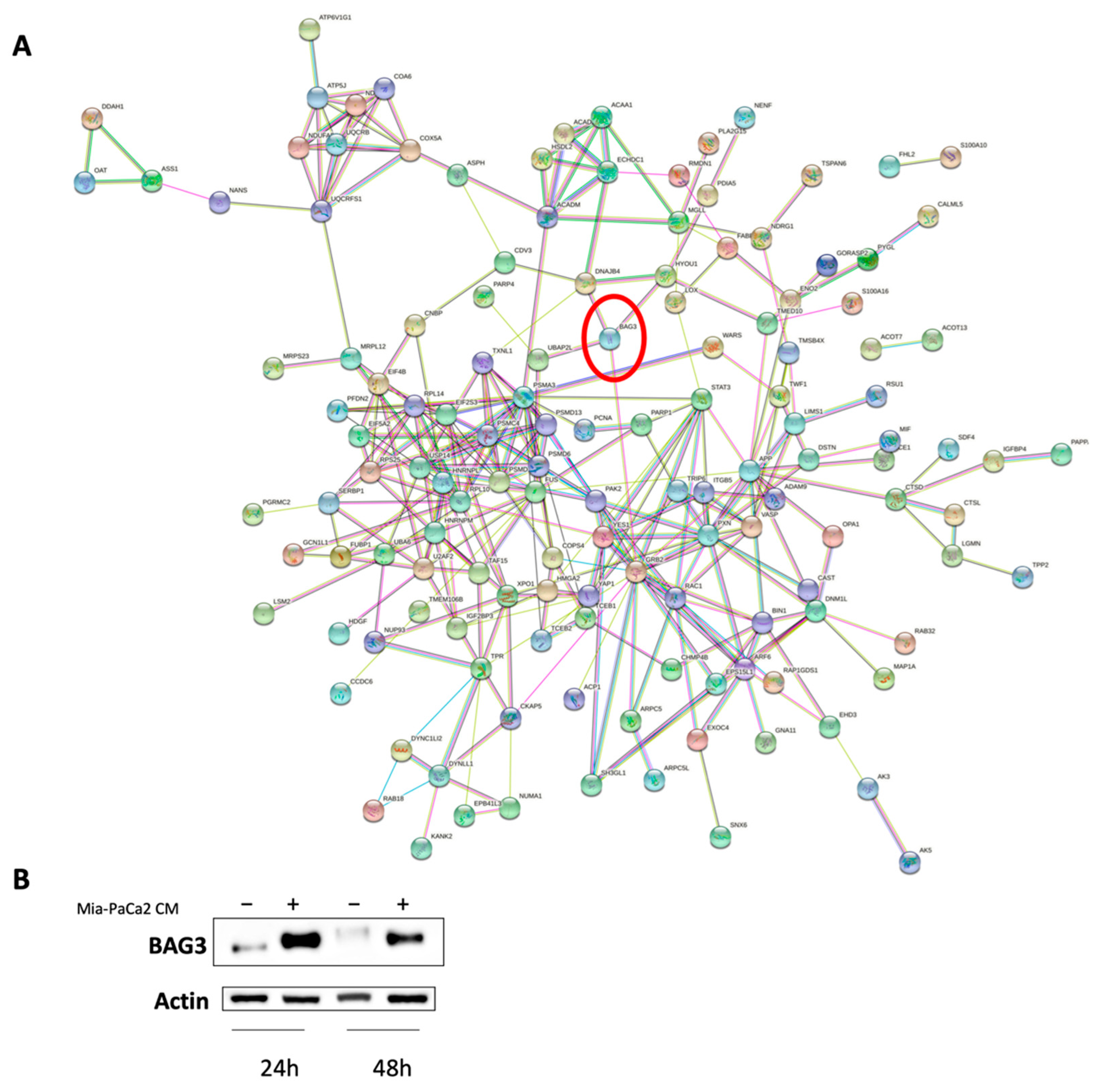
3.3. Regulator Effects Predicted by IPA in HDF Treated Cell Lysates
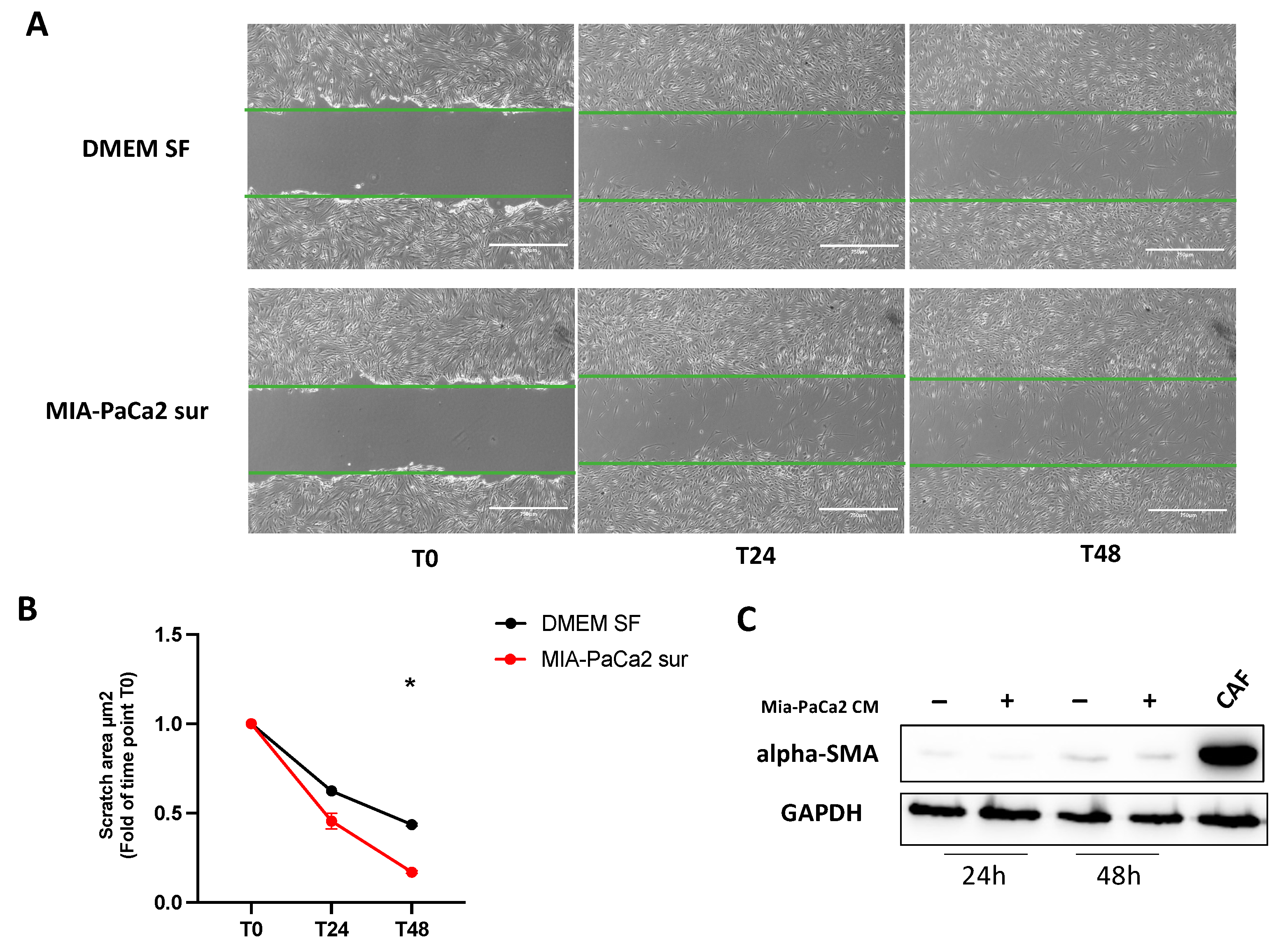
3.4. MIA-PaCa2 CM Increases HDFs Migration
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kleeff, J.; Korc, M.; Apte, M.; La Vecchia, C.; Johnson, C.D.; Biankin, A.V.; Neale, R.E.; Tempero, M.; Tuveson, D.A.; Hruban, R.H.; et al. Pancreatic cancer. Nat. Rev. Dis. Primers 2016, 2, 16022. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef]
- Hidalgo, M.; Cascinu, S.; Kleeff, J.; Labianca, R.; Löhr, J.M.; Neoptolemos, J.; Real, F.X.; Van Laethem, J.L.; Heinemann, V. Addressing the challenges of pancreatic cancer: Future directions for improving outcomes. Pancreatology 2015, 15, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Strobel, O.; Neoptolemos, J.; Jäger, D.; Büchler, M.W. Optimizing the outcomes of pancreatic cancer surgery. Nat. Rev. Clin. Oncol. 2019, 16, 11–26. [Google Scholar] [CrossRef]
- Dougan, S.K. The Pancreatic Cancer Microenvironment. Cancer J. 2017, 23, 321–325. [Google Scholar] [CrossRef]
- Huber, M.; Brehm, C.U.; Gress, T.M.; Buchholz, M.; Alhamwe, B.A.; von Strandmann, E.P.; Slater, E.P.; Bartsch, J.W.; Bauer, C.; Lauth, M. The Immune Microenvironment in Pancreatic Cancer. Int. J. Mol. Sci. 2020, 21, 7307. [Google Scholar] [CrossRef] [PubMed]
- Li, K.Y.; Yuan, J.L.; Trafton, D.; Wang, J.X.; Niu, N.; Yuan, C.H.; Liu, X.B.; Zheng, L. Pancreatic ductal adenocarcinoma immune microenvironment and immunotherapy prospects. Chronic Dis. Transl. Med. 2020, 6, 6–17. [Google Scholar] [CrossRef]
- Von Ahrens, D.; Bhagat, T.D.; Nagrath, D.; Maitra, A.; Verma, A. The role of stromal cancer-associated fibroblasts in pancreatic cancer. J. Hematol. Oncol. 2017, 10, 76. [Google Scholar] [CrossRef] [Green Version]
- Norton, J.; Foster, D.; Chinta, M.; Titan, A.; Longaker, M. Pancreatic Cancer Associated Fibroblasts (CAF): Under-Explored Target for Pancreatic Cancer Treatment. Cancers 2020, 12, 1347. [Google Scholar] [CrossRef]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Truffi, M.; Sorrentino, L.; Corsi, F. Fibroblasts in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 15–29. [Google Scholar] [CrossRef] [PubMed]
- D’Arcangelo, E.; Wu, N.C.; Cadavid, J.L.; McGuigan, A.P. The life cycle of cancer-associated fibroblasts within the tumour stroma and its importance in disease outcome. Br. J. Cancer 2020, 122, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Sperb, N.; Tsesmelis, M.; Wirth, T. Crosstalk between Tumor and Stromal Cells in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 5486. [Google Scholar] [CrossRef] [PubMed]
- Bonollo, F.; Thalmann, G.N.; Kruithof-de Julio, M.; Karkampouna, S. The Role of Cancer-Associated Fibroblasts in Prostate Cancer Tumorigenesis. Cancers 2020, 12, 1887. [Google Scholar] [CrossRef] [PubMed]
- Procacci, P.; Moscheni, C.; Sartori, P.; Sommariva, M.; Gagliano, N. Tumor—Stroma Cross-Talk in Human Pancreatic Ductal Adenocarcinoma: A Focus on the Effect of the Extracellular Matrix on Tumor Cell Phenotype and Invasive Potential. Cells 2018, 7, 158. [Google Scholar] [CrossRef] [Green Version]
- Amrutkar, M.; Aasrum, M.; Verbeke, C.S.; Gladhaug, I.P. Secretion of fibronectin by human pancreatic stellate cells promotes chemoresistance to gemcitabine in pancreatic cancer cells. BMC Cancer 2019, 19, 596. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef]
- Madonna, R.; Moscato, S.; Polizzi, E.; Pieragostino, D.; Cufaro, M.C.; Del Boccio, P.; Bianchi, F.; De Caterina, R.; Mattii, L.; Hills, C.E. Connexin 43 and Connexin 26 Involvement in the Ponatinib-Induced Cardiomyopathy: Sex-Related Differences in a Murine Model. Int. J. Mol. Sci. 2021, 22, 5815. [Google Scholar] [CrossRef]
- Potenza, F.; Cufaro, M.C.; Di Biase, L.; Panella, V.; Di Campli, A.; Ruggieri, A.G.; Dufrusine, B.; Restelli, E.; Pietrangelo, L.; Protasi, F.; et al. Proteomic Analysis of Marinesco-Sjogren Syndrome Fibroblasts Indicates Pro-Survival Metabolic Adaptation to SIL1 Loss. Int. J. Mol. Sci. 2021, 22, 12449. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Landry, N.M.; Rattan, S.G.; Dixon, I.M.C. An Improved Method of Maintaining Primary Murine Cardiac Fibroblasts in Two-Dimensional Cell Culture. Sci. Rep. 2019, 9, 12889. [Google Scholar] [CrossRef] [Green Version]
- Clabaut, A.; Grare, C.; Léger, T.; Hardouin, P.; Broux, O. Variations of secretome profiles according to conditioned medium preparation: The example of human mesenchymal stem cell-derived adipocytes. Electrophoresis 2015, 36, 2587–2593. [Google Scholar] [CrossRef]
- Cufaro, M.C.; Pieragostino, D.; Lanuti, P.; Rossi, C.; Cicalini, I.; Federici, L.; De Laurenzi, V.; Boccio, P. Del Extracellular Vesicles and Their Potential Use in Monitoring Cancer Progression and Therapy: The Contribution of Proteomics. J. Oncol. 2019, 2019, 1639854. [Google Scholar] [CrossRef]
- Simeone, P.; Bologna, G.; Lanuti, P.; Pierdomenico, L.; Guagnano, M.T.; Pieragostino, D.; Del Boccio, P.; Vergara, D.; Marchisio, M.; Miscia, S.; et al. Extracellular Vesicles as Signaling Mediators and Disease Biomarkers across Biological Barriers. Int. J. Mol. Sci. 2020, 21, 2514. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Xue, L.; Zhang, X.; Bu, S.; Zhu, X.; Lai, D. Autophagy protects ovarian cancer-associated fibroblasts against oxidative stress. Cell Cycle 2016, 15, 1376–1385. [Google Scholar] [CrossRef]
- Xiao, L.; Du, Y.; Shen, Y.; He, Y.; Zhao, H.; Li, Z. TGF-beta 1 induced fibroblast proliferation is mediated by the FGF-2/ERK pathway. Front. Biosci. 2012, 17, 2667–2674. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Chen, X.; Shan, T.; Ma, J.; Lin, W.; Li, W.; Kang, Y. MiR-21-mediated Metabolic Alteration of Cancer-associated Fibroblasts and Its Effect on Pancreatic Cancer Cell Behavior. Int. J. Biol. Sci. 2018, 14, 100–110. [Google Scholar] [CrossRef] [Green Version]
- Sapudom, J.; Karaman, S.; Mohamed, W.K.E.; Garcia-Sabaté, A.; Quartey, B.C.; Teo, J.C.M. 3D in vitro M2 macrophage model to mimic modulation of tissue repair. npj Regen. Med. 2021, 6, 83. [Google Scholar] [CrossRef] [PubMed]
- Wipff, P.J.; Rifkin, D.B.; Meister, J.J.; Hinz, B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J. Cell Biol. 2007, 179, 1311–1323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rønnov-Jessen, L.; Petersen, O.W. A function for filamentous alpha-smooth muscle actin: Retardation of motility in fibroblasts. J. Cell Biol. 1996, 134, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, Y.; Saitoh, M.; Miyazawa, K. Smad3-STAT3 crosstalk in pathophysiological contexts. Acta Biochim. Biophys. Sin. 2018, 50, 82–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, B.; Liu, T.; Wu, J.C.; Luo, S.Z.; Chen, R.; Lu, L.G.; Xu, M.Y. STAT3 aggravates TGF-β1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed. Pharmacother. 2018, 98, 214–221. [Google Scholar] [CrossRef]
- O’Leary, E.M.; Tian, Y.; Nigdelioglu, R.; Witt, L.J.; Cetin-Atalay, R.; Meliton, A.Y.; Woods, P.S.; Kimmig, L.M.; Sun, K.A.; Gökalp, G.A.; et al. TGF-β Promotes Metabolic Reprogramming in Lung Fibroblasts via mTORC1-dependent ATF4 Activation. Am. J. Respir. Cell Mol. Biol. 2020, 63, 601–612. [Google Scholar] [CrossRef]
- Marzullo, L.; Turco, M.C.; De Marco, M. The multiple activities of BAG3 protein: Mechanisms. Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129628. [Google Scholar] [CrossRef]
- An, M.X.; Li, S.; Yao, H.B.; Li, C.; Wang, J.M.; Sun, J.; Li, X.Y.; Meng, X.N.; Wang, H.Q. BAG3 directly stabilizes Hexokinase 2 mRNA and promotes aerobic glycolysis in pancreatic cancer cells. J. Cell Biol. 2017, 216, 4091–4105. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, M.; Tanaka, R.; Hishiya, A.; Homma, S.; Reed, J.C.; Takayama, S. BAG3 directly associates with guanine nucleotide exchange factor of Rap1, PDZGEF2, and regulates cell adhesion. Biochem. Biophys. Res. Commun. 2010, 400, 413–418. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.; Oyang, L.; Liang, J.; Yi, P.; Han, Y.; Luo, X.; Xia, L.; Lin, J.; Tan, S.; Hu, J.; et al. Biological Function of HYOU1 in Tumors and Other Diseases. Onco. Targets. Ther. 2021, 14, 1727–1735. [Google Scholar] [CrossRef] [PubMed]
- Giubellino, A.; Burke, T.R.; Bottaro, D.P. Grb2 signaling in cell motility and cancer. Expert Opin. Ther. Targets 2008, 12, 1021–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Huang, Y. Knockdown of ubiquitin associated protein 2-like inhibits the growth and migration of prostate cancer cells. Oncol. Rep. 2014, 32, 1578–1584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Chen, Y.; Cai, L.; Li, Z.; Guo, X. UBAP2L silencing inhibits cell proliferation and G2/M phase transition in breast cancer. Breast Cancer 2018, 25, 224–232. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.Y.; Hsueh, C.M.; Yu, S.L.; Su, C.C.; Shum, W.Y.; Yeh, K.C.; Chang, G.C.; Chen, J.J.W. HLJ1 is a novel caspase-3 substrate and its expression enhances UV-induced apoptosis in non-small cell lung carcinoma. Nucleic Acids Res. 2010, 38, 6148–6158. [Google Scholar] [CrossRef]
- Acun, T.; Doberstein, N.; Habermann, J.K.; Gemoll, T.; Thorns, C.; Oztas, E.; Ried, T. HLJ1 (DNAJB4) Gene Is a Novel Biomarker Candidate in Breast Cancer. OMICS 2017, 21, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhou, J.; Zhang, C.; Fu, W.; Xiao, X.; Ruan, S.; Zhang, Y.; Luo, X.; Tang, M. HLJ1 is a novel biomarker for colorectal carcinoma progression and overall patient survival. Int. J. Clin. Exp. Pathol. 2014, 7, 969. [Google Scholar]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Damiani, V.; Cufaro, M.C.; Fucito, M.; Dufrusine, B.; Rossi, C.; Del Boccio, P.; Federici, L.; Turco, M.C.; Sallese, M.; Pieragostino, D.; et al. Proteomics Approach Highlights Early Changes in Human Fibroblasts-Pancreatic Ductal Adenocarcinoma Cells Crosstalk. Cells 2022, 11, 1160. https://doi.org/10.3390/cells11071160
Damiani V, Cufaro MC, Fucito M, Dufrusine B, Rossi C, Del Boccio P, Federici L, Turco MC, Sallese M, Pieragostino D, et al. Proteomics Approach Highlights Early Changes in Human Fibroblasts-Pancreatic Ductal Adenocarcinoma Cells Crosstalk. Cells. 2022; 11(7):1160. https://doi.org/10.3390/cells11071160
Chicago/Turabian StyleDamiani, Verena, Maria Concetta Cufaro, Maurine Fucito, Beatrice Dufrusine, Claudia Rossi, Piero Del Boccio, Luca Federici, Maria Caterina Turco, Michele Sallese, Damiana Pieragostino, and et al. 2022. "Proteomics Approach Highlights Early Changes in Human Fibroblasts-Pancreatic Ductal Adenocarcinoma Cells Crosstalk" Cells 11, no. 7: 1160. https://doi.org/10.3390/cells11071160
APA StyleDamiani, V., Cufaro, M. C., Fucito, M., Dufrusine, B., Rossi, C., Del Boccio, P., Federici, L., Turco, M. C., Sallese, M., Pieragostino, D., & De Laurenzi, V. (2022). Proteomics Approach Highlights Early Changes in Human Fibroblasts-Pancreatic Ductal Adenocarcinoma Cells Crosstalk. Cells, 11(7), 1160. https://doi.org/10.3390/cells11071160