Incomplete Recovery of Zebrafish Retina Following Cryoinjury
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Statement
2.2. Cryoinjury
2.3. Retina Dissection
2.4. Histology
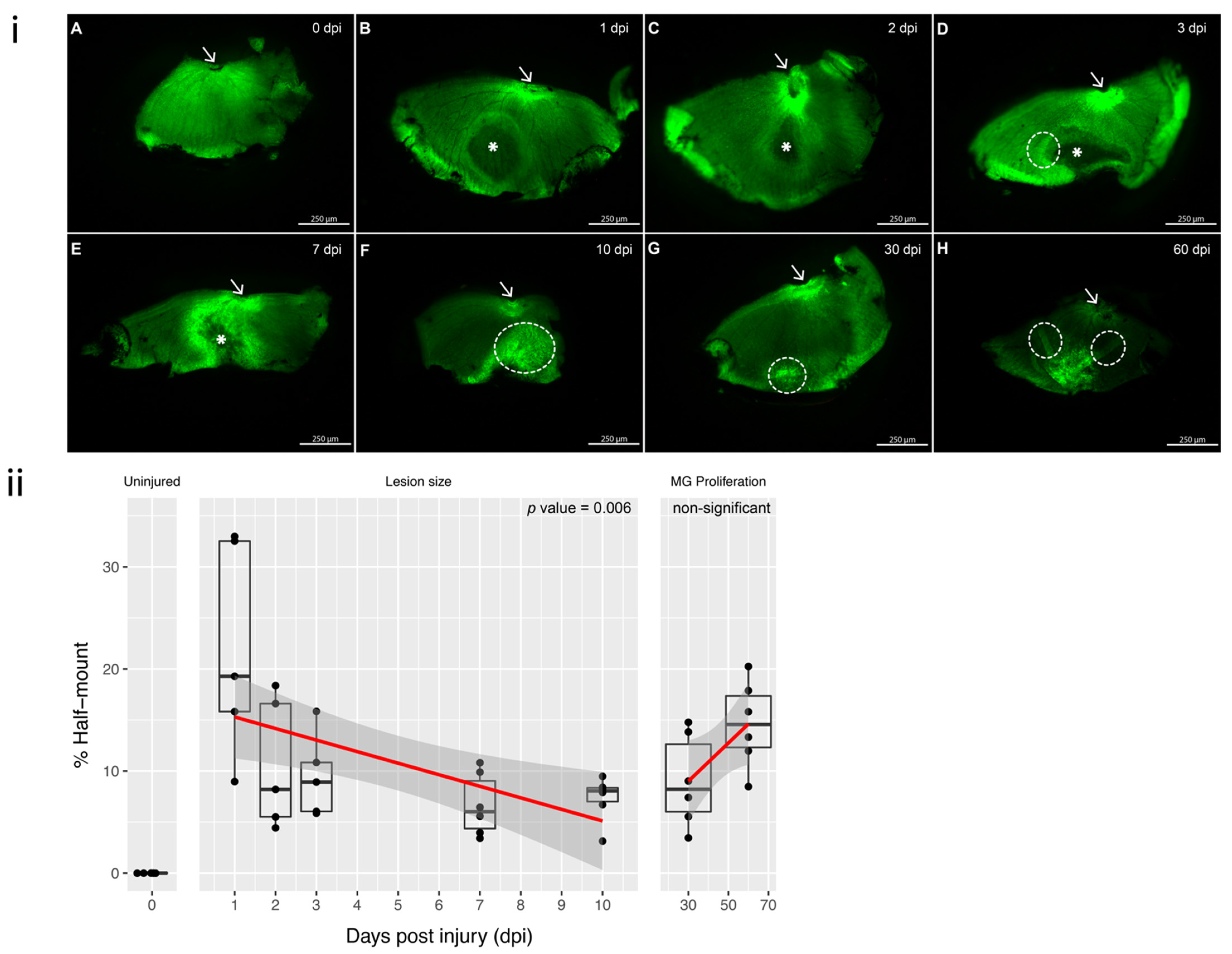
2.5. Progression and the Regression of the Lesion
2.6. Immunofluorescence
2.7. RNA Extraction
2.8. RT-PCR
2.9. RNA Sequencing Library Preparation, Sequencing and Bioinformatics Analysis
3. Results
3.1. Progression of the Induced Lesion with Cryoinjury
3.2. Partial Regeneration Observed by Histology
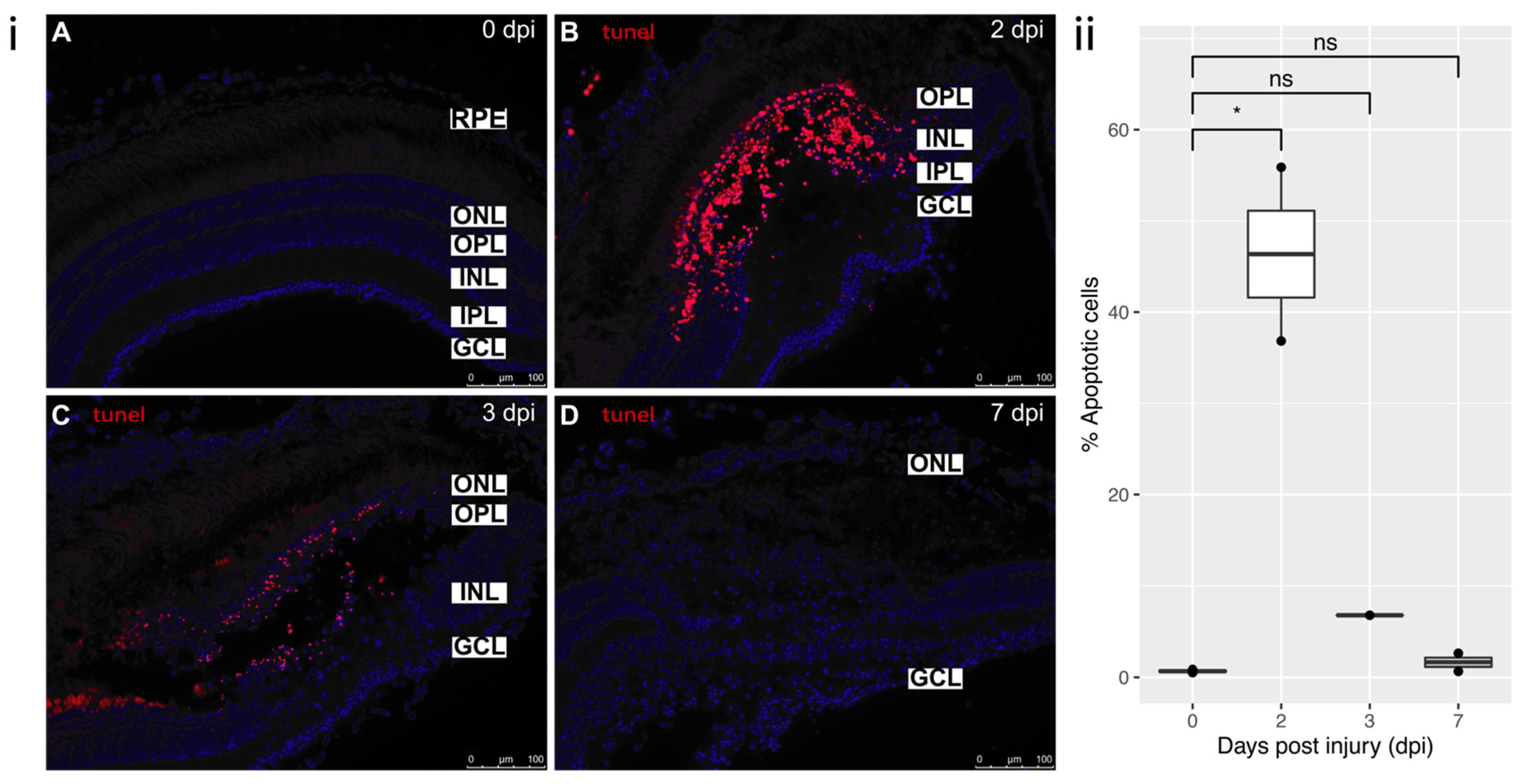
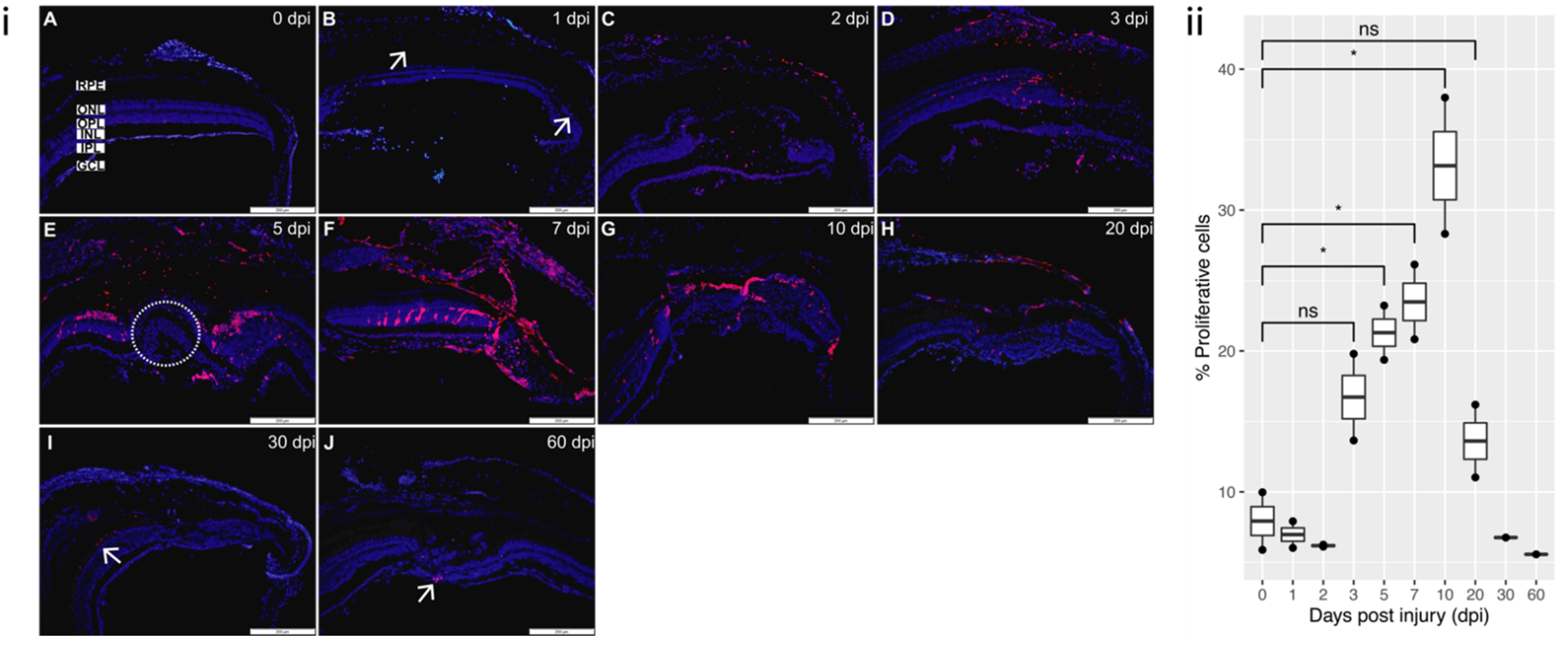
3.3. Cell Death and Proliferation
3.3.1. Cell Death
3.3.2. Proliferation
3.4. Cellular Components of Residual Defect
3.4.1. Red-Green Double Cones
3.4.2. Rods
3.4.3. Horizontal Cells
3.4.4. Müller Glia (MG)
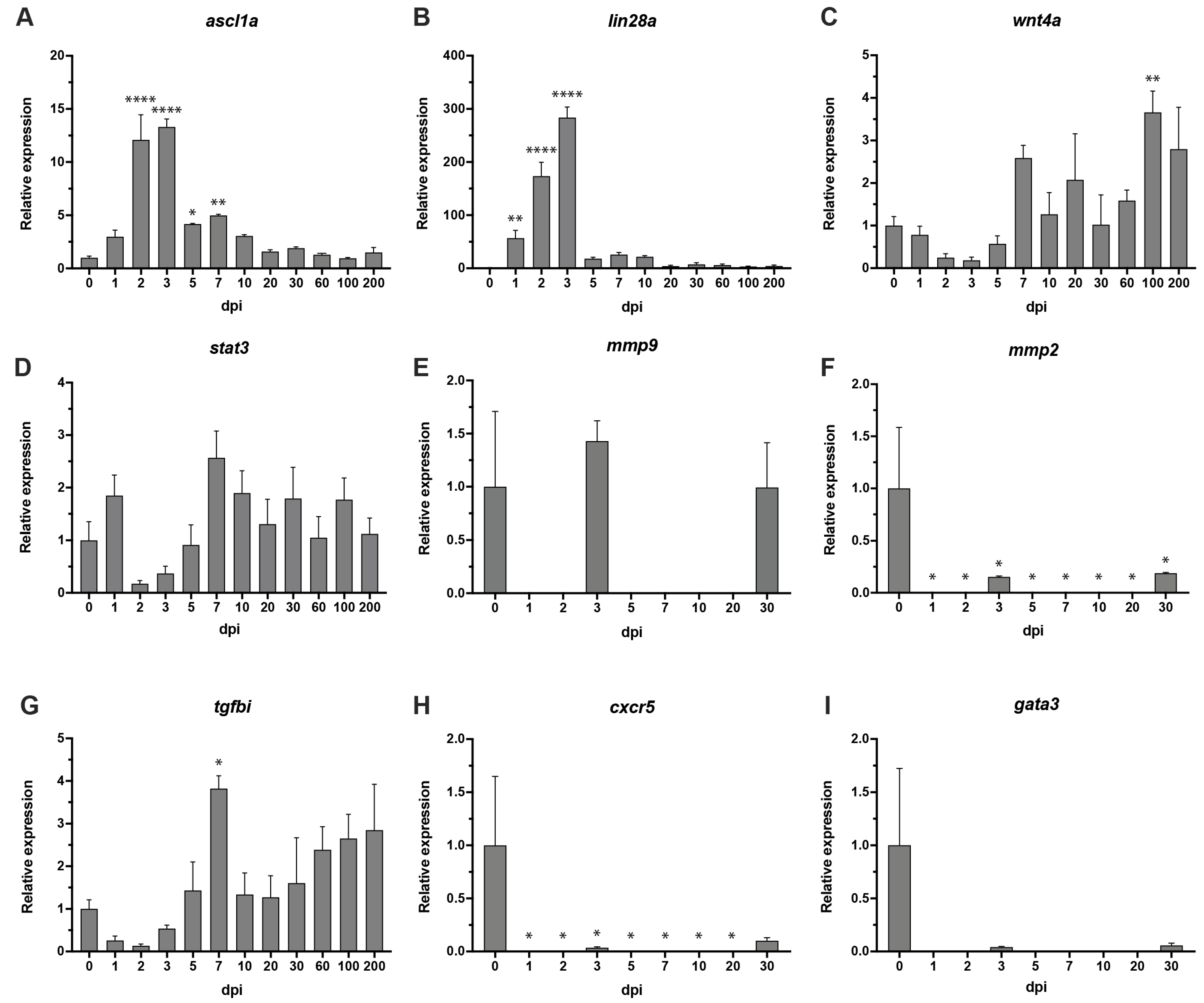
3.5. Modulation of Transcript Levels during the Degeneration, Proliferation and Regeneration Processes
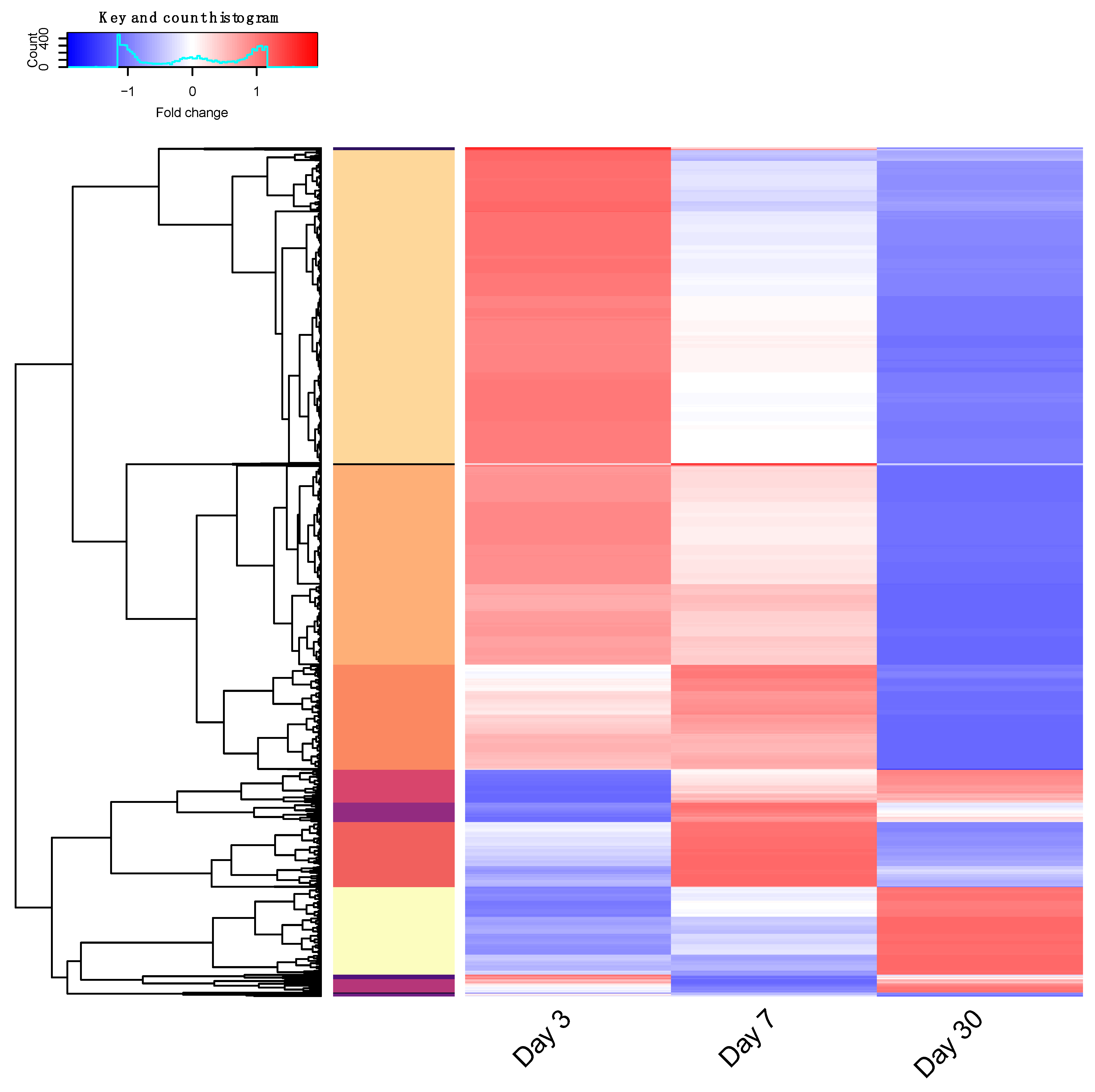
3.6. RNA Sequencing Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ascl1a | achaete–scute homolog 1 |
| CMZ | ciliary marginal zone |
| CNS | central nervous system |
| cxcr5 | C-X-C chemokine receptor type 5 |
| dpi | days post injury |
| ECM | extracellular matrix |
| FC | fold change |
| FDR | false discovery rate |
| gata3 | transcription factor GATA-3 |
| GCL | ganglion cell layer |
| GFAP | glial fibrillary acidic protein |
| GFP | green fluorescent protein |
| H & E | hematoxylin and eosin |
| ILM | inner limiting membrane |
| INL | inner nuclear layer |
| IPL | inner plexiform layer |
| MG | Müller glia |
| mmp2 | matrix metalloproteinase-2 |
| mmp9 | matrix metalloproteinase-9 |
| OLM | outer limiting membrane |
| ONL | outer nuclear layer |
| OPL | outer plexiform layer |
| PBS | phosphate-buffered saline |
| PCNA | proliferating cell nuclear antigen |
| RPE | retinal pigment epithelium |
| RT-PCR | reverse transcriptase polymerase chain reaction |
| SEM | standard error of the mean |
| tgfbi | TGF-β induced |
| VST | variance stabilization transformation |
| wnt4a | wingless-type MMTV integration site family, member 4 |
References
- Zupanc, G.; Sîrbulescu, R. Adult neurogenesis and neuronal regeneration in the central nervous system of teleost fish. Eur. J. Neurosci. 2011, 34, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Kyritsis, N.; Kizil, C.; Brand, M. Neuroinflammation and central nervous system regeneration in vertebrates. Trends Cell Biol. 2014, 24, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Lenkowski, J.R.; Qin, Z.; Sifuentes, C.J.; Thummel, R.; Soto, C.M.; Moens, C.B.; Raymond, P.A. Retinal Regeneration in Adult Zebrafish Requires Regulation of TGFβ Signaling. Glia 2013, 61, 1687–1697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldman, D. Müller glial cell reprogramming and retina regeneration. Nat. Rev. Neurosci. 2014, 15, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Lewis, G.P.; Chapin, E.A.; Luna, G.; Linberg, K.A.; Fisher, S.K. The fate of müller’s glia following experimental retinal detachment: Nuclear migration, cell division, and subretinal glial scar formation. Mol. Vis. 2010, 16, 1361–1372. [Google Scholar]
- Bringmann, A.; Iandiev, I.; Pannicke, T.; Wurm, A.; Hollborn, M.; Wiedemann, P.; Osborne, N.N.; Reichenbach, A. Cellular signaling and factors involved in Müller cell gliosis: Neuroprotective and detrimental effects. Prog. Retin. Eye Res. 2009, 28, 423–451. [Google Scholar] [CrossRef]
- Jorstad, N.L.; Wilken, M.S.; Grimes, W.N.; Wohl, S.G.; VandenBosch, L.S.; Yoshimatsu, T.; Wong, R.O.; Rieke, F.; Reh, T.A. Stimulation of functional neuronal regeneration from Müller glia in adult mice. Nature 2017, 548, 103–107. [Google Scholar] [CrossRef]
- Rueda, E.M.; Hall, B.M.; Hill, M.C.; Swinton, P.G.; Tong, X.; Martin, J.F.; Poché, R.A. The Hippo Pathway Blocks Mammalian Retinal Müller Glial Cell Reprogramming. Cell Rep. 2019, 27, 1637–1649.e6. [Google Scholar] [CrossRef] [Green Version]
- Bernardos, R.L.; Barthel, L.K.; Meyers, J.R.; Raymond, P.A. Late-stage neuronal progenitors in the retina are radial Müller glia that function as retinal stem cells. J. Neurosci. 2007, 27, 7028–7040. [Google Scholar] [CrossRef] [Green Version]
- DiCicco, R.M.; Bell, B.A.; Kaul, C.; Hollyfield, J.G.; Anand-Apte, B.; Perkins, B.D.; Tao, Y.K.; Yuan, A. Retinal regeneration following OCT-guided laser injury in zebrafish. Investig. Ophthalmol. Vis. Sci. 2014, 55, 6281–6288. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, R.; Fausett, B.V.; Goldman, D. Ascl1a regulates Muller glia dedifferentiation and retina regeneration via a Lin-28 dependent, let7 miRNA signaling pathway. Nat. Cell Biol. 2010, 7, 1959–1967. [Google Scholar]
- Raymond, P.A.; Barthel, L.K.; Bernardos, R.L.; Perkowski, J.J. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev. Biol. 2006, 6, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Sherpa, T.; Fimbel, S.M.; Mallory, D.E.; Maaswinkel, H.; Spritzer, S.D.; Sand, J.A.; Li, L.; Hyde, D.R.; Stenkamp, D.L. Ganglion Cell Regeneration Following Whole-Retina Destruction in Zebrafish. Dev. Neurobiol. 2008, 68, 166–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, J.L.; Nelson, C.M.; Luo, X.; Hyde, D.R.; Thummel, R. Characterization of multiple light damage paradigms reveals regional differences in photoreceptor loss. Exp. Eye Res. 2012, 97, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Zou, S.; Tian, C.; Ge, S.; Hu, B. Neurogenesis of Retinal Ganglion Cells Is Not Essential to Visual Functional Recovery after Optic Nerve Injury in Adult Zebrafish. PLoS ONE 2013, 8, e57280. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xiang, X.; Chen, T.; Mao, G.; Deng, L.; Zeng, L.; Zhang, J. In vivo monitoring the dynamic process of acute retinal hemorrhage and repair in zebrafish with spectral-domain optical coherence tomography. J. Biophotonics 2019, 12, e201900235. [Google Scholar] [CrossRef]
- Wang, Y.J.; Cai, S.J.; Cui, J.L.; Chen, Y.; Tang, X.; Li, Y.H. Correlation between photoreceptor injury-regeneration and behavior in a zebrafish model. Neural Regen. Res. 2017, 12, 795–803. [Google Scholar]
- Taylor, S.; Chen, J.; Luo, J.; Hitchcock, P. Light-Induced Photoreceptor Degeneration in the Retina of the Zebrafish. Methods Mol. Biol. 2012, 884, 247–254. [Google Scholar]
- Cameron, D.A. Cellular proliferation and neurogenesis in the injured retina of adult zebrafish. Vis. Neurosci. 2000, 17, 789–797. [Google Scholar] [CrossRef]
- Aleström, P.; D’Angelo, L.; Midtlyng, P.J.; Schorderet, D.F.; Schulte-Merker, S.; Sohm, F.; Warner, S. Zebrafish: Housing and husbandry recommendations. Lab. Anim. 2020, 54, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, S.; Li, W. RSeQC: Quality control of RNA-seq experiments. Bioinformatics 2012, 28, 2184–2185. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Antoine, D.J.; Weemhoff, J.L.; Jenkins, R.E.; Farhood, A.; Park, B.K.; Jaeschke, H. Biomarkers distinguish apoptotic and necrotic cell death during hepatic ischemia/reperfusion injury in mice. Liver Transpl. Off. Publ. Am. Assoc. Study Liver Dis. Int. Liver Transplant. Soc. 2014, 20, 1372–1382. [Google Scholar] [CrossRef]
- Bai, S.; Thummel, R.; Godwin, A.R.; Nagase, H.; Itoh, Y.; Li, L.; Evans, R.; McDermott, J.; Seiki, M.; Sarras, M.P. Matrix metalloproteinase expression and function during fin regeneration in zebrafish: Analysis of MT1-MMP, MMP2 and TIMP2. Matrix Biol. 2005, 24, 247–260. [Google Scholar] [CrossRef]
- Page, L.; Polok, B.; Bustamante, M.; Schorderet, D.F. Bigh3 is upregulated in regenerating zebrafish fin. Zebrafish 2013, 10, 36–42. [Google Scholar] [CrossRef]
- Tucker, B.; Klassen, H.; Yang, L.; Chen, D.F.; Young, M.J. Elevated MMP Expression in the MRL Mouse Retina Creates a Permissive Environment for Retinal Regeneration. Investig. Ophthalmol. Vis. Sci. 2008, 49, 1686–1695. [Google Scholar] [CrossRef]
- Reichenbach, A.; Bringmann, A. New Functions of Müller Cells. Glia 2013, 61, 651–678. [Google Scholar] [CrossRef]
- Chablais, F.; Jaźwińska, A. The regenerative capacity of the zebrafish heart is dependent on TGFβ signaling. Development 2012, 139, 1921–1930. [Google Scholar] [CrossRef] [Green Version]
- Pollak, J.; Wilken, M.S.; Ueki, Y.; Cox, K.E.; Sullivan, J.M.; Taylor, R.J.; Levine, E.M.; Reh, T.A. ASCL1 reprograms mouse Muller glia into neurogenic retinal progenitors. Development 2013, 140, 2619–2631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, C.M.; Gorsuch, R.A.; Bailey, T.J.; Ackerman, K.M.; Kassen, S.C.; Hyde, D.R. Stat3 Defines Three Populations of Müller Glia and Is Required for Initiating Maximal Müller Glia Proliferation in the Regenerating Zebrafish Retina. J. Comp. Neurol 2012, 520, 4294–4311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinhard, J.; Joachim, S.C.; Faissner, A. Extracellular matrix remodeling during retinal development. Exp. Eye Res. 2015, 133, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Kizil, C.; Kyritsis, N.; Dudczig, S.; Kroehne, V.; Freudenreich, D.; Kaslin, J.; Brand, M. Regenerative Neurogenesis from Neural Progenitor Cells Requires Injury-Induced Expression of Gata3. Dev. Cell 2012, 23, 1230–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kizil, C.; Dudczig, S.; Kyritsis, N.; Machate, A.; Blaesche, J.; Kroehne, V.; Brand, M. The chemokine receptor cxcr5 regulates the regenerative neurogenesis response in the adult zebrafish brain. Neural Dev. 2012, 7, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, R.; Zhao, X.-F.; Goldman, D. Ascl1a/Dkk/beta-catenin signaling pathway is necessary and glycogen synthase kinase-3beta inhibition is sufficient for zebrafish retina regeneration. Proc. Natl. Acad. Sci. USA 2011, 108, 15858–15863. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.K.; Sarathchandra, P.; Chester, A.; Yacoub, M.; Brand, T.; Butcher, J.T. Cardiac regeneration following cryoinjury in the adult zebrafish targets a maturation-specific biomechanical remodeling program. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chassot, B.; Pury, D.; Jaźwińska, A. Zebrafish fin regeneration after cryoinjury-induced tissue damage. Biol. Open 2016, 5, 819–828. [Google Scholar] [CrossRef] [Green Version]
- Lenkowski, J.R.; Raymond, P.A. Muller glia: Stem cells for generation and regeneration of retinal neurons in teleost fish. Prog. Retin. Eye Res. 2014, 40, 94–123. [Google Scholar] [CrossRef]
- Gemberling, M.; Bailey, T.J.; Hyde, D.R.; Poss, K.D. The zebrafish as a model for complex tissue regeneration. Trends Genet. 2013, 29, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Godwin, J. The promise of perfect adult tissue repair and regeneration in mammals: Learning from regenerative amphibians and fish. BioEssays 2014, 36, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Vecino, E.; Rodriguez, F.D.; Ruzafa, N.; Pereiro, X.; Sharma, S.C. Glia-neuron interactions in the mammalian retina. Prog. Retin. Eye Res. 2016, 51, 1–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, A.; Pandey, R.K.; Miller, L.J.; Singh, P.K.; Kanwar, M. Müller glia in retinal innate immunity: A perspective on their roles in endophthalmitis. Crit. Rev. Immunol. 2013, 33, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.J.; Fossum, S.L.; Fimbel, S.M.; Montgomery, J.E.; Hyde, D.R. The inhibitor of phagocytosis, O-phospho-l-serine, suppresses Muller glia proliferation and cone cell regeneration in the light-damaged zebrafish retina. Exp. Eye Res. 2010, 91, 601–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, H.; Kotter, M.R.; Franklin, R.J.M. Debris clearance by microglia: An essential link between degeneration and regeneration. Brain 2009, 132, 288–295. [Google Scholar] [CrossRef]
- Stenkamp, D.L.; Cameron, D.A. Cellular pattern formation in the retina: Retinal regeneration as a model system. Mol. Vis. 2002, 8, 280–293. [Google Scholar]
- Hitchcock, P.F.; Myhr, K.J.L.; Easter, S.S.; Mangione-Smith, R.; Jones, D.D. Local regeneration in the retina of the goldfish. J. Neurobiol. 1992, 23, 187–203. [Google Scholar] [CrossRef] [Green Version]
- Bejarano-Escobar, R.; Blasco, M.; Martín-Partido, G.; Francisco-Morcillo, J. Light-induced degeneration and microglial response in the retina of an epibenthonic pigmented teleost: Age-dependent photoreceptor susceptibility to cell death. J. Exp. Biol. 2012, 215, 3799–3812. [Google Scholar] [CrossRef] [Green Version]
- Fimbel, S.M.; Montgomery, J.E.; Burket, C.T.; Hyde, D.R. Regeneration of inner retinal neurons after intravitreal injection of ouabain in zebrafish. J. Neurosci. 2007, 27, 1712–1724. [Google Scholar] [CrossRef] [Green Version]
- Fausett, B.V.; Goldman, D. A role for alpha1 tubulin-expressing Müller glia in regeneration of the injured zebrafish retina. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 6303–6313. [Google Scholar] [CrossRef] [Green Version]
- Fausett, B.V.; Gumerson, J.D.; Goldman, D. The proneural basic helix-loop-helix gene ascl1a is required for retina regeneration. J. Neurosci. Off. J. Soc. Neurosci. 2008, 28, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.L.; Ranski, A.H.; Morgan, G.W.; Thummel, R. Reactive gliosis in the adult zebrafish retina. Exp. Eye Res. 2016, 143, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Mitra, S.; Sharma, P.; Kaur, S.; Khursheed, M.A.; Gupta, S.; Chaudhary, M.; Kurup, A.J.; Ramachandran, R. Dual regulation of lin28a by Myc is necessary during zebrafish retina regeneration. J. Cell Biol. 2019, 218, 489–507. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.M.; Ackerman, K.M.; O’Hayer, P.; Bailey, T.J.; Gorsuch, R.A.; Hyde, D.R. Tumor necrosis factor-alpha is produced by dying retinal neurons and is required for Müller glia proliferation during zebrafish retinal regeneration. J. Neurosci. 2013, 33, 6524–6539. [Google Scholar] [CrossRef] [Green Version]
- Ramachandran, R.; Zhao, X.F.; Goldman, D. Insm1a-mediated gene repression is essential for the formation and differentiation of Müller glia-derived progenitors in the injured retina. Nat. Cell Biol. 2012, 14, 1013–1023. [Google Scholar] [CrossRef] [Green Version]
- Hein, S.J.; Lehmann, L.H.; Kossack, M.; Juergensen, L.; Fuchs, D.; Katus, H.A.; Hassel, D. Advanced echocardiography in adult zebrafish reveals delayed recovery of heart function after myocardial cryoinjury. PLoS ONE 2015, 10, e0122665. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.F.; Wan, J.; Powell, C.; Ramachandran, R.; Myers, M.G.; Goldman, D. Leptin and IL-6 family cytokines synergize to stimulate Müller Glia reprogramming and retina regeneration. Cell Rep. 2014, 9, 272–284. [Google Scholar] [CrossRef] [Green Version]
- Kassen, S.C.; Ramanan, V.; Montgomery, J.E.; TBurket, C.; Liu, C.G.; Vihtelic, T.S.; Hyde, D.R. Time course analysis of gene expression during light-induced photoreceptor cell death and regeneration in albino zebrafish. Dev. Neurobiol 2007, 67, 1009–1031. [Google Scholar] [CrossRef]
- Wan, J.; Zhao, X.F.; Vojtek, A.; Goldman, D. Retinal injury, growth factors and cytokines converge on β- catenin and pStat3 signaling to stimulate retina regeneration. Cell Rep. 2014, 9, 285–297. [Google Scholar] [CrossRef] [Green Version]
- Brown, B.N.; Badylak, S.F. Extracellular matrix as an inductive scaffold for functional tissue reconstruction. Transl. Res. J. Lab. Clin. Med. 2014, 163, 268–285. [Google Scholar] [CrossRef] [Green Version]
- Kato, T.; Miyazaki, K.; Shimizu-Nishikawa, K.; Koshiba, K.; Obara, M.; Mishima, H.K.; Yoshizato, K. Unique expression patterns of matrix metalloproteinases in regenerating newt limbs. Dev. Dyn. 2003, 226, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Thapa, N.; Lee, B.-H.; Kim, I.-S. TGFBIp/βig-h3 protein: A versatile matrix molecule induced by TGF-β. Int. J. Biochem. Cell Biol. 2007, 39, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Kwon, H.J.; Kim, D.S. Matrix metalloproteinase 9 (MMP-9)-dependent processing of βig-h3 protein regulates cell migration, invasion, and adhesion. J. Biol. Chem. 2012, 287, 38957–38969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondragon, A.A.; Betts-Obregon, B.S.; Moritz, R.J.; Parvathaneni, K.; Navarro, M.M.; Kim, H.S.; Lee, C.F.; Lebaron, R.G.; Asmis, R.; Tsin, A.T. BIGH3 protein and macrophages in retinal endothelial cell apoptosis. Apoptosis 2015, 20, 29–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, R.W.D.; Vickaryous, M.K.; Viloria-Petit, A.M. Signalling by transforming growth factor beta isoforms in wound healing and tissue regeneration. J. Dev. Biol. 2016, 4, 21. [Google Scholar] [CrossRef] [Green Version]
- Stella, S.L., Jr.; Geathers, J.S.; Weber, S.R.; Grillo, M.A.; Barber, A.J.; Sundstrom, J.M.; Grillo, S.L. Neurodegeneration, Neuroprotection and Regeneration in the Zebrafish Retina. Cells 2021, 10, 633. [Google Scholar] [CrossRef]
- Gao, H.A.L.; Huang, X.; Chen, X.; Xu, H. Müller Glia-Mediated Retinal Regeneration. Mol. Neurobiol. 2021, 58, 2342–2361. [Google Scholar] [CrossRef]
- Lehman, N.L. Rosettes and Pseudorosettes. In Encyclopedia of the Neurological Sciences, 2nd ed.; Aminoff, M.J., Daroff, R.B., Eds.; Academic Press: Cambridge, MA, USA, 2014; pp. 69–73. [Google Scholar]
- Wang, J.; Karra, R.; Dickson, A.L.; Poss, K.D. Fibronectin is deposited by injury-activated epicardial cells and is necessary for zebrafish heart regeneration. Dev. Biol. 2013, 382, 427–435. [Google Scholar] [CrossRef] [Green Version]
- Tahara, N.; Brush, M.; Kawakami, Y. Cell migration during heart regeneration in zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2016, 245, 774–787. [Google Scholar] [CrossRef] [Green Version]


| GENES | ||||||
| Comparison | FDR < 0.05 | FDR < 0.05 and fold change >2 | ||||
| ALL | UP | DOWN | ALL | UP | DOWN | |
| 3 dpi_vs_0 dpi | 12088 | 6507 | 5581 | 6014 | 4157 | 1857 |
| 7 dpi_vs_0 dpi | 10142 | 5625 | 4517 | 4446 | 3682 | 764 |
| 30 dpi_vs_0 dpi | 2939 | 2489 | 450 | 1500 | 1481 | 19 |
| ISOFORMS | ||||||
| Comparison | FDR < 0.05 | FDR < 0.05 and fold change >2 | ||||
| ALL | UP | DOWN | ALL | UP | DOWN | |
| 3 dpi_vs_0 dpi | 13362 | 7979 | 5383 | 7655 | 5645 | 2010 |
| 7 dpi_vs_0 dpi | 10228 | 6343 | 3885 | 5454 | 4562 | 892 |
| 30 dpi_vs_0 dpi | 2476 | 2205 | 271 | 1673 | 1529 | 144 |
| DEG Cluster * | Annotation Cluster ** | Enrichment Score | Category | Term | Gene Count | % of Pathway | Fold Enrichment | p Value *** | |
|---|---|---|---|---|---|---|---|---|---|
| All significant DEG | 2 | 1 | 12.09 | GOTERM_CC_DIRECT | GO:0005840~ribosome | 34 | 4.21 | 5.59 | 2.71 × 10−13 |
| 2 | 1 | 12.09 | UP_KEYWORDS | Ribosomal protein | 26 | 3.22 | 6.09 | 1.51 × 10−10 | |
| 2 | 1 | 12.09 | KEGG_PATHWAY | dre03010:Ribosome | 31 | 3.84 | 4.25 | 1.34 × 10−09 | |
| 2 | 1 | 12.09 | GOTERM_MF_DIRECT | GO:0003735~structural constituent of ribosome | 31 | 3.84 | 4.21 | 3.49 × 10−08 | |
| 2 | 2 | 4.44 | INTERPRO | IPR002194:Chaperonin TCP-1, conserved site | 6 | 0.74 | 21.61 | 3.39 × 10−3 | |
| 2 | 2 | 4.44 | INTERPRO | IPR027410:TCP-1-like chaperonin intermediate domain | 6 | 0.74 | 19.21 | 7.39 × 10−3 | |
| 2 | 2 | 4.44 | INTERPRO | IPR017998:Chaperone tailless complex polypeptide 1 (TCP-1) | 6 | 0.74 | 19.21 | 7.39 × 10−3 | |
| 2 | 2 | 4.44 | INTERPRO | IPR027409:GroEL-like apical domain | 6 | 0.74 | 14.41 | 4.18 × 10−2 | |
| 2 | 2 | 4.44 | INTERPRO | IPR027413:GroEL-like equatorial domain | 6 | 0.74 | 14.41 | 4.18 × 10−2 | |
| 2 | 3 | 3.55 | INTERPRO | IPR019821:Kinesin, motor region, conserved site | 10 | 1.24 | 6.26 | 3.16 × 10−2 | |
| 2 | 3 | 3.55 | SMART | SM00129:KISc | 10 | 1.24 | 5.96 | 1.00 × 10−2 | |
| 3 | 1 | 26.82 | GOTERM_CC_DIRECT | GO:0005840~ribosome | 42 | 8.55 | 11.27 | 2.72 × 10−29 | |
| 3 | 1 | 26.82 | KEGG_PATHWAY | dre03010:Ribosome | 40 | 8.15 | 11.13 | 1.72 × 10−29 | |
| 3 | 1 | 26.82 | UP_KEYWORDS | Ribosomal protein | 36 | 7.33 | 13.91 | 9.08 × 10−28 | |
| 3 | 1 | 26.82 | GOTERM_MF_DIRECT | GO:0003735~structural constituent of ribosome | 40 | 8.15 | 8.59 | 1.43 × 10−22 | |
| 3 | 1 | 26.82 | UP_KEYWORDS | Ribonucleoprotein | 37 | 7.54 | 9.3 | 5.58 × 10−22 | |
| 3 | 1 | 26.82 | GOTERM_CC_DIRECT | GO:0030529~intracellular ribonucleoprotein complex | 38 | 7.74 | 8.26 | 3.91 × 10−21 | |
| 3 | 2 | 3.55 | GOTERM_CC_DIRECT | GO:0005852~eukaryotic translation initiation factor 3 complex | 6 | 1.22 | 14.48 | 5.29 × 10−3 | |
| 3 | 2 | 3.55 | GOTERM_CC_DIRECT | GO:0016282~eukaryotic 43S preinitiation complex | 5 | 1.02 | 13.58 | 4.98 × 10−2 | |
| 3 | 3 | 3.41 | GOTERM_CC_DIRECT | GO:0005852~eukaryotic translation initiation factor 3 complex | 6 | 1.22 | 14.48 | 5.29 × 10−3 | |
| 4 | 2 | 1.98 | PIR_SUPERFAMILY | PIRSF002285:Op18/stathmin | 3 | 1.2 | 40.88 | 2.97 × 10−2 | |
| 5 | 1 | 8.29 | INTERPRO | IPR008160:Collagen triple helix repeat | 11 | 7.14 | 17.24 | 2.58 × 10−7 | |
| 5 | 1 | 8.29 | UP_KEYWORDS | Collagen | 9 | 5.84 | 20.41 | 1.06 × 10−6 | |
| 5 | 1 | 8.29 | GOTERM_CC_DIRECT | GO:0005581~collagen trimer | 9 | 5.84 | 20.13 | 5.32 × 10−7 | |
| 8 | 1 | 3.51 | GOTERM_BP_DIRECT | GO:0019882~antigen processing and presentation | 5 | 9.43 | 38.96 | 4.36 × 10−4 | |
| DEG significant at all timepoints | 1 | 1 | 5.57 | UP_KEYWORDS | Mitosis | 6 | 10.34 | 38.18 | 2.83 × 10−5 |
| 1 | 1 | 5.57 | UP_KEYWORDS | Cell division | 6 | 10.34 | 24.40 | 2.63 × 10−4 | |
| 1 | 1 | 5.57 | GOTERM_BP_DIRECT | GO:0007067~mitotic nuclear division | 6 | 10.34 | 20.46 | 1.69 × 10−3 | |
| 1 | 2 | 3.33 | UP_KEYWORDS | Proteasome | 4 | 6.90 | 29.22 | 2.00 × 10−2 | |
| 1 | 2 | 3.33 | GOTERM_CC_DIRECT | GO:0000502~proteasome complex | 4 | 6.90 | 25.14 | 2.07 × 10−2 | |
| 1 | 2 | 3.33 | KEGG_PATHWAY | dre03050:Proteasome | 4 | 6.90 | 21.84 | 1.04 × 10−2 | |
| 1 | 3 | 3.18 | INTERPRO | IPR019821:Kinesin, motor region, conserved site | 4 | 6.90 | 33.66 | 3.28 × 10−2 | |
| 1 | 3 | 3.18 | SMART | SM00129:KISc | 4 | 6.90 | 29.22 | 1.03 × 10−2 | |
| 1 | 3 | 3.18 | INTERPRO | IPR001752:Kinesin, motor domain | 4 | 6.90 | 29.22 | 4.96 × 10−2 | |
| 1 | 3 | 3.18 | GOTERM_CC_DIRECT | GO:0005871~kinesin complex | 4 | 6.90 | 23.43 | 2.54 × 10−2 | |
| 1 | 4 | 3.00 | INTERPRO | IPR018525:Mini-chromosome maintenance, conserved site | 3 | 5.17 | 129.05 | 3.40 × 10−2 | |
| 1 | 4 | 3.00 | SMART | SM00350:MCM | 3 | 5.17 | 113.97 | 9.46 × 10−3 | |
| 1 | 4 | 3.00 | INTERPRO | IPR001208:Mini-chromosome maintenance, DNA-dependent ATPase | 3 | 5.17 | 116.14 | 4.23 × 10−2 | |
| 1 | 4 | 3.00 | GOTERM_CC_DIRECT | GO:0042555~MCM complex | 3 | 5.17 | 94.26 | 1.76 × 10−2 | |
| 1 | 4 | 3.00 | GOTERM_MF_DIRECT | GO:0003678~DNA helicase activity | 3 | 5.17 | 84.78 | 3.33 × 10−2 | |
| 1 | 5 | 2.73 | INTERPRO | IPR000243:Peptidase T1A, proteasome beta-subunit | 3 | 5.17 | 116.14 | 4.23 × 10−2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Džulová, D.; Lawless, D.; Pinton, G.G.; Renner, N.A.; Schorderet, D.F. Incomplete Recovery of Zebrafish Retina Following Cryoinjury. Cells 2022, 11, 1373. https://doi.org/10.3390/cells11081373
Džulová D, Lawless D, Pinton GG, Renner NA, Schorderet DF. Incomplete Recovery of Zebrafish Retina Following Cryoinjury. Cells. 2022; 11(8):1373. https://doi.org/10.3390/cells11081373
Chicago/Turabian StyleDžulová, Denisa, Dylan Lawless, Gaëtan G. Pinton, Nicole A. Renner, and Daniel F. Schorderet. 2022. "Incomplete Recovery of Zebrafish Retina Following Cryoinjury" Cells 11, no. 8: 1373. https://doi.org/10.3390/cells11081373






