Genetic Variations and mRNA Expression of Goat DNAH1 and Their Associations with Litter Size
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Isolation and Total RNA Extraction form Goat Tissues
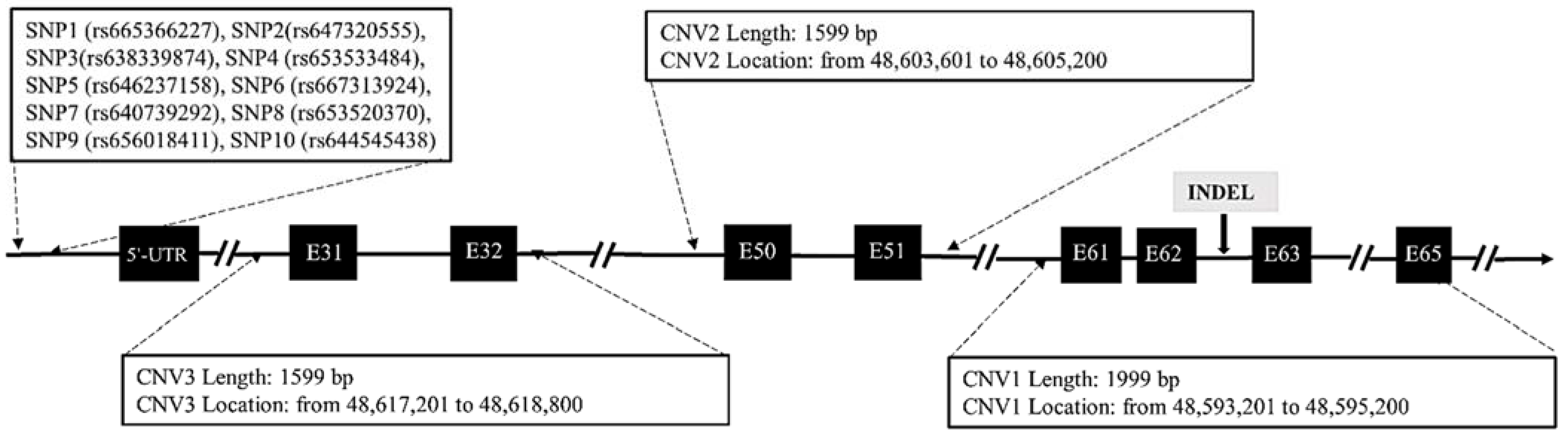
2.2. Identification of Candidate Mutations and Primer Design
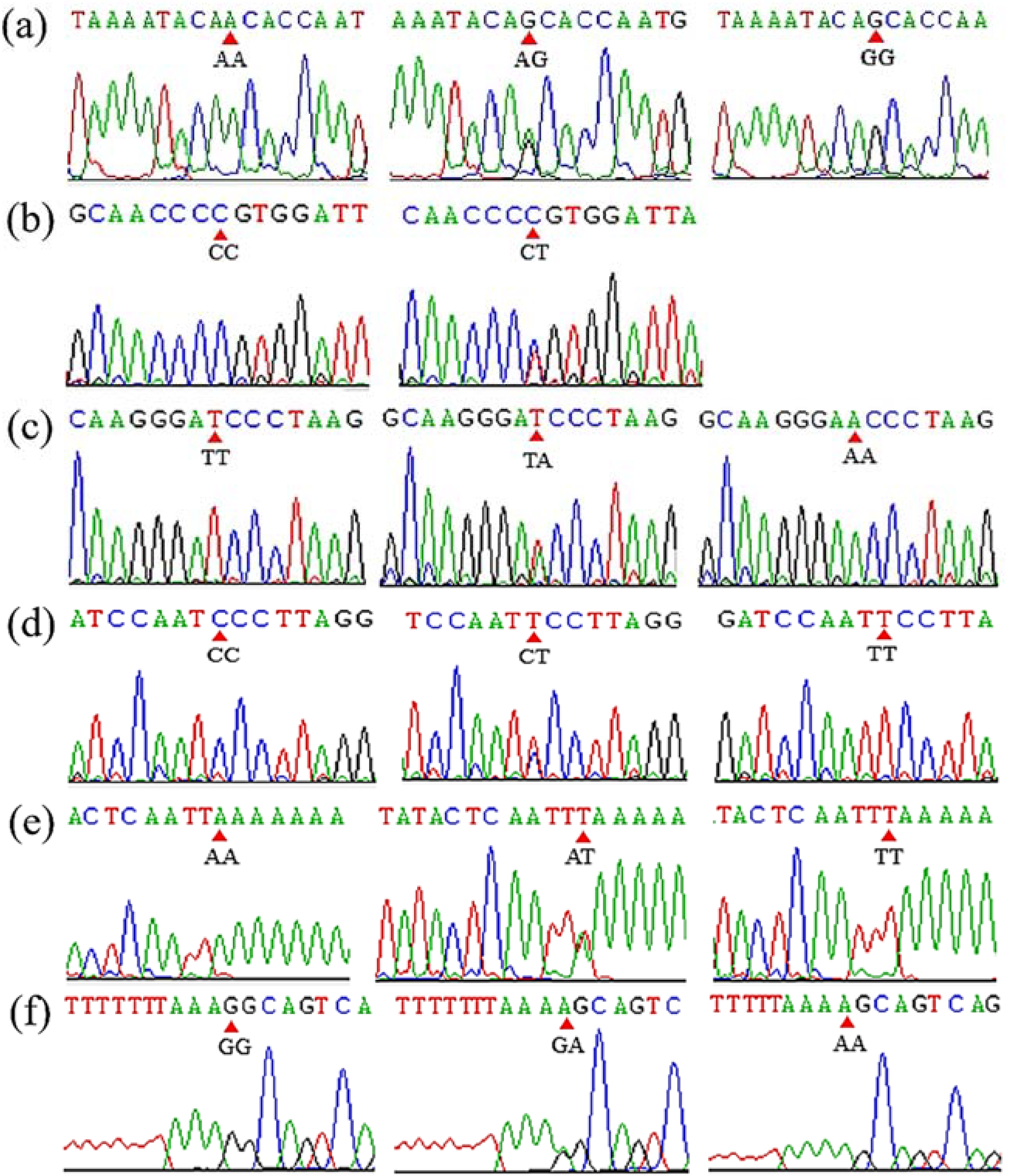
2.3. Detection of SNP, InDel and CNV Mutations of DNAH1 Gene
2.4. Detection of mRNA Expression of DNAH1 Gene
2.5. Statistical Analyses
3. Results
3.1. Identification of SNP, InDel and CNV Polymorphisms within the DNAH1 Gene
3.2. Allele and Genotype Frequencies of SNP, InDel and CNV Variations in DNAH1 Gene
3.3. Association Analysis between SNP, InDel and CNV Variations with Goat Litter Size
3.4. Relationship between Combination Genotypes and Goat Litter Size
3.5. mRNA Expression of DNAH1 in Ovary of Goats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, K.; Kang, Z.; Jiang, E.; Yan, H.; Zhu, H.; Liu, J.; Qu, L.; Lan, X.; Pan, C. Genetic effects of DSCAML1 identified in genome-wide association study revealing strong associations with litter size and semen quality in goat (Capra hircus). Theriogenology 2020, 146, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, S.; Tang, Q.; Jiang, E.; Wang, K.; Lan, X.; Pan, C. Goat sperm associated antigen 17 protein gene (SPAG17): Small and large fragment genetic variation detection, association analysis, and mRNA expression in gonads. Genomics 2020, 112, 5115–5121. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Feng, W.; Kang, Y.; Wang, K.; Yang, Y.; Qu, L.; Chen, H.; Lan, X.; Pan, C. Detection of mRNA expression and copy number variations within the goat FecB gene associated with litter size. Front. Vet. Sci. 2021, 8, 758705. [Google Scholar] [CrossRef] [PubMed]
- Shaat, I.; Mäki-Tanila, A. Variation in direct and maternal genetic effects for meat production traits in Egyptian Zaraibi goats. J. Anim. Breed. Genet. 2009, 126, 198–208. [Google Scholar] [CrossRef]
- Knorst, V.; Byrne, S.; Yates, S.; Asp, T.; Widmer, F.; Studer, B.; Kölliker, R. Pooled DNA sequencing to identify SNPs associated with a major QTL for bacterial wilt resistance in Italian ryegrass (Lolium multiflorum Lam.). Theor. Appl. Genet. 2019, 132, 947–958. [Google Scholar] [CrossRef] [Green Version]
- Jiang, E.; Kang, Z.; Wang, X.; Liu, Y.; Liu, X.; Wang, Z.; Li, X.; Lan, X. Detection of insertions/deletions (InDels) within the goat Runx2 gene and their association with litter size and growth traits. Anim. Biotech. 2021, 32, 169–177. [Google Scholar] [CrossRef]
- Wang, X.; Yang, Q.; Wang, K.; Yan, H.; Pan, C.; Chen, H.; Liu, J.; Zhu, H.; Qu, L.; Lan, X. Two strongly linked single nucleotide polymorphisms (Q320P and V397I) in GDF9 gene are associated with litter size in cashmere goats. Theriogenology 2019, 125, 115–121. [Google Scholar] [CrossRef]
- Ren, F.; Yu, S.; Chen, R.; Lv, X.; Pan, C. Identification of a novel 12-bp insertion/deletion (indel) of iPS-related Oct4 gene and its association with reproductive traits in male piglets. Anim. Reprod. Sci. 2017, 178, 55–60. [Google Scholar] [CrossRef]
- Baccetti, B.; Collodel, G.; Estenoz, M.; Manca, D.; Moretti, E.; Piomboni, P. Gene deletions in an infertile man with sperm fibrous sheath dysplasia. Hum. Reprod. 2005, 20, 2790–2794. [Google Scholar] [CrossRef] [Green Version]
- Coutton, C.; Escoffier, J.; Martinez, G.; Arnoult, C.; Ray, P.F. Teratozoospermia: Spotlight on the main genetic actors in the human. Hum. Reprod. Update 2015, 21, 455–485. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Zhu, D.; Zhang, H.; Jiang, Y.; Hu, X.; Geng, D.; Wang, R.; Liu, R. Associations between DNAH1 gene polymorphisms and male infertility: A retrospective study. Medicine 2018, 97, e13493. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, F.; Li, F.; Jiang, X.; Yang, Y.; Li, X.; Li, W.; Wang, X.; Cheng, J.; Liu, M.; et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat. Commun. 2019, 10, 433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Khelifa, M.; Coutton, C.; Zouari, R.; Karaouzène, T.; Rendu, J.; Bidart, M.; Yassine, S.; Pierre, V.; Delaroche, J.; Hennebicq, S.; et al. Mutations in DNAH1, which encodes an inner arm heavy chain dynein, lead to male infertility from multiple morphological abnormalities of the sperm flagella. Am. J. Hum. Genet. 2014, 94, 95–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merveille, A.C.; Davis, E.E.; Becker-Heck, A.; Legendre, M.; Amirav, I.; Bataille, G.; Belmont, J.; Beydon, N.; Billen, F.; Clément, A.; et al. CCDC39 is required for assembly of inner dynein arms and the dynein regulatory complex and for normal ciliary motility in humans and dogs. Nat. Genet. 2011, 43, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Pausch, H.; Venhoranta, H.; Wurmser, C.; Hakala, K.; Iso-Touru, T.; Sironen, A.; Vingborg, R.K.; Lohi, H.; Söderquist, L.; Fries, R.; et al. A frameshift mutation in ARMC3 is associated with a tail stump sperm defect in Swedish Red (Bos taurus) cattle. BMC Genet. 2016, 17, 49. [Google Scholar] [CrossRef] [Green Version]
- Zariwala, M.A.; Omran, H.; Ferkol, T.W. The emerging genetics of primary ciliary dyskinesia. Proc. Am. Thorac. Soc. 2011, 8, 430–433. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Huang, S.; Zhao, X.; Wu, F.; Zhu, D.; Zhai, X.; Wang, A. Successful live birth following natural cycle oocyte retrieval in a woman with primary infertility and atypical primary ovarian insufficiency with a DNAH1 gene deletion mutation. Genet. Test. Mol. Biomarkers 2021, 25, 668–673. [Google Scholar] [CrossRef]
- Imtiaz, F.; Allam, R.; Ramzan, K.; Al-Sayed, M. Variation in DNAH1 may contribute to primary ciliary dyskinesia. BMC Med. Genet. 2015, 16, 14. [Google Scholar] [CrossRef] [Green Version]
- Emiralioğlu, N.; Taşkıran, E.Z.; Koşukcu, C.; Bilgiç, E.; Atilla, P.; Kaya, B.; Günaydın, Ö.; Yüzbaşıoğlu, A.; Tuğcu, G.D.; Ademhan, D.; et al. Genotype and phenotype evaluation of patients with primary ciliary dyskinesia: First results from Turkey. Pediatr. Pulmonol. 2020, 55, 383–393. [Google Scholar] [CrossRef]
- Wang, Z.; Pan, Y.; He, L.; Song, X.; Chen, H.; Pan, C.; Qu, L.; Zhu, H.; Lan, X. Multiple morphological abnormalities of the sperm flagella (MMAF)-associated genes: The relationships between genetic variation and litter size in goats. Gene 2020, 753, 144778. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Zhang, X.; Jiang, E.; Yan, H.; Zhu, H.; Chen, H.; Liu, J.; Qu, L.; Pan, C.; Lan, X. InDels within caprine IGF2BP1 intron 2 and the 3’-untranslated regions are associated with goat growth traits. Anim. Genet. 2020, 51, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Aljanabi, S.M.; Martinez, I. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic. Acids Res. 1997, 25, 4692–4693. [Google Scholar] [CrossRef] [PubMed]
- Müllenbach, R.; Lagoda, P.J.; Welter, C. An efficient salt-chloroform extraction of DNA from blood and tissues. Trends Genet. 1989, 5, 391. [Google Scholar] [PubMed]
- Fontanesi, L.; Beretti, F.; Riggio, V.; Gómez González, E.; Dall’Olio, S.; Davoli, R.; Russo, V.; Portolano, B. Copy number variation and missense mutations of the agouti signaling protein (ASIP) gene in goat breeds with different coat colors. Cytogenet. Genome Res. 2009, 126, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reich, D.E.; Cargill, M.; Bolk, S.; Ireland, J.; Sabeti, P.C.; Richter, D.J.; Lavery, T.; Kouyoumjian, R.; Farhadian, S.F.; Ward, R.; et al. Linkage disequilibrium in the human genome. Nature 2001, 411, 199–204. [Google Scholar] [CrossRef]
- Cui, Y.; Yan, H.; Wang, K.; Xu, H.; Zhang, X.; Zhu, H.; Liu, J.; Qu, L.; Lan, X.; Pan, C. Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat. Front. Genet. 2018, 9, 91. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, X.; Cai, H.; Pan, C.; Lei, C.; Chen, H.; Lan, X. Genetic variants and effects on milk traits of the caprine paired-like homeodomain transcription factor 2 (PITX2) gene in dairy goats. Gene 2013, 532, 203–210. [Google Scholar] [CrossRef]
- Jiang, R.; Cheng, J.; Cao, X.K.; Ma, Y.L.; Chaogetu, B.; Huang, Y.Z.; Lan, X.Y.; Lei, C.Z.; Hu, L.Y.; Chen, H. Copy number variation of the SHE gene in sheep and its association with economic traits. Animals 2019, 9, 531. [Google Scholar] [CrossRef] [Green Version]
- Hui, Y.; Zhang, Y.; Wang, K.; Pan, C.; Chen, H.; Qu, L.; Song, X.; Lan, X. Goat DNMT3B: An indel mutation detection, association analysis with litter size and mRNA expression in gonads. Theriogenology 2020, 147, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Maiti, A.K.; Mattéi, M.G.; Jorissen, M.; Volz, A.; Zeigler, A.; Bouvagnet, P. Identification, tissue specific expression, and chromosomal localisation of several human dynein heavy chain genes. Eur. J. Hum. Genet. 2000, 8, 923–932. [Google Scholar] [CrossRef] [PubMed]
- Wambergue, C.; Zouari, R.; Fourati Ben Mustapha, S.; Martinez, G.; Devillard, F.; Hennebicq, S.; Satre, V.; Brouillet, S.; Halouani, L.; Marrakchi, O.; et al. Patients with multiple morphological abnormalities of the sperm flagella due to DNAH1 mutations have a good prognosis following intracytoplasmic sperm injection. Hum. Reprod. 2016, 31, 1164–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, C.; Nie, H.; Meng, L.; Wang, W.; Li, H.; Yuan, S.; Cheng, D.; He, W.; Liu, G.; Du, J.; et al. Novel mutations in SPEF2 causing different defects between flagella and cilia bridge: The phenotypic link between MMAF and PCD. Hum. Genet. 2020, 139, 257–271. [Google Scholar] [CrossRef]
- Zhuang, B.-J.; Xu, S.-Y.; Dong, L.; Zhang, P.-H.; Huang, X.-P.; Li, G.-S.; You, Y.-D.; Chen, D.; Yu, X.-J.; Chang, D.-G. Novel DNAH1 Mutation Loci Lead to Multiple Morphological Abnormalities of the Sperm Flagella and Literature Review. World J. Men’s Heal. 2022, 40, e27. [Google Scholar] [CrossRef]
- Cartharius, K.; Frech, K.; Grote, K.; Klocke, B.; Haltmeier, M.; Klingenhoff, A.; Frisch, M.; Bayerlein, M.; Werner, T. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics 2005, 21, 2933–2942. [Google Scholar] [CrossRef] [Green Version]
- Kalkan, G.; Karakus, N.; Baş, Y.; Takçı, Z.; Ozuğuz, P.; Ateş, O.; Yigit, S. The association between Interleukin (IL)-4 gene intron 3 VNTR polymorphism and alopecia areata (AA) in Turkish population. Gene 2013, 527, 565–569. [Google Scholar] [CrossRef]
- Vaz-Drago, R.; Custódio, N.; Carmo-Fonseca, M. Deep intronic mutations and human disease. Hum. Genet. 2017, 136, 1093–1111. [Google Scholar] [CrossRef]
- Xiang, G.; Ren, J.; Hai, T.; Fu, R.; Yu, D.; Wang, J.; Li, W.; Wang, H.; Zhou, Q. Editing porcine IGF2 regulatory element improved meat production in Chinese Bama pigs. Cell Mol. Life Sci. 2018, 75, 4619–4628. [Google Scholar] [CrossRef]
- Van Laere, A.S.; Nguyen, M.; Braunschweig, M.; Nezer, C.; Collette, C.; Moreau, L.; Archibald, A.L.; Haley, C.S.; Buys, N.; Tally, M.; et al. A regulatory mutation in IGF2 causes a major QTL effect on muscle growth in the pig. Nature 2003, 425, 832–836. [Google Scholar] [CrossRef]
- Gibson, J.; Tapper, W.; Ennis, S.; Collins, A. Exome-based linkage disequilibrium maps of individual genes: Functional clustering and relationship to disease. Hum. Genet. 2013, 132, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; Xu, S.; Maruki, T.; Jiang, X.; Pfaffelhuber, P.; Haubold, B. Genome-wide linkage-disequilibrium profiles from single individuals. Genetics 2014, 198, 269–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Litter Size Traits | Sample Size |
|---|---|
| Mothers of single kid | 550 |
| Mothers of double kids | 498 |
| Mothers of three kids | 14 |
| Mothers of four kids | 1 |
| No record | 38 |
| Loci (Size) | Genotypes (Size) | Frequency | Ho | He | PIC | χ2 (p-Value) | |
|---|---|---|---|---|---|---|---|
| Genotypes | Alleles | ||||||
| SNP1 | AA (146) | 0.559 | 0.736 (A) | 0.611 | 0.389 | 0.313 | 2.294 |
| (n = 261) | AG (92) | 0.353 | 0.264 (G) | (p = 0.130) | |||
| GG (23) | 0.088 | ||||||
| SNP3 | GG (234) | 0.777 | 0.889 (G) | 0.802 | 0.198 | 0.178 | 4.721 |
| (n = 301) | GA (67) | 0.223 | 0.111 (A) | (p = 0.030) | |||
| AA (0) | 0 | ||||||
| SNP4 | CC (204) | 0.872 | 0.932 (C) | 0.873 | 0.127 | 0.119 | 0.864 |
| (n = 234) | CT (28) | 0.120 | 0.068 (T) | (p = 0.352) | |||
| TT (2) | 0.009 | ||||||
| SNP5 | TT (124) | 0.527 | 0.715 (T) | 0.592 | 0.408 | 0.325 | 1.556 |
| (n = 235) | TA (88) | 0.374 | 0.285 (A) | (p = 0.212) | |||
| AA (23) | 0.099 | ||||||
| SNP7 | CC (191) | 0.608 | 0.756 (C) | 0.631 | 0.369 | 0.301 | 0.143 |
| (n = 314) | CT (93) | 0.296 | 0.244 (T) | (p = 0.705) | |||
| TT (30) | 0.096 | ||||||
| SNP9 | AA (189) | 0.601 | 0.779 (A) | 0.655 | 0.345 | 0.285 | 0.205 |
| (n = 314) | AT (111) | 0.354 | 0.221 (T) | (p = 0.651) | |||
| TT (14) | 0.045 | ||||||
| SNP10 | GG (180) | 0.583 | 0.754 (G) | 0.629 | 0.371 | 0.302 | 1.746 |
| (n = 309) | GA (106) | 0.343 | 0.246 (A) | (p = 0.186) | |||
| AA (23) | 0.074 | ||||||
| 27 bp | II (n = 920) | 0.928 | I (0.964) | 0.930 | 0.070 | 0.068 | 0.0780 |
| (n = 991) | ID (n = 70) | 0.071 | D (0.036) | (p = 0.780) | |||
| DD (n = 1) | 0.001 | ||||||
| 15 bp | II (n = 0) | 0 | I (0.022) | 0.958 | 0.042 | 0.042 | 0.169 |
| (n = 346) | ID (n = 15) | 0.043 | D (0.978) | (p = 0.680) | |||
| DD (n = 331) | 0.957 | ||||||
| Loci | Sizes | MSL Genotypes | MML Genotypes | p-Values | ||||
|---|---|---|---|---|---|---|---|---|
| Wild Genotype | Heterozygous Genotype | Mutant Genotype | Wild Genotype | Heterozygous Genotype | Mutant Genotype | |||
| SNP1 | 261 | 70 | 61 | 13 | 76 | 31 | 10 | 0.021 |
| SNP3 | 301 | 129 | 28 | 0 | 105 | 39 | 0 | 0.054 |
| SNP4 | 234 | 112 | 15 | 2 | 92 | 13 | 0 | 0.436 |
| SNP5 | 235 | 60 | 58 | 13 | 64 | 30 | 10 | 0.041 |
| SNP7 | 314 | 87 | 59 | 20 | 104 | 34 | 10 | 0.005 |
| SNP9 | 314 | 104 | 56 | 6 | 85 | 55 | 8 | 0.555 |
| SNP10 | 309 | 85 | 64 | 13 | 95 | 42 | 10 | 0.091 |
| InDel-P1 | 991 | 478 | 56 | 1 | 442 | 14 | 0 | 0.000022 |
| InDel-P3 | 346 | 220 | 9 | 0 | 111 | 6 | 0 | 0.605 |
| Loci | Sizes | MSL Genotypes | MML Genotypes | p-Values | ||||
|---|---|---|---|---|---|---|---|---|
| Loss | Medium | Gain | Loss | Medium | Gain | |||
| CNV1 | 296 | 104 | 38 | 8 | 66 | 50 | 30 | 0.000011 |
| CNV2 | 301 | 71 | 37 | 42 | 46 | 36 | 69 | 0.003 |
| CNV3 | 294 | 126 | 16 | 7 | 92 | 35 | 18 | 0.000187 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Wang, R.; Pan, C.; Chen, H.; Qu, L.; Wu, L.; Guo, Z.; Zhu, H.; Lan, X. Genetic Variations and mRNA Expression of Goat DNAH1 and Their Associations with Litter Size. Cells 2022, 11, 1371. https://doi.org/10.3390/cells11081371
Wang Z, Wang R, Pan C, Chen H, Qu L, Wu L, Guo Z, Zhu H, Lan X. Genetic Variations and mRNA Expression of Goat DNAH1 and Their Associations with Litter Size. Cells. 2022; 11(8):1371. https://doi.org/10.3390/cells11081371
Chicago/Turabian StyleWang, Zhen, Ruolan Wang, Chuanying Pan, Hong Chen, Lei Qu, Lian Wu, Zhengang Guo, Haijing Zhu, and Xianyong Lan. 2022. "Genetic Variations and mRNA Expression of Goat DNAH1 and Their Associations with Litter Size" Cells 11, no. 8: 1371. https://doi.org/10.3390/cells11081371






