Identification, Structural, and Expression Analyses of SPX Genes in Giant Duckweed (Spirodela polyrhiza) Reveals Its Role in Response to Low Phosphorus and Nitrogen Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of SPX Genes
2.2. Physiochemical Properties, Subcellular Localization and SSRs (Simple Sequence Repeats) Identification
2.3. Phylogenetic Classification, Gene Structures, and Motif Analysis
2.4. Cis-Acting Elements, Chromosomal Localization, Gene Duplication, and Synteny Analysis
2.5. Molecular Interaction Networks and Protein Structure Prediction for SpSPX Proteins
2.6. Expression Profiles of SpSPX Genes under Low P/N Stress
2.6.1. Expression Analysis of SpSPX Genes by RNA-Seq Data
2.6.2. Expression Analysis of SpSPX Genes by qRT–PCR (Quantitative Real-Time PCR)
3. Results
3.1. Identification and Phylogenetic Analysis of SPX Genes in 28 Species
3.2. Gene Structure and Conserved Motifs of SPX Genes
3.3. Cis-Regulatory Element Analysis of SPX Genes
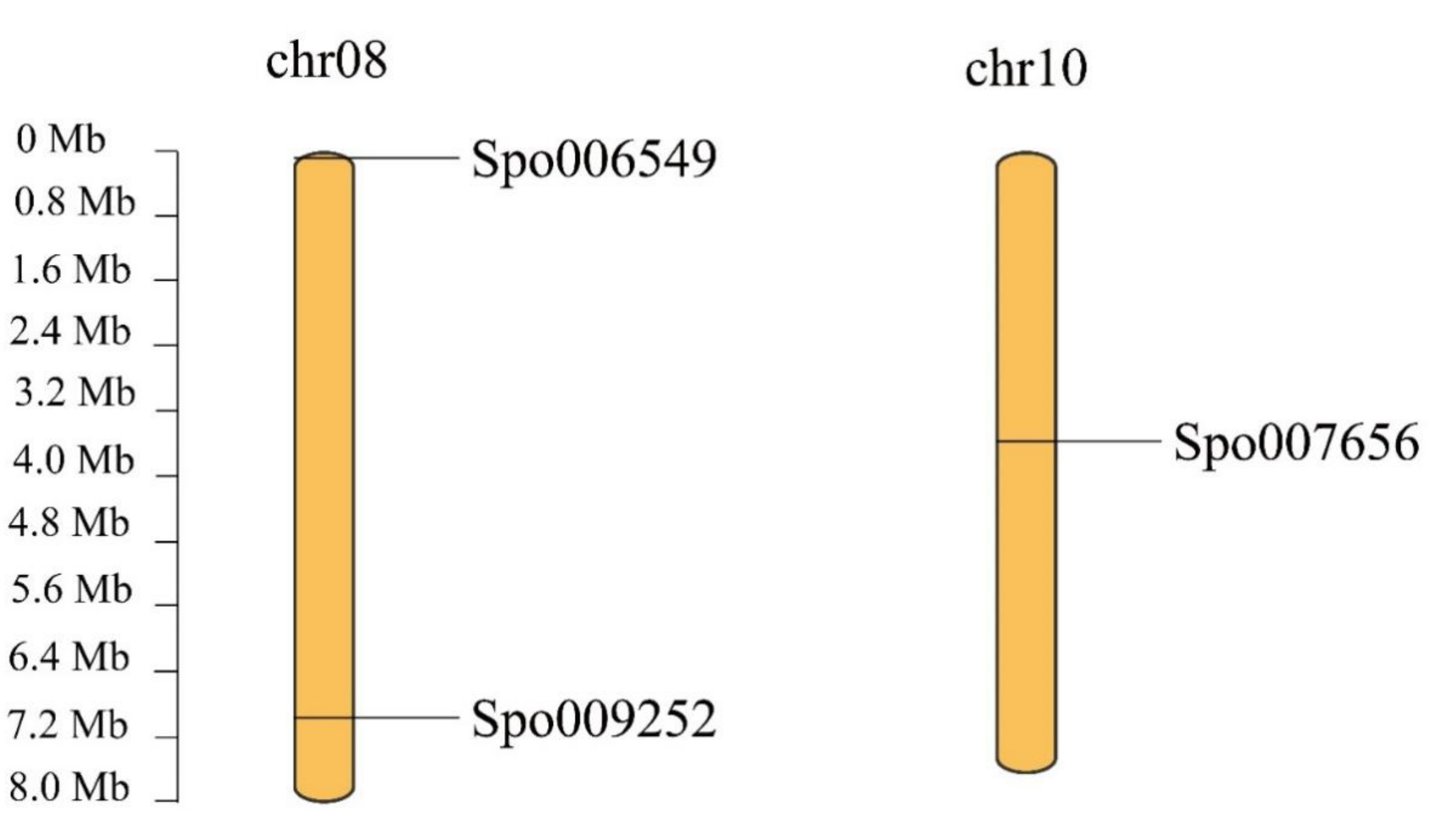
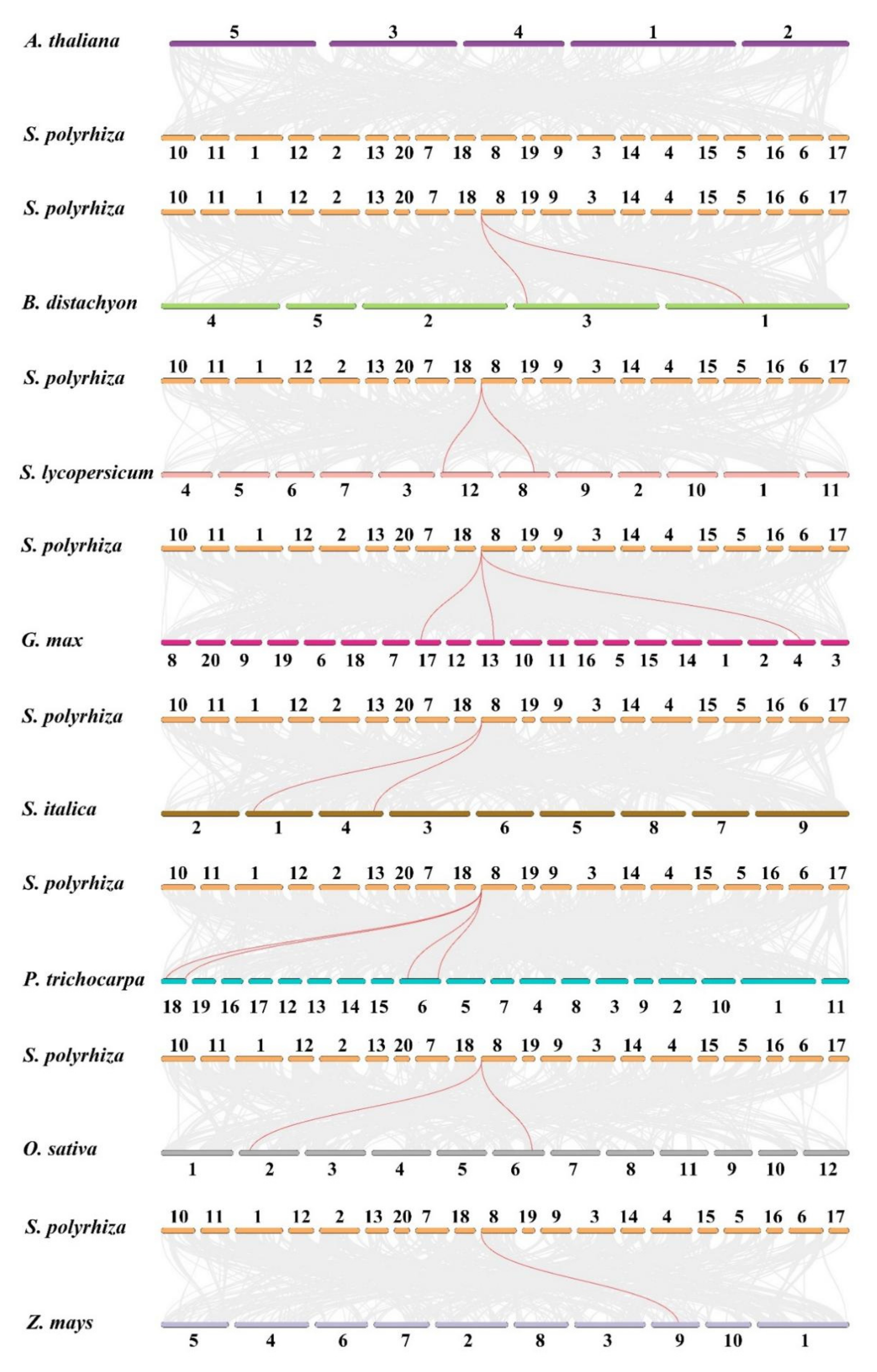
3.4. Chromosomal Localization, Gene Duplication and Synteny Analysis of SPX Genes in S. polyrhiza
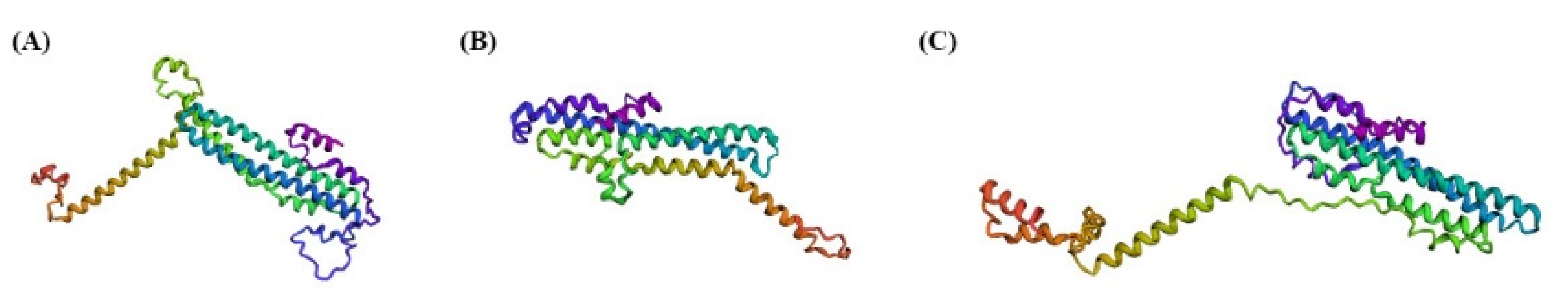
3.5. Molecular Interaction Networks and Protein Structure Prediction for SpSPXs
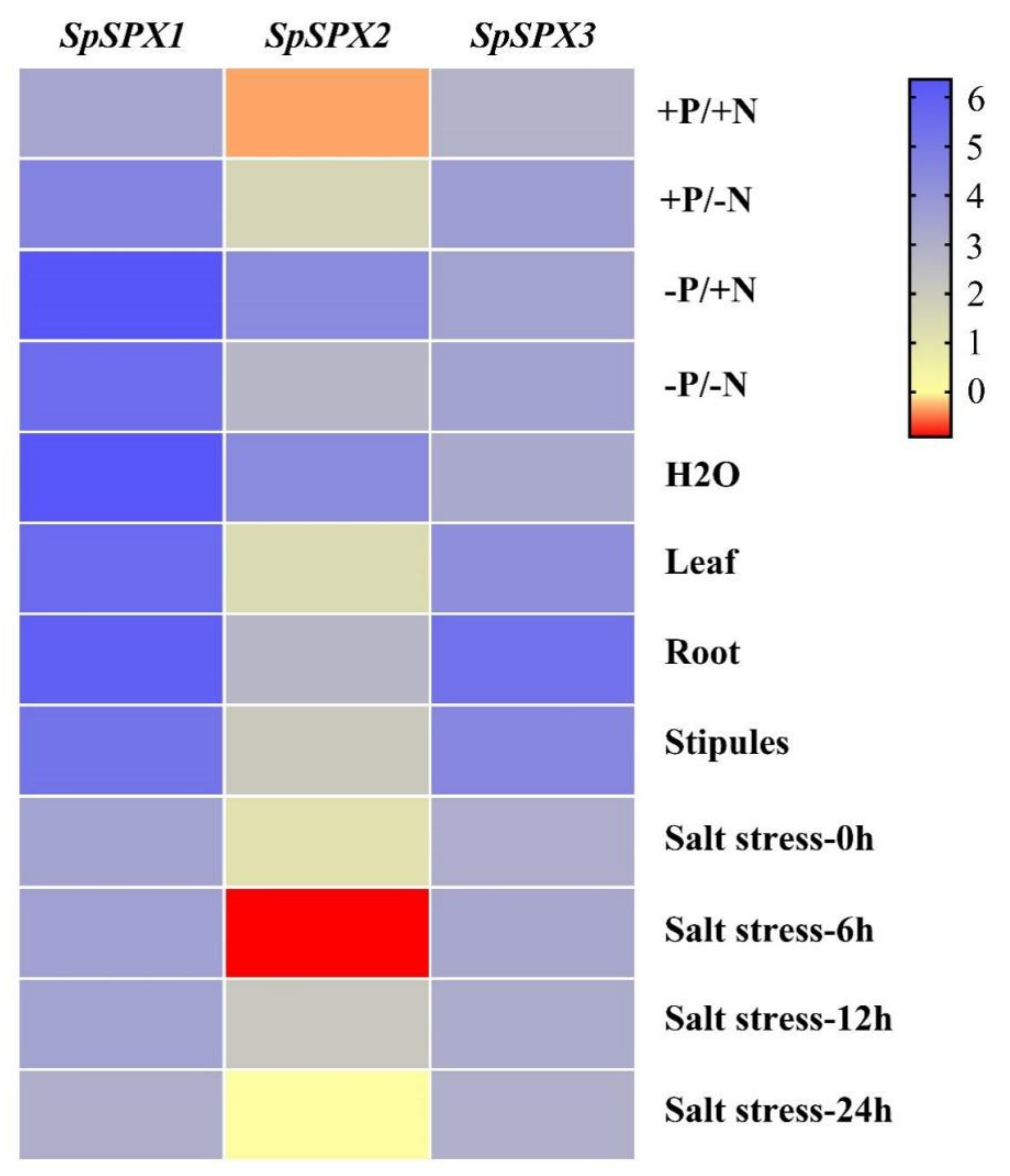
3.6. Transcriptional Patterns of SpSPX Genes in S. polyrhiza
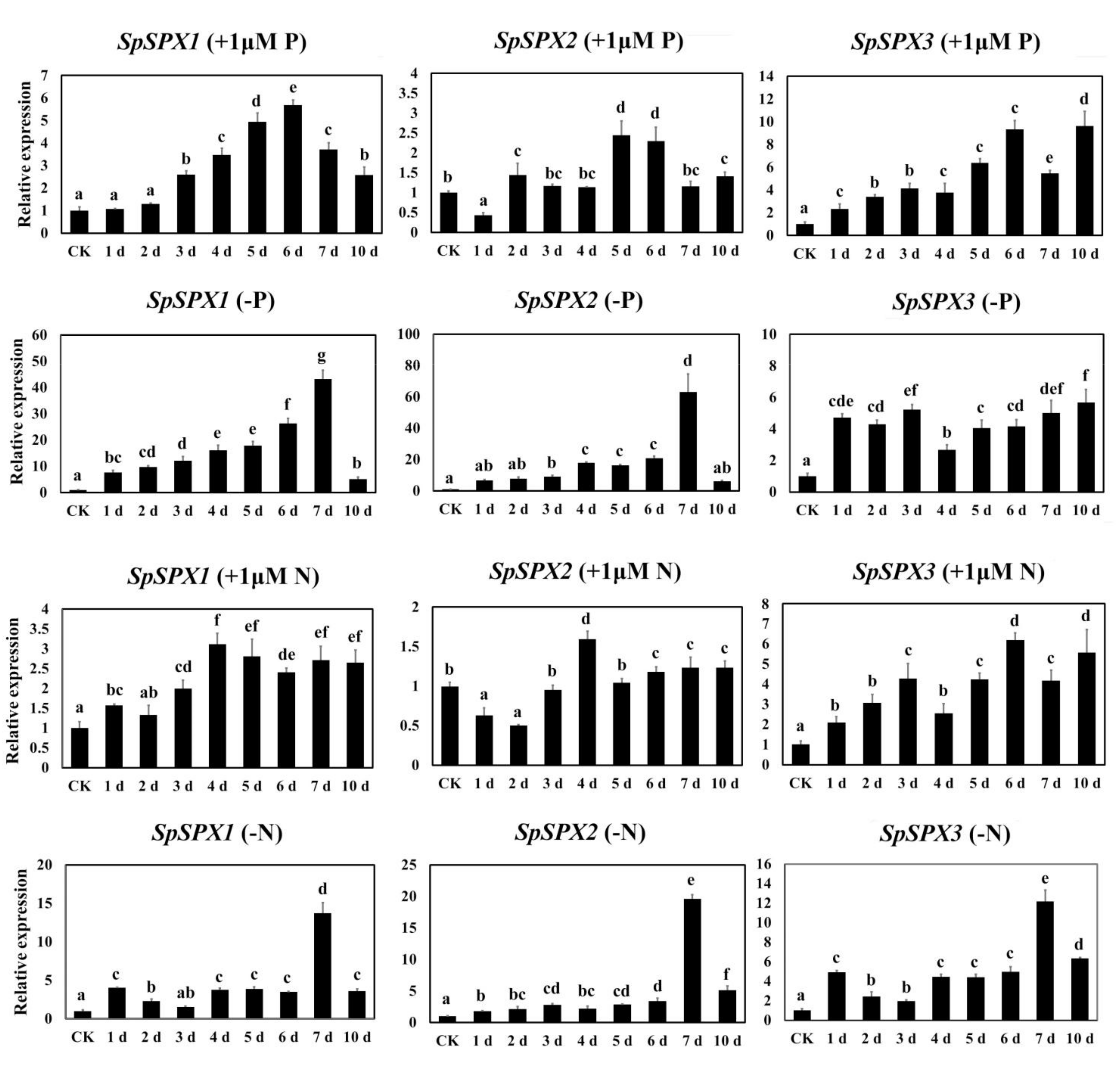
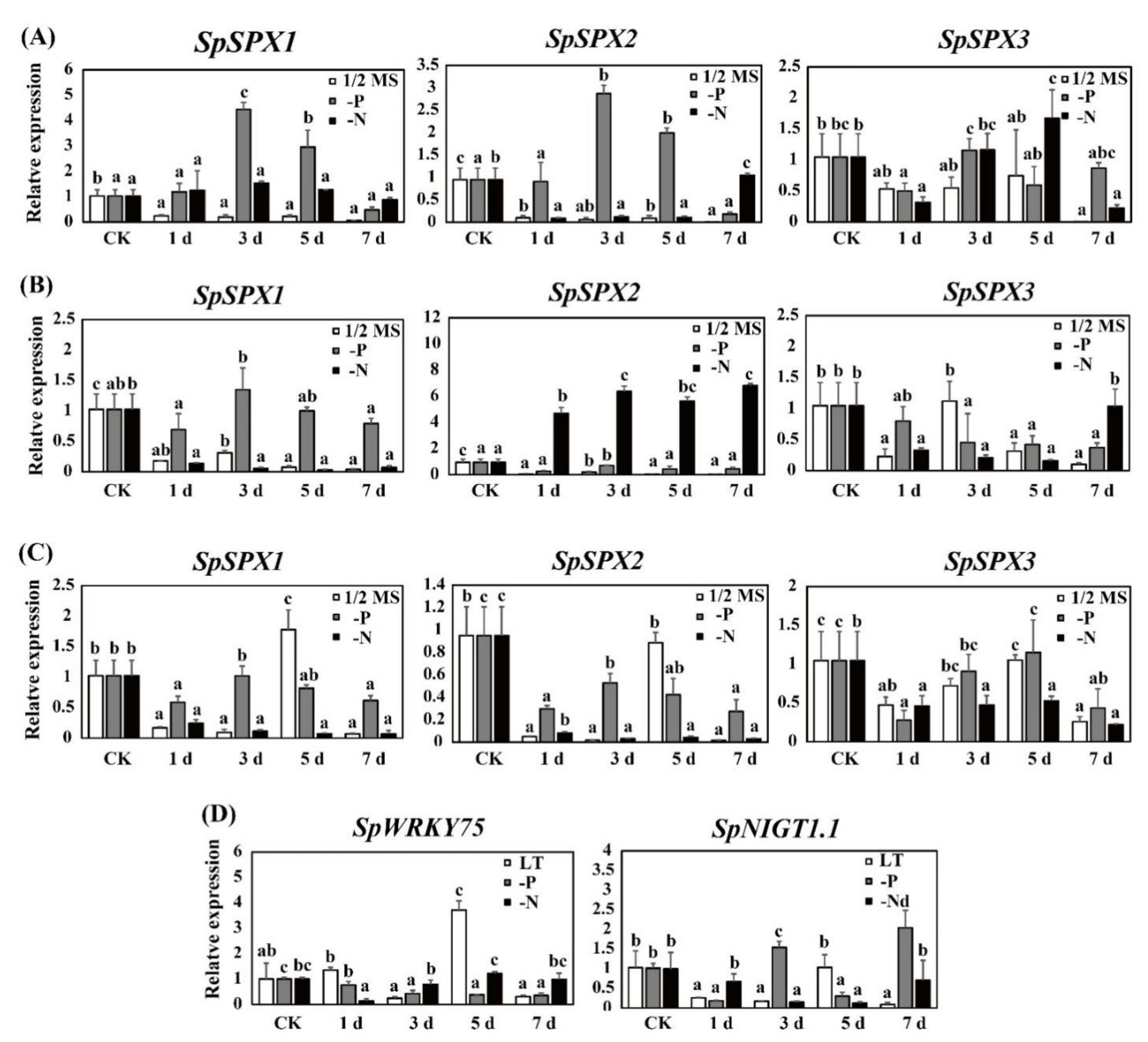
3.7. Expression Analysis of SpSPX Genes in Response to low P/N Stress by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heuer, S.; Gaxiola, R.; Schilling, R.; Herrera-Estrella, L.; López-Arredondo, D.; Wissuwa, M.; Delhaize, E.; Rouached, H. Improving phosphorus use efficiency: A complex trait with emerging opportunities. Plant J. 2017, 90, 868–885. [Google Scholar] [CrossRef] [PubMed]
- Holford, I.C.R. Soil phosphorus: Its measurement, and its uptake by plants. Aust. J. Soil Res. 1997, 35, 227–240. [Google Scholar] [CrossRef]
- Gilbert, N. Environment: The disappearing nutrient. Nature 2009, 461, 716–718. [Google Scholar] [CrossRef] [PubMed]
- López-Arredondo, D.L.; Leyva-González, M.A.; González-Morales, S.I.; López-Bucio, J.; Herrera-Estrella, L. Phosphate nutrition: Improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 2014, 65, 95–123. [Google Scholar] [CrossRef]
- Rubio, V.; Linhares, F.; Solano, R.; Martin, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed]
- Puga, M.I.; Rojas-Triana, M.; De Lorenzo, L.; Leyva, A.; Rubio, V.; Paz-Ares, J. Novel signals in the regulation of Pi starvation responses in plants: Facts and promises. Curr. Opin. Plant Biol. 2017, 39, 40–49. [Google Scholar] [CrossRef]
- Gonzalez, E.; Solano, R.; Rubio, V.; Leyva, A.; Paz-Ares, J. Phosphate Transporter Traffic Facilitator1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell. 2005, 17, 3500–3512. [Google Scholar] [CrossRef]
- Bari, R.; Pant, B.D.; Stitt, M.; Scheible, W. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006, 141, 988–999. [Google Scholar] [CrossRef]
- Lin, W.; Huang, T.; Chiou, T. Nitrogen limitation adaption, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell. 2013, 25, 4061–4074. [Google Scholar] [CrossRef]
- Wu, P.; Shou, H.; Xu, G.; Lian, X. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 2013, 16, 205–212. [Google Scholar] [CrossRef]
- Dong, J.; Ma, G.; Wei, M.; Satheesh, V.; Zhang, R.; Ge, S.; Li, J.; Zhang, T.; Wittwer, C.; Jessen, H.J.; et al. Inositol Pyrophosphate InsP8 acts as an intracellular phosphate signal in Arabidopsis. Mol. Plant. 2019, 12, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Arpat, B.A.; Wang, Z.; Poirier, Y.; Tyerman, S.D.; Wu, P.; Shou, H.; Whelan, J. The emerging importance of the SPX domain-containing proteins in phosphate homeostasis. New Phytol. 2012, 193, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yang, L.; Luan, M.; Wang, Y.; Zhang, C.; Zhang, B.; Shi, J.; Zhao, F.G.; Lan, W.; Luan, S. A vacuolar phosphate transporter essential for phosphate homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E6571–E6578. [Google Scholar] [CrossRef]
- Wege, S.; Khan, G.A.; Jung, J.; Vogiatzaki, E.; Pradervand, S.; Aller, I.; Meyer, A.J.; Poirier, Y. The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 2016, 170, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Yi, K.; Dang, L.; Huang, H.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef]
- Puga, M.I.; Mateos, I.; Charukesi, R.; Wang, Z.; Franco-Zorrilla, J.M.; De Lorenzo, L.; Irigoyen, M.L.; Masiero, S.; Bustos, R.; Rodríguez, J.; et al. SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14947–14952. [Google Scholar] [CrossRef]
- Wang, Z.; Ruan, W.; Shi, J.; Zhang, L.; Xiang, D.; Yang, C.; Li, C.; Wu, Z.; Liu, Y.; Shou, H.; et al. Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 14953–14958. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, Y.; Guo, J.; Zhu, X.; Shi, J.; He, Q.; Liu, Y.; Wu, Y.; Zhang, L.; Lv, Q.; et al. Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 2018, 219, 135–148. [Google Scholar] [CrossRef]
- Shi, J.; Hu, H.; Zhang, K.; Zhang, W.; Yu, Y.; Wu, Z.; Wu, P. The paralogous SPX3 and SPX5 genes redundantly modulate Pi homeostasis in rice. J. Exp. Bot. 2014, 65, 859–870. [Google Scholar] [CrossRef]
- Lv, Q.; Zhong, Y.; Wang, Y.; Wang, Z.; Zhang, L.; Shi, J.; Wu, Z.; Liu, Y.; Mao, C.; Yi, K.; et al. SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 2014, 26, 1586–1597. [Google Scholar] [CrossRef]
- Hu, B.; Jiang, Z.; Wang, W.; Qiu, Y.; Zhang, Z.; Liu, Y.; Li, A.; Gao, X.; Liu, L.; Qian, Y.; et al. Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat. Plants 2019, 5, 401–413. [Google Scholar] [CrossRef] [PubMed]
- Bog, M.; Sree, K.S.; Fuchs, J.; Hoang, P.T.N.; Schubert, I.; Kuever, J.; Rabenstein, A.; Paolacci, S.; Jansen, M.A.K.; Appenroth, K.J. A taxonomic revision of Lemna sect. Uninerves (Lemnaceae). Taxon 2020, 69, 56–66. [Google Scholar] [CrossRef]
- Acosta, K.; Appenroth, K.J.; Borisjuk, L.; Edelman, M.; Heinig, U.; Jansen, M.A.K.; Oyama, T.; Pasaribu, B.; Schubert, I.; Sorrels, S.; et al. Return of the Lemnaceae: Duckweed as a model plant system in the genomics and postgenomics era. Plant Cell. 2021, 33, 3207–3234. [Google Scholar] [CrossRef] [PubMed]
- An, D.; Zhou, Y.; Li, C.; Xiao, Q.; Wang, T.; Zhang, Y.; Wu, Y.; Li, Y.; Chao, D.; Messing, J.; et al. Plant evolution and environmental adaptation unveiled by long-read whole-genome sequencing of Spirodela. Proc. Natl. Acad. Sci. USA 2019, 116, 18893–18899. [Google Scholar] [CrossRef]
- Yang, J.J.; Li, G.J.; Bishopp, A.; Heenatigala, P.P.M.; Hu, S.; Chen, Y.; Wu, Z.; Kumar, S.; Duan, P.; Yao, L.; et al. A Comparison of growth on mercuric chloride for three Lemnaceae species reveals differences in growth dynamics that effect their suitability for use in either monitoring or remediating ecosystems contaminated with mercury. Front Chem. 2018, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Cheng, J.J.; Appenroth, K. Growing duckweed for biofuel production: A review. Plant Biol. 2015, 1, 16–23. [Google Scholar] [CrossRef]
- Zhang, Z.; Xin, J.; Cai, S.; Xiao, Q. Study on growth of duckweed and its removal efficiency of phosphorus under different P-concentrations. Acta Agric. Jiangxi 2011, 23, 160–162. [Google Scholar]
- Toyama, T.; Hanaoka, T.; Tanaka, Y.; Morikawa, M.; Mori, K. Comprehensive evaluation of nitrogen removal rate and biomass, ethanol, and methane production yields by combination of four major duckweeds and three types of wastewater effluent. Bioresour. Technol. 2018, 250, 464–473. [Google Scholar] [CrossRef]
- Wang, C.; Ying, S.; Huang, H.; Li, K.; Wu, P.; Shou, H. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. 2009, 57, 895–904. [Google Scholar] [CrossRef]
- Wang, Z.; Hu, H.; Huang, H.; Duan, K.; Wu, Z.; Wu, P. Regulation of OsSPX1 and OsSPX3 on expression of OsSPX domain genes and Pi-starvation signaling in rice. J. Integr. Plant Biol. 2009, 51, 663–674. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Xu, Y.; Yao, M.; Xie, F.; Gai, J.; Li, Y. Soybean SPX1 is an important component of the response to phosphate deficiency for phosphorus homeostasis. Plant Sci. 2016, 248, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Yang, C.; Din, G.; Shi, L.; Xu, F. Genome-wide identification and characterization of SPX domain-containing members and their responses to phosphate deficiency in Brassica napus. Front Plant Sci. 2017, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, M.; Gahlaut, V.; Nagaraju, M.; Ghaudhary, S.; Kumar, A.; Tyagi, P.; Gajula, M.N.V.P.; Singh, K.P. Genome-wide identification, characterization, and expression profiling of SPX gene family in wheat. Int. J. Biol. Macromol. 2019, 140, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Shang, W.; Li, C.; Jia, L.; Wang, X.; Xing, G.; Zheng, W. Evolution of the SPX gene family in plants and its role in the response mechanism to phosphorus stress. Open Biol. 2018, 8, 170231. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. Nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Wu, Z.C.; Xiao, X.; Chou, K.C. iLoc-Plant: A multi-label classifier for predicting the subcellular localization of plant proteins with both single and multiple sites. Mol. Biosyst. 2011, 7, 3287–3297. [Google Scholar] [CrossRef]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef]
- Savojardo, C.; Martelli, P.L.; Fariselli, P.; Profiti, G.; Casadio, R. BUSCA: An integrative web server to predict subcellular localization of proteins. Nucleic Acids Res. 2018, 46, W459–W466. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef] [PubMed]
- Li, K.B. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics 2003, 19, 1585–1586. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.P.; Guo, A.Y.; Zhang, H.; Luo, J.C.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef]
- Chen, Y.; Zhao, X.; Li, G.; Kumar, S.; Sun, Z.; Li, Y.; Guo, W.; Yang, J.; Hou, H. Genome-wide identification of the Nramp gene family in Spirodela polyrhiza and expression analysis under cadmium stress. Int. J. Mol. Sci. 2021, 22, 6414. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Santos, A.; Mering, V.C.; Jensen, L.J.; Bork, P.; Kuhn, M. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016, 44, D380–D384. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, K.; Dustin Schaeffer, R.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Su, H.; Wang, W.; Ye, L.; Wei, H.; Peng, Z.; Anishchenko, I.; Baker, D.; Yang, J. The trRosetta server for fast and accurate protein structure prediction. Nat. Protoc. 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zhao, X.Y.; Yang, J.J.; Li, G.J.; Sun, Z.L.; Hu, S.Q.; Chen, Y.; Guo, W.J.; Hou, H.W. Genome-wide identification and comparative analysis of the WRKY gene family in aquatic plants and their response to abiotic stresses in giant duckweed (Spirodela polyrhiza). Genomics 2021, 113, 1761–1777. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Ueda, Y.; Kiba, T.; Yanagisawa, S. Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signaling via transcriptional regulation of SPX genes. Plant J. 2020, 102, 448–466. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Li, F.; Fan, K.; Ma, F.; Yue, E.; Bibi, N.; Wang, M.; Shen, H.; Hasan, M.M.; Wang, X. Genomic identification and comparative expansion analysis of the non-specific lipid transfer protein gene family in Gossypium. Sci. Rep. 2016, 6, 38948. [Google Scholar] [CrossRef]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; PérezPérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, S.; Li, H.; Wang, L.; Liu, Y.; Niu, L.; Yang, Q.; Meng, D.; Fu, Y. Genome-wide analysis and characterization of R2R3-MYB family in pigeon pea (Cajanus cajan) and their functional identification in phenylpropanoids biosynthesis. Planta 2021, 254, 64. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 2014, 345, 1251788. [Google Scholar] [CrossRef] [PubMed]
- O’Hare, M.T.; Baattrup-Pedersen, A.; Baumgarte, I.; Freeman, A.; Gunn, I.D.M.; Lazar, A.N.; Sinclair, R.; Wade, A.J.; Bowers, M.J. Responses of aquatic plants to eutrophication in rivers: A revised conceptual model. Front. Plant Sci. 2018, 9, 451. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Zhang, X.; Yao, W.; Zhao, K.; Lin, L.; Gao, F.; Zhou, B.; Jiang, T. Genome-wide search and structural and functional analyses for late embryogenesis-abundant (LEA) gene family in poplar. BMC Plant Biol. 2021, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Biłas, R.; Szafran, K.; Hnatuszko-Konka, K.; Kononowicz, A.K. Cis-regulatory elements used to control gene expression in plants. Plant Cell Tiss. Org. 2016, 127, 269–287. [Google Scholar] [CrossRef]
- Li, Y.; Wu, H.; Fan, H.; Zhao, T.; Ling, H.Q. Characterization of the AtSPX3 promoter elucidates its complex regulation in response to phosphorus deficiency. Plant Cell Physiol. 2016, 57, 1767–1778. [Google Scholar] [CrossRef][Green Version]
- Osorio, M.B.; Ng, S.; Berkowitz, O.; De Clercq, I.; Mao, C.; Shou, H.; Whelan, J.; Jost, R. SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 2019, 181, 332–352. [Google Scholar] [CrossRef]
- Yan, H.; Sheng, M.; Wang, C.; Liu, Y.; Yang, J.; Liu, F.; Xu, W.; Su, Z. AtSPX1-mediated transcriptional regulation during leaf senescence in Arabidopsis thaliana. Plant Sci. 2019, 283, 238–246. [Google Scholar] [CrossRef]
- Ruan, W.; Guo, M.; Xu, L.; Wang, X.; Zhao, H.; Wang, K.Y. An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 2018, 30, 853–870. [Google Scholar] [CrossRef]
- Ruan, W.; Guo, M.; Wang, X.; Guo, Z.; Xu, Z.; Xu, L.; Zhao, H.; Sun, H.; Yan, C.; Keke, Y. Two RING-finger ubiquitin E3 ligases regulate the degradation of SPX4, an internal phosphate sensor, for phosphate homeostasis and signaling in rice. Mol. Plant 2019, 12, 1060–1074. [Google Scholar] [CrossRef]
- Poirier, Y. Post-translational regulation of SPX Proteins for coordinated nutrient signaling. Mol. Plant 2019, 12, 1041–1043. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]




| Species | Type of Organism | Group A | Group B | Group C | Group D | Group E | Total |
|---|---|---|---|---|---|---|---|
| C. reinhardtii | Yeast | 1 | 1 | ||||
| S. cerevisiae | Algae | 3 | 6 | 9 | |||
| P. patens | Moss | 7 | 7 | ||||
| A.filiculoides | Fern | 2 | 1 | 1 | 4 | ||
| S. cucullata | Fern | 2 | 1 | 1 | 4 | ||
| S. moellendorffii | Fern | 4 | 2 | 6 | |||
| B. distachyon | Monocot | 2 | 3 | 5 | |||
| N. nucifera | Monocot | 1 | 3 | 4 | |||
| O. sativa | Monocot | 2 | 4 | 6 | |||
| S. italic | Monocot | 2 | 3 | 5 | |||
| S. polyrhiza | Monocot | 1 | 2 | 3 | |||
| T. aestivum | Monocot | 3 | 12 | 15 | |||
| W. australiana | Monocot | 1 | 1 | 2 | |||
| Z. mays | Monocot | 2 | 4 | 6 | |||
| Z. marina | Monocot | 1 | 2 | 3 | |||
| A. thaliana | Dicot | 2 | 2 | 4 | |||
| A. trichopoda | Dicot | 1 | 2 | 3 | |||
| B. napus | Dicot | 5 | 6 | 11 | |||
| C. papaya | Dicot | 1 | 2 | 2 | 5 | ||
| C. clementine | Dicot | 1 | 2 | 2 | 5 | ||
| E. grandis | Dicot | 3 | 2 | 5 | |||
| G. max | Dicot | 7 | 2 | 9 | |||
| G. hirsutum | Dicot | 1 | 6 | 5 | 12 | ||
| I. triloba | Dicot | 3 | 3 | ||||
| P. trichocarpa | Dicot | 1 | 4 | 2 | 7 | ||
| P. persica | Dicot | 2 | 2 | 4 | |||
| R. communis | Dicot | 2 | 2 | 2 | 6 | ||
| S. lycopersicum | Dicot | 1 | 2 | 3 | 6 | ||
| Total | 1 | 16 | 8 | 61 | 74 | 160 | |
| 0.625% | 10% | 5% | 38.125% | 46.25% |
| SSR Motif Length | Number | Percentage/% |
|---|---|---|
| 1 | 2523 | 35.95 |
| 2 | 1237 | 17.63 |
| 3 | 610 | 8.69 |
| 4 | 1632 | 23.25 |
| 5 | 602 | 8.58 |
| 6 | 309 | 4.40 |
| 7 | 66 | 0.94 |
| 8 | 23 | 0.33 |
| 9 | 11 | 0.16 |
| 10 | 5 | 0.07 |
| Species | Species | Colinear Gene Pairs |
|---|---|---|
| A. thaliana | S. polyrhiza | - |
| S. polyrhiza | B. distachyon | SpSPX1, BdSPX3, BdSPX5 |
| S. polyrhiza | S. lycopersicum | SpSPX1, SlSPX5, SlSPX6 |
| S. polyrhiza | G. max | SpSPX1, GmSPX3, GmSPX7, GmSPX8 |
| S. polyrhiza | S. italic | SpSPX1, SiSPX5, SiSPX4 |
| S. polyrhiza | P. trichocarpa | SpSPX1, PtSPX3, PtSPX4, PtSPX6, PtSPX7 |
| S. polyrhiza | O. sativa | SpSPX1, OsSPX2, OsSPX1 |
| S. polyrhiza | Z. mays | SpSPX1, ZmaySPX5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, J.; Zhao, X.; Chen, Y.; Li, G.; Li, X.; Xia, M.; Sun, Z.; Chen, Y.; Li, Y.; Yao, L.; et al. Identification, Structural, and Expression Analyses of SPX Genes in Giant Duckweed (Spirodela polyrhiza) Reveals Its Role in Response to Low Phosphorus and Nitrogen Stresses. Cells 2022, 11, 1167. https://doi.org/10.3390/cells11071167
Yang J, Zhao X, Chen Y, Li G, Li X, Xia M, Sun Z, Chen Y, Li Y, Yao L, et al. Identification, Structural, and Expression Analyses of SPX Genes in Giant Duckweed (Spirodela polyrhiza) Reveals Its Role in Response to Low Phosphorus and Nitrogen Stresses. Cells. 2022; 11(7):1167. https://doi.org/10.3390/cells11071167
Chicago/Turabian StyleYang, Jingjing, Xuyao Zhao, Yan Chen, Gaojie Li, Xiaozhe Li, Manli Xia, Zuoliang Sun, Yimeng Chen, Yixian Li, Lunguang Yao, and et al. 2022. "Identification, Structural, and Expression Analyses of SPX Genes in Giant Duckweed (Spirodela polyrhiza) Reveals Its Role in Response to Low Phosphorus and Nitrogen Stresses" Cells 11, no. 7: 1167. https://doi.org/10.3390/cells11071167
APA StyleYang, J., Zhao, X., Chen, Y., Li, G., Li, X., Xia, M., Sun, Z., Chen, Y., Li, Y., Yao, L., & Hou, H. (2022). Identification, Structural, and Expression Analyses of SPX Genes in Giant Duckweed (Spirodela polyrhiza) Reveals Its Role in Response to Low Phosphorus and Nitrogen Stresses. Cells, 11(7), 1167. https://doi.org/10.3390/cells11071167






