MicroRNAs as Regulators of Phagocytosis
Abstract
:1. MicroRNAs—Biogenesis, Genomics, Regulation, Mechanisms of Action and Biological Functions
2. The Immune System and MicroRNAs
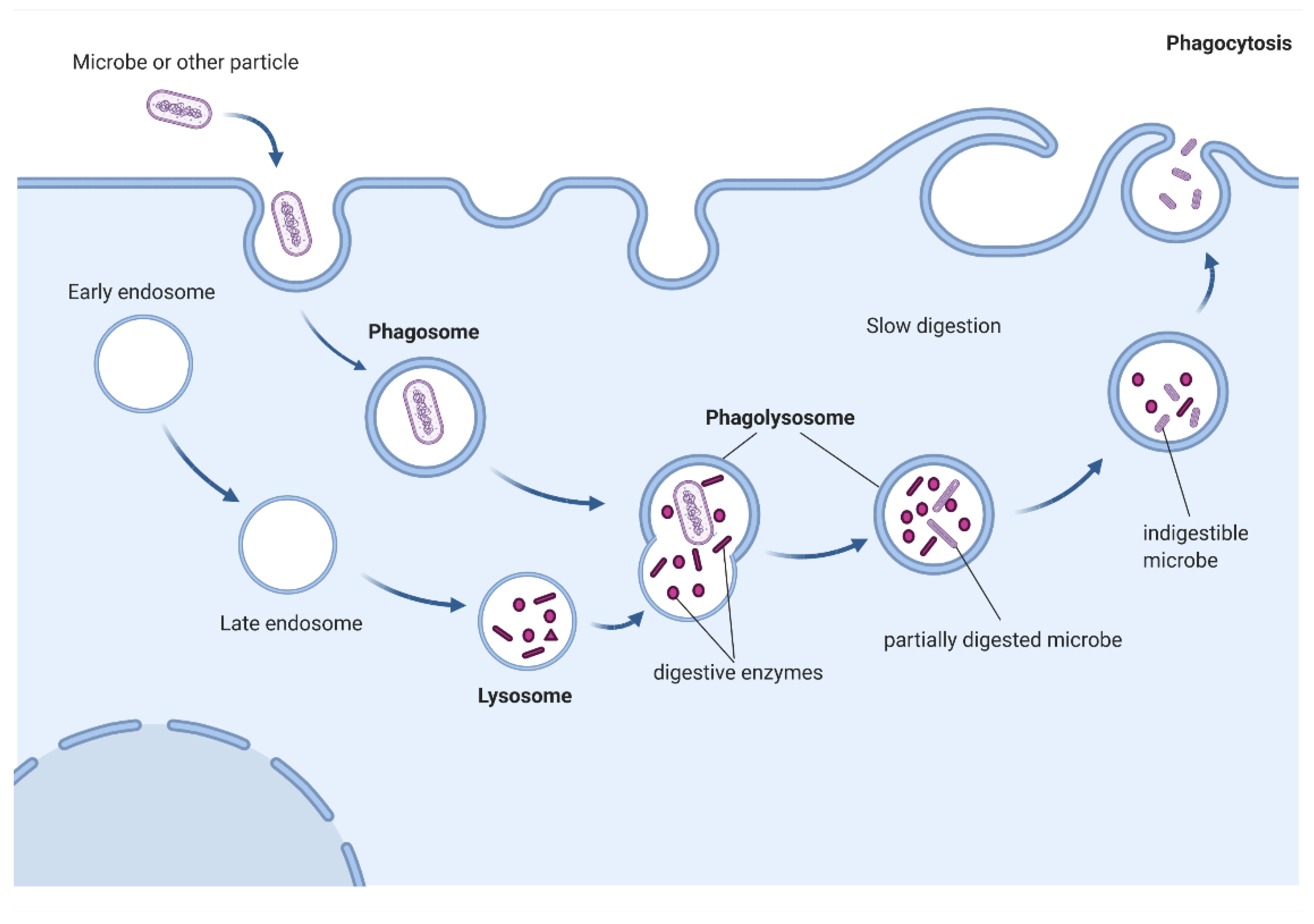
3. Phagocytosis—Overview
Examples of Cells with Phagocytic Capacity
4. Role of MicroRNA in Regulation of Different Stages of Phagocytosis Performed by Macrophages
4.1. Differentiation
4.2. Polarization
4.3. Recognition of Pathogen-Associated Molecular Patterns (PAMPs) by Pattern-Recognition Receptors (PRRs) Expressed on/in Macrophages
4.4. Phagocytosis—Uptake
4.5. Modulation of Phagosomal Maturation
4.6. Modulation of Reactive Oxygen Species (ROS) Production inside of Phagosomes and Phagolysosomes
4.7. Modulation of Lysosomal Activity
4.8. Antigen Presenting
4.9. Resolving Inflammation: Conversion to Anti-Inflammatory Phenotype to Terminate Anti-Infectious Response and to Promote Tissue Repair
5. Microglia
6. Osteoclasts
7. Neutrophils
8. Retinal Pigment Epithelium
9. Vascular Smooth Muscle Cells
10. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [Green Version]
- Ellwanger, D.C.; Büttner, F.A.; Mewes, H.W.; Stümpflen, V. The sufficient minimal set of miRNA seed types. Bioinformatics 2011, 27, 1346–1350. [Google Scholar] [CrossRef]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.H.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Mol. Cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [Green Version]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Krek, A.; Grün, D.; Poy, M.N.; Wolf, R.; Rosenberg, L.; Epstein, E.J.; MacMenamin, P.; da Piedade, I.; Gunsalus, K.C.; Stoffel, M.; et al. Combinatorial microRNA target predictions. Nat. Genet. 2005, 37, 495–500. [Google Scholar] [CrossRef]
- Loeb, G.B.; Khan, A.A.; Canner, D.; Hiatt, J.B.; Shendure, J.; Darnell, R.B.; Leslie, C.S.; Rudensky, A.Y. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell 2012, 48, 760–770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.A.; Jo, M.H.; Choi, Y.G.; Park, J.; Kwon, S.C.; Hohng, S.; Kim, V.N.; Woo, J.S. Functional Anatomy of the Human Microprocessor. Cell 2015, 161, 1374–1387. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Hutvágner, G.; McLachlan, J.; Pasquinelli, A.E.; Bálint, E.; Tuschl, T.; Zamore, P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 2001, 293, 834–838. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef]
- Suzuki, H.I.; Katsura, A.; Yasuda, T.; Ueno, T.; Mano, H.; Sugimoto, K.; Miyazono, K. Small-RNA asymmetry is directly driven by mammalian Argonautes. Nat. Struct. Mol. Biol. 2015, 22, 512–521. [Google Scholar] [CrossRef]
- Jazdzewski, K.; Liyanarachchi, S.; Swierniak, M.; Pachucki, J.; Ringel, M.D.; Jarzab, B.; de la Chapelle, A. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc. Natl. Acad. Sci. USA 2009, 106, 1502–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gantier, M.P.; McCoy, C.E.; Rusinova, I.; Saulep, D.; Wang, D.; Xu, D.; Irving, A.T.; Behlke, M.A.; Hertzog, P.J.; Mackay, F.; et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011, 39, 5692–5703. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutvágner, G.; Simard, M.J.; Mello, C.C.; Zamore, P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004, 2, E98. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Kauppinen, S.; Lund, A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006, 372, 137–141. [Google Scholar] [CrossRef]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar] [CrossRef]
- Xie, J.; Ameres, S.L.; Friedline, R.; Hung, J.H.; Zhang, Y.; Xie, Q.; Zhong, L.; Su, Q.; He, R.; Li, M.; et al. Long-term, efficient inhibition of microRNA function in mice using rAAV vectors. Nat. Methods 2012, 9, 403–409. [Google Scholar] [CrossRef]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [Green Version]
- Armand-Labit, V.; Pradines, A. Circulating cell-free microRNAs as clinical cancer biomarkers. Biomol. Concepts 2017, 8, 61–81. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Liu, D.; Chen, X.; Li, J.; Li, L.; Bian, Z.; Sun, F.; Lu, J.; Yin, Y.; Cai, X.; et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 2010, 39, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, A.; Yan, I.K.; Foye, C.; Parasramka, M.; Patel, T. MicroRNAs as paracrine signaling mediators in cancers and metabolic diseases. Best Pract. Research. Clin. Endocrinol. Metab. 2016, 30, 577–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, M.; Hu, R.; Runtsch, M.C.; Kagele, D.A.; Mosbruger, T.L.; Tolmachova, T.; Seabra, M.C.; Round, J.L.; Ward, D.M.; O’Connell, R.M. Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 2015, 6, 7321. [Google Scholar] [CrossRef] [Green Version]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [Green Version]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Reviews. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef]
- Chevillet, J.R.; Kang, Q.; Ruf, I.K.; Briggs, H.A.; Vojtech, L.N.; Hughes, S.M.; Cheng, H.H.; Arroyo, J.D.; Meredith, E.K.; Gallichotte, E.N.; et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA 2014, 111, 14888–14893. [Google Scholar] [CrossRef] [Green Version]
- Monticelli, S.; Natoli, G. Transcriptional determination and functional specificity of myeloid cells: Making sense of diversity. Nat. Reviews. Immunol. 2017, 17, 595–607. [Google Scholar] [CrossRef]
- Jia, Y.; Wei, Y. Modulators of MicroRNA Function in the Immune System. Int. J. Mol. Sci. 2020, 21, 2357. [Google Scholar] [CrossRef]
- Jia, X.; Yuan, S.; Wang, Y.; Fu, Y.; Ge, Y.; Ge, Y.; Lan, X.; Feng, Y.; Qiu, F.; Li, P.; et al. The role of alternative polyadenylation in the antiviral innate immune response. Nat. Commun. 2017, 8, 14605. [Google Scholar] [CrossRef]
- Ledwith, B.J.; Manam, S.; Troilo, P.J.; Barnum, A.B.; Pauley, C.J.; Griffiths, T.G., 2nd; Harper, L.B.; Beare, C.M.; Bagdon, W.J.; Nichols, W.W. Plasmid DNA vaccines: Investigation of integration into host cellular DNA following intramuscular injection in mice. Intervirology 2000, 43, 258–272. [Google Scholar] [CrossRef]
- Stephenson, M.L.; Zamecnik, P.C. Inhibition of Rous sarcoma viral RNA translation by a specific oligodeoxyribonucleotide. Proc. Natl. Acad. Sci. USA 1978, 75, 285–288. [Google Scholar] [CrossRef] [Green Version]
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468. [Google Scholar] [CrossRef]
- Damase, T.R.; Sukhovershin, R.; Boada, C.; Taraballi, F.; Pettigrew, R.I.; Cooke, J.P. The Limitless Future of RNA Therapeutics. Front. Bioeng. Biotechnol. 2021, 9, 628137. [Google Scholar] [CrossRef]
- Lindow, M.; Kauppinen, S. Discovering the first microRNA-targeted drug. J. Cell Biol. 2012, 199, 407–412. [Google Scholar] [CrossRef]
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Investig. New Drugs 2017, 35, 180–188. [Google Scholar] [CrossRef]
- Yu, A.M.; Choi, Y.H.; Tu, M.J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020, 72, 862–898. [Google Scholar] [CrossRef]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Zhao, J.L.; Rao, D.S. MicroRNA function in myeloid biology. Blood 2011, 118, 2960–2969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Baltimore, D. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012, 30, 295–312. [Google Scholar] [CrossRef]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Reviews. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Fagundes, C.T.; Yang, G.; Palsson-McDermott, E.M.; Wochal, P.; McGettrick, A.F.; Foley, N.H.; Early, J.O.; Chen, L.; Zhang, H.; et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl. Acad. Sci. USA 2015, 112, 7231–7236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Zhang, H.; Rodriguez, S.; Cao, L.; Parish, J.; Mumaw, C.; Zollman, A.; Kamoka, M.M.; Mu, J.; Chen, D.Z.; et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-κB-dependent manner. Cell Stem Cell 2014, 15, 51–65. [Google Scholar] [CrossRef] [Green Version]
- Androulidaki, A.; Iliopoulos, D.; Arranz, A.; Doxaki, C.; Schworer, S.; Zacharioudaki, V.; Margioris, A.N.; Tsichlis, P.N.; Tsatsanis, C. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 2009, 31, 220–231. [Google Scholar] [CrossRef] [Green Version]
- Taganov, K.D.; Boldin, M.P.; Chang, K.J.; Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Boldin, M.P.; Taganov, K.D.; Nicoll, J.; Paquette, R.L.; Baltimore, D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008, 205, 585–594. [Google Scholar] [CrossRef]
- Marques, S.C.; Laursen, M.B.; Bødker, J.S.; Kjeldsen, M.K.; Falgreen, S.; Schmitz, A.; Bøgsted, M.; Johnsen, H.E.; Dybkaer, K. MicroRNAs in B-cells: From normal differentiation to treatment of malignancies. Oncotarget 2015, 6, 7–25. [Google Scholar] [CrossRef] [Green Version]
- Podshivalova, K.; Salomon, D.R. MicroRNA regulation of T-lymphocyte immunity: Modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit. Rev. Immunol. 2013, 33, 435–476. [Google Scholar] [CrossRef]
- Dooley, J.; Linterman, M.A.; Liston, A. MicroRNA regulation of T-cell development. Immunol. Rev. 2013, 253, 53–64. [Google Scholar] [CrossRef] [PubMed]
- de Yébenes, V.G.; Bartolomé-Izquierdo, N.; Ramiro, A.R. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013, 253, 25–39. [Google Scholar] [CrossRef] [Green Version]
- Jeker, L.T.; Bluestone, J.A. MicroRNA regulation of T-cell differentiation and function. Immunol. Rev. 2013, 253, 65–81. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Wan, Y.; Ji, Q.; Fang, Y.; Wu, Y. The role of microRNAs in B-cell development and function. Cell. Mol. Immunol. 2013, 10, 107–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Underhill, D.M.; Gordon, S.; Imhof, B.A.; Núñez, G.; Bousso, P. Élie Metchnikoff (1845–1916): Celebrating 100 years of cellular immunology and beyond. Nat. Reviews. Immunol. 2016, 16, 651–656. [Google Scholar] [CrossRef] [PubMed]
- Heifets, L. Centennial of Metchnikoff’s discovery. J. Reticuloendothel. Soc. 1982, 31, 381–391. [Google Scholar]
- Aderem, A.; Underhill, D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999, 17, 593–623. [Google Scholar] [CrossRef]
- Leijh, P.C.; van den Barselaar, M.T.; Daha, M.R.; van Furth, R. Participation of immunoglobulins and complement components in the intracellular killing of Staphylococcus aureus and Escherichia coli by human granulocytes. Infect. Immun. 1981, 33, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Flannagan, R.S.; Jaumouillé, V.; Grinstein, S. The cell biology of phagocytosis. Annu. Rev. Pathol. 2012, 7, 61–98. [Google Scholar] [CrossRef]
- Freeman, S.A.; Grinstein, S. Phagocytosis: Receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014, 262, 193–215. [Google Scholar] [CrossRef] [PubMed]
- Bohdanowicz, M.; Cosío, G.; Backer, J.M.; Grinstein, S. Class I and class III phosphoinositide 3-kinases are required for actin polymerization that propels phagosomes. J. Cell Biol. 2010, 191, 999–1012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levin, R.; Grinstein, S.; Canton, J. The life cycle of phagosomes: Formation, maturation, and resolution. Immunol. Rev. 2016, 273, 156–179. [Google Scholar] [CrossRef]
- Balce, D.R.; Yates, R.M. Fluorometric Approaches to Measuring Reductive and Oxidative Events in Phagosomes. Methods Mol. Biol. 2017, 1519, 215–225. [Google Scholar] [CrossRef]
- Weiss, G.; Schaible, U.E. Macrophage defense mechanisms against intracellular bacteria. Immunol. Rev. 2015, 264, 182–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturgill-Koszycki, S.; Schlesinger, P.H.; Chakraborty, P.; Haddix, P.L.; Collins, H.L.; Fok, A.K.; Allen, R.D.; Gluck, S.L.; Heuser, J.; Russell, D.G. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton-ATPase. Science 1994, 263, 678–681. [Google Scholar] [CrossRef]
- Aberdein, J.D.; Cole, J.; Bewley, M.A.; Marriott, H.M.; Dockrell, D.H. Alveolar macrophages in pulmonary host defence the unrecognized role of apoptosis as a mechanism of intracellular bacterial killing. Clin. Exp. Immunol. 2013, 174, 193–202. [Google Scholar] [CrossRef]
- Mortimer, P.M.; Mc Intyre, S.A.; Thomas, D.C. Beyond the Extra Respiration of Phagocytosis: NADPH Oxidase 2 in Adaptive Immunity and Inflammation. Front. Immunol. 2021, 12, 733918. [Google Scholar] [CrossRef]
- Canton, J.; Khezri, R.; Glogauer, M.; Grinstein, S. Contrasting phagosome pH regulation and maturation in human M1 and M2 macrophages. Mol. Biol. Cell 2014, 25, 3330–3341. [Google Scholar] [CrossRef]
- El Chemaly, A.; Nunes, P.; Jimaja, W.; Castelbou, C.; Demaurex, N. Hv1 proton channels differentially regulate the pH of neutrophil and macrophage phagosomes by sustaining the production of phagosomal ROS that inhibit the delivery of vacuolar ATPases. J. Leukoc. Biol. 2014, 95, 827–839. [Google Scholar] [CrossRef]
- Savina, A.; Amigorena, S. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 2007, 219, 143–156. [Google Scholar] [CrossRef]
- Jubrail, J.; Morris, P.; Bewley, M.A.; Stoneham, S.; Johnston, S.A.; Foster, S.J.; Peden, A.A.; Read, R.C.; Marriott, H.M.; Dockrell, D.H. Inability to sustain intraphagolysosomal killing of Staphylococcus aureus predisposes to bacterial persistence in macrophages. Cell. Microbiol. 2016, 18, 80–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuart, L.M.; Ezekowitz, R.A. Phagocytosis and comparative innate immunity: Learning on the fly. Nat. Reviews. Immunol. 2008, 8, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, K.S. Find-me and eat-me signals in apoptotic cell clearance: Progress and conundrums. J. Exp. Med. 2010, 207, 1807–1817. [Google Scholar] [CrossRef]
- Ravichandran, K.S.; Lorenz, U. Engulfment of apoptotic cells: Signals for a good meal. Nat. Reviews. Immunol. 2007, 7, 964–974. [Google Scholar] [CrossRef]
- Nordenfelt, P.; Tapper, H. Phagosome dynamics during phagocytosis by neutrophils. J. Leukoc. Biol. 2011, 90, 271–284. [Google Scholar] [CrossRef]
- Ip, W.K.; Sokolovska, A.; Charriere, G.M.; Boyer, L.; Dejardin, S.; Cappillino, M.P.; Yantosca, L.M.; Takahashi, K.; Moore, K.J.; Lacy-Hulbert, A.; et al. Phagocytosis and phagosome acidification are required for pathogen processing and MyD88-dependent responses to Staphylococcus aureus. J. Immunol. 2010, 184, 7071–7081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaumouillé, V.; Waterman, C.M. Physical Constraints and Forces Involved in Phagocytosis. Front. Immunol. 2020, 11, 1097. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Lu, J.H. Autophagy and Macrophage Functions: Inflammatory Response and Phagocytosis. Cells 2019, 9, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canton, J. Phagosome maturation in polarized macrophages. J. Leukoc. Biol. 2014, 96, 729–738. [Google Scholar] [CrossRef]
- Niida, S.; Kaku, M.; Amano, H.; Yoshida, H.; Kataoka, H.; Nishikawa, S.; Tanne, K.; Maeda, N.; Nishikawa, S.; Kodama, H. Vascular endothelial growth factor can substitute for macrophage colony-stimulating factor in the support of osteoclastic bone resorption. J. Exp. Med. 1999, 190, 293–298. [Google Scholar] [CrossRef] [Green Version]
- Paolicelli, R.C.; Bolasco, G.; Pagani, F.; Maggi, L.; Scianni, M.; Panzanelli, P.; Giustetto, M.; Ferreira, T.A.; Guiducci, E.; Dumas, L.; et al. Synaptic pruning by microglia is necessary for normal brain development. Science 2011, 333, 1456–1458. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.; Lobov, I.B.; Vallance, J.E.; Tsujikawa, K.; Shiojima, I.; Akunuru, S.; Walsh, K.; Benjamin, L.E.; Lang, R.A. Obligatory participation of macrophages in an angiopoietin 2-mediated cell death switch. Development 2007, 134, 4449–4458. [Google Scholar] [CrossRef] [Green Version]
- Fantin, A.; Vieira, J.M.; Gestri, G.; Denti, L.; Schwarz, Q.; Prykhozhij, S.; Peri, F.; Wilson, S.W.; Ruhrberg, C. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood 2010, 116, 829–840. [Google Scholar] [CrossRef] [Green Version]
- Edwards, J.P.; Zhang, X.; Frauwirth, K.A.; Mosser, D.M. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol. 2006, 80, 1298–1307. [Google Scholar] [CrossRef] [Green Version]
- Sindrilaru, A.; Peters, T.; Wieschalka, S.; Baican, C.; Baican, A.; Peter, H.; Hainzl, A.; Schatz, S.; Qi, Y.; Schlecht, A.; et al. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J. Clin. Investig. 2011, 121, 985–997. [Google Scholar] [CrossRef] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Beyer, M.; Mallmann, M.R.; Xue, J.; Staratschek-Jox, A.; Vorholt, D.; Krebs, W.; Sommer, D.; Sander, J.; Mertens, C.; Nino-Castro, A.; et al. High-resolution transcriptome of human macrophages. PLoS ONE 2012, 7, e45466. [Google Scholar] [CrossRef]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of monocytes, macrophages, and dendritic cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef] [Green Version]
- Greenlee-Wacker, M.C. Clearance of apoptotic neutrophils and resolution of inflammation. Immunol. Rev. 2016, 273, 357–370. [Google Scholar] [CrossRef] [Green Version]
- Gierlikowska, B.; Gierlikowski, W.; Demkow, U. Alantolactone Enhances the Phagocytic Properties of Human Macrophages and Modulates Their Proinflammatory Functions. Front. Pharmacol. 2020, 11, 1339. [Google Scholar] [CrossRef]
- Kaufmann, S.H.E.; Dorhoi, A. Molecular Determinants in Phagocyte-Bacteria Interactions. Immunity 2016, 44, 476–491. [Google Scholar] [CrossRef] [Green Version]
- Seider, K.; Brunke, S.; Schild, L.; Jablonowski, N.; Wilson, D.; Majer, O.; Barz, D.; Haas, A.; Kuchler, K.; Schaller, M.; et al. The facultative intracellular pathogen Candida glabrata subverts macrophage cytokine production and phagolysosome maturation. J. Immunol. 2011, 187, 3072–3086. [Google Scholar] [CrossRef] [Green Version]
- Wahl, S.M.; Swisher, J.; McCartney-Francis, N.; Chen, W. TGF-beta: The perpetrator of immune suppression by regulatory T cells and suicidal T cells. J. Leukoc. Biol. 2004, 76, 15–24. [Google Scholar] [CrossRef]
- Kevany, B.M.; Palczewski, K. Phagocytosis of retinal rod and cone photoreceptors. Physiology 2010, 25, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Sorokin, V.; Vickneson, K.; Kofidis, T.; Woo, C.C.; Lin, X.Y.; Foo, R.; Shanahan, C.M. Role of Vascular Smooth Muscle Cell Plasticity and Interactions in Vessel Wall Inflammation. Front. Immunol. 2020, 11, 599415. [Google Scholar] [CrossRef]
- Zhang, L.; Hao, C.; Yao, S.; Tang, R.; Guo, W.; Cong, H.; Li, J.; Bao, L.; Wang, D.; Li, Y.; et al. Exosomal miRNA Profiling to Identify Nanoparticle Phagocytic Mechanisms. Small Weinh. Der Bergstr. Ger. 2018, 14, e1704008. [Google Scholar] [CrossRef]
- Tripathi, A.; Srivastava, V.; Singh, B.N. hsa-let-7b-5p facilitates Mycobacterium tuberculosis survival in THP-1 human macrophages by Fas downregulation. FEMS Microbiol. Lett. 2018, 365, fny040. [Google Scholar] [CrossRef]
- Rong, J.; Xu, L.; Hu, Y.; Liu, F.; Yu, Y.; Guo, H.; Ni, X.; Huang, Y.; Zhao, L.; Wang, Z. Inhibition of let-7b-5p contributes to an anti-tumorigenic macrophage phenotype through the SOCS1/STAT pathway in prostate cancer. Cancer Cell Int. 2020, 20, 470. [Google Scholar] [CrossRef]
- Banerjee, S.; Xie, N.; Cui, H.; Tan, Z.; Yang, S.; Icyuz, M.; Abraham, E.; Liu, G. MicroRNA let-7c regulates macrophage polarization. J. Immunol. 2013, 190, 6542–6549. [Google Scholar] [CrossRef]
- Liu, C.; Wang, J.; Zhang, X. The involvement of MiR-1-clathrin pathway in the regulation of phagocytosis. PLoS ONE 2014, 9, e98747. [Google Scholar] [CrossRef]
- Bazzoni, F.; Rossato, M.; Fabbri, M.; Gaudiosi, D.; Mirolo, M.; Mori, L.; Tamassia, N.; Mantovani, A.; Cassatella, M.A.; Locati, M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc. Natl. Acad. Sci. USA 2009, 106, 5282–5287. [Google Scholar] [CrossRef] [Green Version]
- Moon, H.G.; Yang, J.; Zheng, Y.; Jin, Y. miR-15a/16 regulates macrophage phagocytosis after bacterial infection. J. Immunol. 2014, 193, 4558–4567. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Pan, C.; Li, L.; Bian, Z.; Lv, Z.; Shi, L.; Zhang, J.; Li, D.; Gu, H.; Zhang, C.Y.; et al. MicroRNA-17/20a/106a modulate macrophage inflammatory responses through targeting signal-regulatory protein α. J. Allergy Clin. Immunol. 2013, 132, 426–436.e428. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Liu, X.; Wang, W.; Cai, Y.; Li, S.; Chen, Q.; Liao, M.; Zhang, M.; Zeng, G.; Zhou, B.; et al. Down-regulation of miR-20a-5p triggers cell apoptosis to facilitate mycobacterial clearance through targeting JNK2 in human macrophages. Cell Cycle 2016, 15, 2527–2538. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Yi, Z.; Fu, Y. Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by Mcl-1 upregulation. J. Cell. Biochem. 2019, 120, 5889–5896. [Google Scholar] [CrossRef]
- Madhyastha, R.; Madhyastha, H.; Nurrahmah, Q.I.; Purbasari, B.; Maruyama, M.; Nakajima, Y. MicroRNA 21 Elicits a Pro-inflammatory Response in Macrophages, with Exosomes Functioning as Delivery Vehicles. Inflammation 2021, 44, 1274–1287. [Google Scholar] [CrossRef]
- Wang, Z.; Brandt, S.; Medeiros, A.; Wang, S.; Wu, H.; Dent, A.; Serezani, C.H. MicroRNA 21 is a homeostatic regulator of macrophage polarization and prevents prostaglandin E2-mediated M2 generation. PLoS ONE 2015, 10, e0115855. [Google Scholar] [CrossRef]
- Johnston, D.G.W.; Kearney, J.; Zasłona, Z.; Williams, M.A.; O’Neill, L.A.J.; Corr, S.C. MicroRNA-21 Limits Uptake of Listeria monocytogenes by Macrophages to Reduce the Intracellular Niche and Control Infection. Front. Cell. Infect. Microbiol. 2017, 7, 201. [Google Scholar] [CrossRef]
- Sheedy, F.J.; Palsson-McDermott, E.; Hennessy, E.J.; Martin, C.; O’Leary, J.J.; Ruan, Q.; Johnson, D.S.; Chen, Y.; O’Neill, L.A. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 2010, 11, 141–147. [Google Scholar] [CrossRef]
- Das, A.; Ganesh, K.; Khanna, S.; Sen, C.K.; Roy, S. Engulfment of apoptotic cells by macrophages: A role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 2014, 192, 1120–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.C.; Lee, C.P.; Hsiao, C.C.; Hsu, P.Y.; Wang, T.Y.; Wu, C.C.; Chao, T.Y.; Leung, S.Y.; Chang, Y.P.; Lin, M.C. MicroRNA-23a-3p Down-Regulation in Active Pulmonary Tuberculosis Patients with High Bacterial Burden Inhibits Mononuclear Cell Function and Phagocytosis through TLR4/TNF-α/TGF-β1/IL-10 Signaling via Targeting IRF1/SP1. Int. J. Mol. Sci. 2020, 21, 8587. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.R.; Fordham, J.B.; Nares, S. MicroRNA target Fc receptors to regulate Ab-dependent Ag uptake in primary macrophages and dendritic cells. Innate Immun. 2016, 22, 510–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fordham, J.B.; Naqvi, A.R.; Nares, S. miR-24 Regulates Macrophage Polarization and Plasticity. J. Clin. Cell. Immunol. 2015, 6, 362. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, A.R.; Fordham, J.B.; Nares, S. miR-24, miR-30b, and miR-142-3p regulate phagocytosis in myeloid inflammatory cells. J. Immunol. 2015, 194, 1916–1927. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.K.; Kumar, M.; Chakraborty, S.; Banerjee, S.K.; Kumar, R.; Gupta, P.; Jana, K.; Gupta, U.D.; Ghosh, Z.; Kundu, M.; et al. MicroRNA 26a (miR-26a)/KLF4 and CREB-C/EBPβ regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 2017, 13, e1006410. [Google Scholar] [CrossRef]
- de Couto, G.; Jaghatspanyan, E.; DeBerge, M.; Liu, W.; Luther, K.; Wang, Y.; Tang, J.; Thorp, E.B.; Marbán, E. Mechanism of Enhanced MerTK-Dependent Macrophage Efferocytosis by Extracellular Vesicles. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 2082–2096. [Google Scholar] [CrossRef]
- Saha, B.; Momen-Heravi, F.; Kodys, K.; Szabo, G. MicroRNA Cargo of Extracellular Vesicles from Alcohol-exposed Monocytes Signals Naive Monocytes to Differentiate into M2 Macrophages. J. Biol. Chem. 2016, 291, 149–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valverde, A.; Nares, S.; Naqvi, A.R. Impaired cell migration and structural defects in myeloid cells overexpressing miR-30b and miR-142-3p. Biochim. Et Biophys. Acta. Gene Regul. Mech. 2020, 1863, 194628. [Google Scholar] [CrossRef]
- Hu, Z.Q.; Rao, C.L.; Tang, M.L.; Zhang, Y.; Lu, X.X.; Chen, J.G.; Mao, C.; Deng, L.; Li, Q.; Mao, X.H. Rab32 GTPase, as a direct target of miR-30b/c, controls the intracellular survival of Burkholderia pseudomallei by regulating phagosome maturation. PLoS Pathog. 2019, 15, e1007879. [Google Scholar] [CrossRef]
- Fernandes, J.C.R.; Aoki, J.I.; Maia Acuña, S.; Zampieri, R.A.; Markus, R.P.; Floeter-Winter, L.M.; Muxel, S.M. Melatonin and Leishmania amazonensis Infection Altered miR-294, miR-30e, and miR-302d Impacting on Tnf, Mcp-1, and Nos2 Expression. Front. Cell. Infect. Microbiol. 2019, 9, 60. [Google Scholar] [CrossRef]
- Ouimet, M.; Ediriweera, H.N.; Gundra, U.M.; Sheedy, F.J.; Ramkhelawon, B.; Hutchison, S.B.; Rinehold, K.; van Solingen, C.; Fullerton, M.D.; Cecchini, K.; et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Investig. 2015, 125, 4334–4348. [Google Scholar] [CrossRef] [Green Version]
- McCubbrey, A.L.; Nelson, J.D.; Stolberg, V.R.; Blakely, P.K.; McCloskey, L.; Janssen, W.J.; Freeman, C.M.; Curtis, J.L. MicroRNA-34a Negatively Regulates Efferocytosis by Tissue Macrophages in Part via SIRT1. J. Immunol. 2016, 196, 1366–1375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lai, L.; Song, Y.; Liu, Y.; Chen, Q.; Han, Q.; Chen, W.; Pan, T.; Zhang, Y.; Cao, X.; Wang, Q. MicroRNA-92a negatively regulates Toll-like receptor (TLR)-triggered inflammatory response in macrophages by targeting MKK4 kinase. J. Biol. Chem. 2013, 288, 7956–7967. [Google Scholar] [CrossRef] [Green Version]
- Tserel, L.; Runnel, T.; Kisand, K.; Pihlap, M.; Bakhoff, L.; Kolde, R.; Peterson, H.; Vilo, J.; Peterson, P.; Rebane, A. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J. Biol. Chem. 2011, 286, 26487–26495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pires, D.; Bernard, E.M.; Pombo, J.P.; Carmo, N.; Fialho, C.; Gutierrez, M.G.; Bettencourt, P.; Anes, E. Mycobacterium tuberculosis Modulates miR-106b-5p to Control Cathepsin S Expression Resulting in Higher Pathogen Survival and Poor T-Cell Activation. Front. Immunol. 2017, 8, 1819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herdoiza Padilla, E.; Crauwels, P.; Bergner, T.; Wiederspohn, N.; Förstner, S.; Rinas, R.; Ruf, A.; Kleemann, M.; Handrick, R.; Tuckermann, J.; et al. mir-124-5p Regulates Phagocytosis of Human Macrophages by Targeting the Actin Cytoskeleton via the ARP2/3 Complex. Front. Immunol. 2019, 10, 2210. [Google Scholar] [CrossRef]
- Banerjee, S.; Cui, H.; Xie, N.; Tan, Z.; Yang, S.; Icyuz, M.; Thannickal, V.J.; Abraham, E.; Liu, G. miR-125a-5p regulates differential activation of macrophages and inflammation. J. Biol. Chem. 2013, 288, 35428–35436. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Saucedo, D.; Ruíz-Rosado, J.D.; Terrazas, C.; Callejas, B.E.; Satoskar, A.R.; Partida-Sánchez, S.; Terrazas, L.I. Taenia crassiceps-Excreted/Secreted Products Induce a Defined MicroRNA Profile that Modulates Inflammatory Properties of Macrophages. J. Immunol. Res. 2019, 2019, 2946713. [Google Scholar] [CrossRef]
- Huleihel, L.; Bartolacci, J.G.; Dziki, J.L.; Vorobyov, T.; Arnold, B.; Scarritt, M.E.; Pineda Molina, C.; LoPresti, S.T.; Brown, B.N.; Naranjo, J.D.; et al. Matrix-Bound Nanovesicles Recapitulate Extracellular Matrix Effects on Macrophage Phenotype. Tissue Eng. Part A 2017, 23, 1283–1294. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.A.; et al. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef]
- Rajaram, M.V.; Ni, B.; Morris, J.D.; Brooks, M.N.; Carlson, T.K.; Bakthavachalu, B.; Schoenberg, D.R.; Torrelles, J.B.; Schlesinger, L.S. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. USA 2011, 108, 17408–17413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaudhuri, A.A.; So, A.Y.; Sinha, N.; Gibson, W.S.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. MicroRNA-125b potentiates macrophage activation. J. Immunol. 2011, 187, 5062–5068. [Google Scholar] [CrossRef]
- Xi, Q.; Chen, Y.; Yang, G.Z.; Zhang, J.Y.; Zhang, L.J.; Guo, X.D.; Zhao, J.Y.; Xue, Z.Y.; Li, Y.; Zhang, R. miR-128 Regulates Tumor Cell CD47 Expression and Promotes Anti-tumor Immunity in Pancreatic Cancer. Front. Immunol. 2020, 11, 890. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, P.; Marion, S.; Pires, D.; Santos, L.F.; Lastrucci, C.; Carmo, N.; Blake, J.; Benes, V.; Griffiths, G.; Neyrolles, O.; et al. Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: The case of N-Wasp and miR-142-3p. Front. Cell. Infect. Microbiol. 2013, 3, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Murray, S.C.; Staitieh, B.S.; Spearman, P.; Guidot, D.M. HIV Impairs Alveolar Macrophage Function via MicroRNA-144-Induced Suppression of Nrf2. Am. J. Med. Sci. 2021, 361, 90–97. [Google Scholar] [CrossRef]
- Huang, Y.; Du, K.L.; Guo, P.Y.; Zhao, R.M.; Wang, B.; Zhao, X.L.; Zhang, C.Q. IL-16 regulates macrophage polarization as a target gene of mir-145-3p. Mol. Immunol. 2019, 107, 1–9. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, A.; Xia, C.; Lin, Q.; Chen, C. A single nucleotide polymorphism in primary-microRNA-146a reduces the expression of mature microRNA-146a in patients with Alzheimer’s disease and is associated with the pathogenesis of Alzheimer’s disease. Mol. Med. Rep. 2015, 12, 4037–4042. [Google Scholar] [CrossRef] [Green Version]
- Yang, K.; He, Y.S.; Wang, X.Q.; Lu, L.; Chen, Q.J.; Liu, J.; Sun, Z.; Shen, W.F. MiR-146a inhibits oxidized low-density lipoprotein-induced lipid accumulation and inflammatory response via targeting toll-like receptor 4. FEBS Lett. 2011, 585, 854–860. [Google Scholar] [CrossRef] [Green Version]
- Lochhead, R.B.; Ma, Y.; Zachary, J.F.; Baltimore, D.; Zhao, J.L.; Weis, J.H.; O’Connell, R.M.; Weis, J.J. MicroRNA-146a provides feedback regulation of lyme arthritis but not carditis during infection with Borrelia burgdorferi. PLoS Pathog. 2014, 10, e1004212. [Google Scholar] [CrossRef]
- Peng, L.; Zhang, H.; Hao, Y.; Xu, F.; Yang, J.; Zhang, R.; Lu, G.; Zheng, Z.; Cui, M.; Qi, C.F.; et al. Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine 2016, 14, 83–96. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Liu, X.J.; Zhou, Q.; Xie, J.; Ma, T.T.; Meng, X.M.; Li, J. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int. Immunopharmacol. 2016, 32, 46–54. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Mukherjee, S.; Ali, N. Super enhancer-mediated transcription of miR146a-5p drives M2 polarization during Leishmania donovani infection. PLoS Pathog. 2021, 17, e1009343. [Google Scholar] [CrossRef] [PubMed]
- Pauley, K.M.; Stewart, C.M.; Gauna, A.E.; Dupre, L.C.; Kuklani, R.; Chan, A.L.; Pauley, B.A.; Reeves, W.H.; Chan, E.K.; Cha, S. Altered miR-146a expression in Sjögren’s syndrome and its functional role in innate immunity. Eur. J. Immunol. 2011, 41, 2029–2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Yan, H.; Li, Z.; Jing, T.; Zhu, W.; Ge, J.; Zheng, X.; Pan, X.; Yan, H.; Zhu, J. MicroRNA-155 regulates lipid uptake, adhesion/chemokine marker secretion and SCG2 expression in oxLDL-stimulated dendritic cells/macrophages. Int. J. Cardiol. 2011, 147, 446–447. [Google Scholar] [CrossRef] [PubMed]
- Jia, C.; Chen, H.; Wei, M.; Chen, X.; Zhang, Y.; Cao, L.; Yuan, P.; Wang, F.; Yang, G.; Ma, J. Gold nanoparticle-based miR155 antagonist macrophage delivery restores the cardiac function in ovariectomized diabetic mouse model. Int. J. Nanomed. 2017, 12, 4963–4979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, L.; Cai, S.Y.; Sun, J.; Chen, J. MicroRNA-155 promotes pro-inflammatory functions and augments apoptosis of monocytes/macrophages during Vibrio anguillarum infection in ayu, Plecoglossus altivelis. Fish Shellfish. Immunol. 2019, 86, 70–81. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yao, H.; Zhang, H.; Lan, K.; Wang, H.; Su, Y.; Li, D.; Song, Z.; Cui, F.; Yin, Y.; Zhang, X. Purified Streptococcus pneumoniae Endopeptidase O (PepO) Enhances Particle Uptake by Macrophages in a Toll-Like Receptor 2- and miR-155-Dependent Manner. Infect. Immun. 2017, 85, e01012-16. [Google Scholar] [CrossRef] [Green Version]
- Nazari-Jahantigh, M.; Wei, Y.; Noels, H.; Akhtar, S.; Zhou, Z.; Koenen, R.R.; Heyll, K.; Gremse, F.; Kiessling, F.; Grommes, J.; et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Investig. 2012, 122, 4190–4202. [Google Scholar] [CrossRef] [Green Version]
- Srinoun, K.; Nopparatana, C.; Wongchanchailert, M.; Fucharoen, S. MiR-155 enhances phagocytic activity of β-thalassemia/HbE monocytes via targeting of BACH1. Int. J. Hematol. 2017, 106, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, K.; Zhou, L.; Minhaowu; Wu, Y.; Zhu, M.; Lai, X.; Chen, T.; Feng, L.; Li, M.; et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 2013, 9, e1003697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Wu, M.; Li, M.; Li, D.; Peng, A.; Nie, X.; Sun, M.; Wang, J.; Wu, Y.; Deng, Q.; et al. miR-155 suppresses bacterial clearance in Pseudomonas aeruginosa-induced keratitis by targeting Rheb. J. Infect. Dis. 2014, 210, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Zeng, X.; Zhao, L.; Wei, Q.; Yu, L.; Wang, X.; Yu, Z.; Cao, Y.; Shan, F.; Wei, M. miR-181a Induces Macrophage Polarized to M2 Phenotype and Promotes M2 Macrophage-mediated Tumor Cell Metastasis by Targeting KLF6 and C/EBPα. Mol. Therapy. Nucleic Acids 2016, 5, e368. [Google Scholar] [CrossRef] [Green Version]
- Pierdomenico, A.M.; Recchiuti, A.; Simiele, F.; Codagnone, M.; Mari, V.C.; Davì, G.; Romano, M. MicroRNA-181b regulates ALX/FPR2 receptor expression and proresolution signaling in human macrophages. J. Biol. Chem. 2015, 290, 3592–3600. [Google Scholar] [CrossRef] [Green Version]
- Pierdomenico, A.M.; Patruno, S.; Codagnone, M.; Simiele, F.; Mari, V.C.; Plebani, R.; Recchiuti, A.; Romano, M. microRNA-181b is increased in cystic fibrosis cells and impairs lipoxin A(4) receptor-dependent mechanisms of inflammation resolution and antimicrobial defense. Sci. Rep. 2017, 7, 13519. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Q.; Xi, X.; Jiao, J.; Xu, W.; Huang, J.; Lai, Z. High serum miR-183 level is associated with the bioactivity of macrophage derived from tuberculosis patients. Int. J. Clin. Exp. Pathol. 2015, 8, 655–659. [Google Scholar]
- Muraleedharan, C.K.; McClellan, S.A.; Barrett, R.P.; Li, C.; Montenegro, D.; Carion, T.; Berger, E.; Hazlett, L.D.; Xu, S. Inactivation of the miR-183/96/182 Cluster Decreases the Severity of Pseudomonas aeruginosa-Induced Keratitis. Investig. Ophthalmol. Vis. Sci. 2016, 57, 1506–1517. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Zhao, Z.; Chou, F.; Zuo, L.; Liu, T.; Yeh, S.; Bushinsky, D.; Zeng, G.; Chang, C. Loss of the androgen receptor suppresses intrarenal calcium oxalate crystals deposition via altering macrophage recruitment/M2 polarization with change of the miR-185-5p/CSF-1 signals. Cell Death Dis. 2019, 10, 275. [Google Scholar] [CrossRef]
- Zhao, Y.; Yu, X.; Tang, H.; Han, R.; Wang, X.; Wang, J.; Wang, K.; Li, G. MicroRNA-200a Promotes Phagocytosis of Macrophages and Suppresses Cell Proliferation, Migration, and Invasion in Nasopharyngeal Carcinoma by Targeting CD47. BioMed Res. Int. 2020, 2020, 3723781. [Google Scholar] [CrossRef] [Green Version]
- Zhuang, G.; Meng, C.; Guo, X.; Cheruku, P.S.; Shi, L.; Xu, H.; Li, H.; Wang, G.; Evans, A.R.; Safe, S.; et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012, 125, 2892–2903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, X.; Zhang, C.; Han, W.; Zhao, H.; Zhang, H.; Jiao, J. MicroRNA-223 Is Upregulated in Active Tuberculosis Patients and Inhibits Apoptosis of Macrophages by Targeting FOXO3. Genet. Test. Mol. Biomark. 2015, 19, 650–656. [Google Scholar] [CrossRef] [PubMed]
- Tay, H.L.; Kaiko, G.E.; Plank, M.; Li, J.; Maltby, S.; Essilfie, A.T.; Jarnicki, A.; Yang, M.; Mattes, J.; Hansbro, P.M.; et al. Antagonism of miR-328 increases the antimicrobial function of macrophages and neutrophils and rapid clearance of non-typeable Haemophilus influenzae (NTHi) from infected lung. PLoS Pathog. 2015, 11, e1004549. [Google Scholar] [CrossRef]
- Xi, Q.; Zhang, J.; Yang, G.; Zhang, L.; Chen, Y.; Wang, C.; Zhang, Z.; Guo, X.; Zhao, J.; Xue, Z.; et al. Restoration of miR-340 controls pancreatic cancer cell CD47 expression to promote macrophage phagocytosis and enhance antitumor immunity. J. Immunother. Cancer 2020, 8, e000253. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, X.; Wang, J.; Song, N.; Zhu, A.; Jia, L. miR-378a Modulates Macrophage Phagocytosis and Differentiation through Targeting CD47-SIRPα Axis in Atherosclerosis. Scand. J. Immunol. 2019, 90, e12766. [Google Scholar] [CrossRef]
- Shi, M.M.; Zhu, Y.G.; Yan, J.Y.; Rouby, J.J.; Summah, H.; Monsel, A.; Qu, J.M. Role of miR-466 in mesenchymal stromal cell derived extracellular vesicles treating inoculation pneumonia caused by multidrug-resistant Pseudomonas aeruginosa. Clin. Transl. Med. 2021, 11, e287. [Google Scholar] [CrossRef]
- Liu, Y.; Jiang, J.; Wang, X.; Zhai, F.; Cheng, X. miR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS ONE 2013, 8, e78381. [Google Scholar] [CrossRef] [Green Version]
- He, P.P.; OuYang, X.P.; Li, Y.; Lv, Y.C.; Wang, Z.B.; Yao, F.; Xie, W.; Tan, Y.L.; Li, L.; Zhang, M.; et al. MicroRNA-590 Inhibits Lipoprotein Lipase Expression and Prevents Atherosclerosis in apoE Knockout Mice. PLoS ONE 2015, 10, e0138788. [Google Scholar] [CrossRef]
- Jiang, A.; Zhang, S.; Li, Z.; Liang, R.; Ren, S.; Li, J.; Pu, Y.; Yang, J. miR-615-3p promotes the phagocytic capacity of splenic macrophages by targeting ligand-dependent nuclear receptor corepressor in cirrhosis-related portal hypertension. Exp. Biol. Med. 2011, 236, 672–680. [Google Scholar] [CrossRef]
- Tan, W.; Tang, H.; Jiang, X.; Ye, F.; Huang, L.; Shi, D.; Li, L.; Huang, X.; Li, L.; Xie, X.; et al. Metformin mediates induction of miR-708 to inhibit self-renewal and chemoresistance of breast cancer stem cells through targeting CD47. J. Cell. Mol. Med. 2019, 23, 5994–6004. [Google Scholar] [CrossRef]
- Li, W.T.; Zhang, Q. MicroRNA-708-5p regulates mycobacterial vitality and the secretion of inflammatory factors in Mycobacterium tuberculosis-infected macrophages by targeting TLR4. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8028–8038. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.; Wang, S.; Guo, X.; Wang, J.; Zhang, Z.; Qiu, W.; Gao, X.; Chen, Z.; Xu, J.; Zhao, R.; et al. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene 2020, 39, 428–442. [Google Scholar] [CrossRef] [PubMed]
- Pagliari, M.; Munari, F.; Toffoletto, M.; Lonardi, S.; Chemello, F.; Codolo, G.; Millino, C.; Della Bella, C.; Pacchioni, B.; Vermi, W.; et al. Helicobacter pylori Affects the Antigen Presentation Activity of Macrophages Modulating the Expression of the Immune Receptor CD300E through miR-4270. Front. Immunol. 2017, 8, 1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Chen, M.T.; Zhang, X.H.; Yin, X.L.; Ning, H.M.; Su, R.; Lin, H.S.; Song, L.; Wang, F.; Ma, Y.N.; et al. The PU.1-Modulated MicroRNA-22 Is a Regulator of Monocyte/Macrophage Differentiation and Acute Myeloid Leukemia. PLoS Genet. 2016, 12, e1006259. [Google Scholar] [CrossRef]
- Pospisil, V.; Vargova, K.; Kokavec, J.; Rybarova, J.; Savvulidi, F.; Jonasova, A.; Necas, E.; Zavadil, J.; Laslo, P.; Stopka, T. Epigenetic silencing of the oncogenic miR-17-92 cluster during PU.1-directed macrophage differentiation. EMBO J. 2011, 30, 4450–4464. [Google Scholar] [CrossRef]
- Rosa, A.; Ballarino, M.; Sorrentino, A.; Sthandier, O.; De Angelis, F.G.; Marchioni, M.; Masella, B.; Guarini, A.; Fatica, A.; Peschle, C.; et al. The interplay between the master transcription factor PU.1 and miR-424 regulates human monocyte/macrophage differentiation. Proc. Natl. Acad. Sci. USA 2007, 104, 19849–19854. [Google Scholar] [CrossRef] [Green Version]
- Kohno, K.; Koya-Miyata, S.; Harashima, A.; Tsukuda, T.; Katakami, M.; Ariyasu, T.; Ushio, S.; Iwaki, K. Inflammatory M1-like macrophages polarized by NK-4 undergo enhanced phenotypic switching to an anti-inflammatory M2-like phenotype upon co-culture with apoptotic cells. J. Inflamm. 2021, 18, 2. [Google Scholar] [CrossRef]
- Murray, P.J.; Smale, S.T. Restraint of inflammatory signaling by interdependent strata of negative regulatory pathways. Nat. Immunol. 2012, 13, 916–924. [Google Scholar] [CrossRef]
- Wang, Y.; Han, C.C.; Cui, D.; Li, Y.; Ma, Y.; Wei, W. Is macrophage polarization important in rheumatoid arthritis? Int. Immunopharmacol. 2017, 50, 345–352. [Google Scholar] [CrossRef]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 79, 541–566. [Google Scholar] [CrossRef] [PubMed]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef] [PubMed]
- Rőszer, T. Understanding the Mysterious M2 Macrophage through Activation Markers and Effector Mechanisms. Mediat. Inflamm. 2015, 2015, 816460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neaga, A.; Bagacean, C.; Tempescul, A.; Jimbu, L.; Mesaros, O.; Blag, C.; Tomuleasa, C.; Bocsan, C.; Gaman, M.; Zdrenghea, M. MicroRNAs Associated With a Good Prognosis of Acute Myeloid Leukemia and Their Effect on Macrophage Polarization. Front. Immunol. 2020, 11, 582915. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.W.; Dickson, A.M.; Clay, G.; McCaffrey, A.P.; Wilson, M.E. Identifying functional microRNAs in macrophages with polarized phenotypes. J. Biol. Chem. 2012, 287, 21816–21825. [Google Scholar] [CrossRef] [Green Version]
- Cobos Jiménez, V.; Bradley, E.J.; Willemsen, A.M.; van Kampen, A.H.; Baas, F.; Kootstra, N.A. Next-generation sequencing of microRNAs uncovers expression signatures in polarized macrophages. Physiol. Genom. 2014, 46, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Ying, W.; Tseng, A.; Chang, R.C.; Morin, A.; Brehm, T.; Triff, K.; Nair, V.; Zhuang, G.; Song, H.; Kanameni, S.; et al. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Investig. 2015, 125, 4149–4159. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Ge, B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett. 2018, 431, 22–30. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H.; Liu, Y.; Wang, K.; Feng, Y.; Liu, M.; Xiao, X. KLF4 regulates the expression of interleukin-10 in RAW264.7 macrophages. Biochem. Biophys. Res. Commun. 2007, 362, 575–581. [Google Scholar] [CrossRef]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [CrossRef]
- Yang, J.; Wise, L.; Fukuchi, K.I. TLR4 Cross-Talk With NLRP3 Inflammasome and Complement Signaling Pathways in Alzheimer’s Disease. Front. Immunol. 2020, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Takeda, K.; Akira, S. Toll-like receptors in innate immunity. Int. Immunol. 2005, 17, 1–14. [Google Scholar] [CrossRef]
- Lee, J.W.; Nam, H.; Kim, L.E.; Jeon, Y.; Min, H.; Ha, S.; Lee, Y.; Kim, S.Y.; Lee, S.J.; Kim, E.K.; et al. TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy 2019, 15, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Madadi, S.; Schwarzenbach, H.; Saidijam, M.; Mahjub, R.; Soleimani, M. Potential microRNA-related targets in clearance pathways of amyloid-β: Novel therapeutic approach for the treatment of Alzheimer’s disease. Cell Biosci. 2019, 9, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Li, Y.J.; Wu, X.Y.; Hong, Z.; Wei, W.S. MicroRNA-181c negatively regulates the inflammatory response in oxygen-glucose-deprived microglia by targeting Toll-like receptor 4. J. Neurochem. 2015, 132, 713–723. [Google Scholar] [CrossRef]
- Guha, M.; Mackman, N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J. Biol. Chem. 2002, 277, 32124–32132. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Hou, J.; Ma, F.; Wang, P.; Liu, X.; Li, N.; Wang, J.; Wang, Q.; Cao, X. Type I IFN inhibits innate IL-10 production in macrophages through histone deacetylase 11 by downregulating microRNA-145. J. Immunol. 2013, 191, 3896–3904. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Friggeri, A.; Yang, Y.; Park, Y.J.; Tsuruta, Y.; Abraham, E. miR-147, a microRNA that is induced upon Toll-like receptor stimulation, regulates murine macrophage inflammatory responses. Proc. Natl. Acad. Sci. USA 2009, 106, 15819–15824. [Google Scholar] [CrossRef] [Green Version]
- Ghorpade, D.S.; Leyland, R.; Kurowska-Stolarska, M.; Patil, S.A.; Balaji, K.N. MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol. Cell. Biol. 2012, 32, 2239–2253. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Hou, J.; Lin, L.; Wang, C.; Liu, X.; Li, D.; Ma, F.; Wang, Z.; Cao, X. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010, 185, 6226–6233. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.Q.; Li, L.M.; Guo, Y.L.; Bai, R.; Wang, C.; Bian, Z.; Zhang, C.Y.; Zen, K. Signal regulatory protein alpha negatively regulates beta2 integrin-mediated monocyte adhesion, transendothelial migration and phagocytosis. PLoS ONE 2008, 3, e3291. [Google Scholar] [CrossRef] [Green Version]
- Bogdan, C. Nitric oxide and the immune response. Nat. Immunol. 2001, 2, 907–916. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Chen, Y.; Xu, Y.; Tang, M.; Zhang, X. Morphine contributed to the deterioration of cancer via miR-543/MARCKS/FcγR-mediated phagocytosis pathway. J. Pharm. Pharmacol. 2019, 71, 1584–1598. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, J.; Wang, L.; Du, F.; Zhao, J.; Fang, R. Expression profile of microRNAs in porcine alveolar macrophages after Toxoplasma gondii infection. Parasites Vectors 2019, 12, 65. [Google Scholar] [CrossRef] [Green Version]
- He, J.; Gao, Y.; Wu, G.; Lei, X.; Zhang, Y.; Pan, W.; Yu, H. Bioinformatics analysis of microarray data to reveal the pathogenesis of brain ischemia. Mol. Med. Rep. 2018, 18, 333–341. [Google Scholar] [CrossRef]
- El Kebir, D.; Filep, J.G. Modulation of Neutrophil Apoptosis and the Resolution of Inflammation through β2 Integrins. Front. Immunol. 2013, 4, 60. [Google Scholar] [CrossRef] [Green Version]
- Seimon, T.; Tabas, I. Mechanisms and consequences of macrophage apoptosis in atherosclerosis. J. Lipid Res. 2009, 50, S382–S387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suresh Babu, S.; Thandavarayan, R.A.; Joladarashi, D.; Jeyabal, P.; Krishnamurthy, S.; Bhimaraj, A.; Youker, K.A.; Krishnamurthy, P. MicroRNA-126 overexpression rescues diabetes-induced impairment in efferocytosis of apoptotic cardiomyocytes. Sci. Rep. 2016, 6, 36207. [Google Scholar] [CrossRef] [Green Version]
- Boldin, M.P.; Taganov, K.D.; Rao, D.S.; Yang, L.; Zhao, J.L.; Kalwani, M.; Garcia-Flores, Y.; Luong, M.; Devrekanli, A.; Xu, J.; et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011, 208, 1189–1201. [Google Scholar] [CrossRef]
- Zhao, J.L.; Rao, D.S.; Boldin, M.P.; Taganov, K.D.; O’Connell, R.M.; Baltimore, D. NF-kappaB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA 2011, 108, 9184–9189. [Google Scholar] [CrossRef] [Green Version]
- Nahid, M.A.; Pauley, K.M.; Satoh, M.; Chan, E.K. miR-146a is critical for endotoxin-induced tolerance: IMPLICATION IN INNATE IMMUNITY. J. Biol. Chem. 2009, 284, 34590–34599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, J.; Wang, P.; Lin, L.; Liu, X.; Ma, F.; An, H.; Wang, Z.; Cao, X. MicroRNA-146a feedback inhibits RIG-I-dependent Type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 2009, 183, 2150–2158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossato, M.; Curtale, G.; Tamassia, N.; Castellucci, M.; Mori, L.; Gasperini, S.; Mariotti, B.; De Luca, M.; Mirolo, M.; Cassatella, M.A.; et al. IL-10-induced microRNA-187 negatively regulates TNF-α, IL-6, and IL-12p40 production in TLR4-stimulated monocytes. Proc. Natl. Acad. Sci. USA 2012, 109, E3101–E3110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Dalli, J.; Chiang, N.; Baron, R.M.; Quintana, C.; Serhan, C.N. Plasticity of leukocytic exudates in resolving acute inflammation is regulated by MicroRNA and proresolving mediators. Immunity 2013, 39, 885–898. [Google Scholar] [CrossRef] [Green Version]
- Colonna, M.; Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu. Rev. Immunol. 2017, 35, 441–468. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Moore, C.S.; Cialic, R.; Lanser, A.J.; Gabriely, G.; Koeglsperger, T.; Dake, B.; Wu, P.M.; Doykan, C.E.; et al. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014, 17, 131–143. [Google Scholar] [CrossRef] [Green Version]
- Freilich, R.W.; Woodbury, M.E.; Ikezu, T. Integrated expression profiles of mRNA and miRNA in polarized primary murine microglia. PLoS ONE 2013, 8, e79416. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Han, Z.; Hu, T.; Zhang, S.; Ge, X.; Huang, S.; Wang, L.; Yu, J.; Li, W.; Wang, Y.; et al. Neuron-derived exosomes with high miR-21-5p expression promoted polarization of M1 microglia in culture. Brain Behav. Immun. 2020, 83, 270–282. [Google Scholar] [CrossRef]
- Bhattacharjee, S.; Zhao, Y.; Dua, P.; Rogaev, E.I.; Lukiw, W.J. microRNA-34a-Mediated Down-Regulation of the Microglial-Enriched Triggering Receptor and Phagocytosis-Sensor TREM2 in Age-Related Macular Degeneration. PLoS ONE 2016, 11, e0150211. [Google Scholar] [CrossRef]
- Su, W.; Hopkins, S.; Nesser, N.K.; Sopher, B.; Silvestroni, A.; Ammanuel, S.; Jayadev, S.; Möller, T.; Weinstein, J.; Garden, G.A. The p53 transcription factor modulates microglia behavior through microRNA-dependent regulation of c-Maf. J. Immunol. 2014, 192, 358–366. [Google Scholar] [CrossRef]
- Yang, J.; Cao, L.L.; Wang, X.P.; Guo, W.; Guo, R.B.; Sun, Y.Q.; Xue, T.F.; Cai, Z.Y.; Ji, J.; Cheng, H.; et al. Neuronal extracellular vesicle derived miR-98 prevents salvageable neurons from microglial phagocytosis in acute ischemic stroke. Cell Death Dis. 2021, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Ponomarev, E.D.; Veremeyko, T.; Barteneva, N.; Krichevsky, A.M.; Weiner, H.L. MicroRNA-124 promotes microglia quiescence and suppresses EAE by deactivating macrophages via the C/EBP-α-PU.1 pathway. Nat. Med. 2011, 17, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Yip, P.K.; Bowes, A.L.; Hall, J.C.E.; Burguillos, M.A.; Ip, T.H.R.; Baskerville, T.; Liu, Z.H.; Mohamed, M.; Getachew, F.; Lindsay, A.D.; et al. Docosahexaenoic acid reduces microglia phagocytic activity via miR-124 and induces neuroprotection in rodent models of spinal cord contusion injury. Hum. Mol. Genet. 2019, 28, 2427–2448. [Google Scholar] [CrossRef] [PubMed]
- Svahn, A.J.; Giacomotto, J.; Graeber, M.B.; Rinkwitz, S.; Becker, T.S. miR-124 Contributes to the functional maturity of microglia. Dev. Neurobiol. 2016, 76, 507–518. [Google Scholar] [CrossRef]
- Talebi, F.; Ghorbani, S.; Chan, W.F.; Boghozian, R.; Masoumi, F.; Ghasemi, S.; Vojgani, M.; Power, C.; Noorbakhsh, F. MicroRNA-142 regulates inflammation and T cell differentiation in an animal model of multiple sclerosis. J. Neuroinflammation 2017, 14, 55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, B.; Yang, R.; Xu, B.; Fu, J.; Qu, X.; Li, L.; Dai, M.; Tan, C.; Chen, H.; Wang, X. miR-155 and miR-146a collectively regulate meningitic Escherichia coli infection-mediated neuroinflammatory responses. J. Neuroinflammation 2021, 18, 114. [Google Scholar] [CrossRef]
- Cardoso, A.L.; Guedes, J.R.; Pereira de Almeida, L.; Pedroso de Lima, M.C. miR-155 modulates microglia-mediated immune response by down-regulating SOCS-1 and promoting cytokine and nitric oxide production. Immunology 2012, 135, 73–88. [Google Scholar] [CrossRef]
- Butovsky, O.; Jedrychowski, M.P.; Cialic, R.; Krasemann, S.; Murugaiyan, G.; Fanek, Z.; Greco, D.J.; Wu, P.M.; Doykan, C.E.; Kiner, O.; et al. Targeting miR-155 restores abnormal microglia and attenuates disease in SOD1 mice. Ann. Neurol. 2015, 77, 75–99. [Google Scholar] [CrossRef]
- Aloi, M.S.; Prater, K.E.; Sopher, B.; Davidson, S.; Jayadev, S.; Garden, G.A. The pro-inflammatory microRNA miR-155 influences fibrillar β-Amyloid(1) (-42) catabolism by microglia. Glia 2021, 69, 1736–1748. [Google Scholar] [CrossRef]
- Galloway, D.A.; Blandford, S.N.; Berry, T.; Williams, J.B.; Stefanelli, M.; Ploughman, M.; Moore, C.S. miR-223 promotes regenerative myeloid cell phenotype and function in the demyelinated central nervous system. Glia 2019, 67, 857–869. [Google Scholar] [CrossRef]
- Bao, Y.; Zhu, Y.; He, G.; Ni, H.; Liu, C.; Ma, L.; Zhang, L.; Shi, D. Dexmedetomidine Attenuates Neuroinflammation In LPS-Stimulated BV2 Microglia Cells Through Upregulation Of miR-340. Drug Des. Dev. Ther. 2019, 13, 3465–3475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunha, C.; Gomes, C.; Vaz, A.R.; Brites, D. Exploring New Inflammatory Biomarkers and Pathways during LPS-Induced M1 Polarization. Mediat. Inflamm. 2016, 2016, 6986175. [Google Scholar] [CrossRef] [PubMed]
- Pareek, S.; Roy, S.; Kumari, B.; Jain, P.; Banerjee, A.; Vrati, S. MiR-155 induction in microglial cells suppresses Japanese encephalitis virus replication and negatively modulates innate immune responses. J. Neuroinflammation 2014, 11, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, S.; Zhang, H.W.; Lu, M.H.; He, X.H.; Li, Y.; Gu, H.; Liu, M.F.; Wang, E.D. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef] [Green Version]
- Worm, J.; Stenvang, J.; Petri, A.; Frederiksen, K.S.; Obad, S.; Elmén, J.; Hedtjärn, M.; Straarup, E.M.; Hansen, J.B.; Kauppinen, S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009, 37, 5784–5792. [Google Scholar] [CrossRef]
- Pena-Philippides, J.C.; Caballero-Garrido, E.; Lordkipanidze, T.; Roitbak, T. In vivo inhibition of miR-155 significantly alters post-stroke inflammatory response. J. Neuroinflammation 2016, 13, 287. [Google Scholar] [CrossRef] [Green Version]
- Caballero-Garrido, E.; Pena-Philippides, J.C.; Lordkipanidze, T.; Bragin, D.; Yang, Y.; Erhardt, E.B.; Roitbak, T. In Vivo Inhibition of miR-155 Promotes Recovery after Experimental Mouse Stroke. J. Neurosci. Off. J. Soc. Neurosci. 2015, 35, 12446–12464. [Google Scholar] [CrossRef] [Green Version]
- Moore, C.S.; Rao, V.T.; Durafourt, B.A.; Bedell, B.J.; Ludwin, S.K.; Bar-Or, A.; Antel, J.P. miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization. Ann. Neurol. 2013, 74, 709–720. [Google Scholar] [CrossRef]
- Junker, A.; Krumbholz, M.; Eisele, S.; Mohan, H.; Augstein, F.; Bittner, R.; Lassmann, H.; Wekerle, H.; Hohlfeld, R.; Meinl, E. MicroRNA profiling of multiple sclerosis lesions identifies modulators of the regulatory protein CD47. Brain A J. Neurol. 2009, 132, 3342–3352. [Google Scholar] [CrossRef] [Green Version]
- Murugaiyan, G.; Beynon, V.; Mittal, A.; Joller, N.; Weiner, H.L. Silencing microRNA-155 ameliorates experimental autoimmune encephalomyelitis. J. Immunol. 2011, 187, 2213–2221. [Google Scholar] [CrossRef] [Green Version]
- Lei, P.; Li, Y.; Chen, X.; Yang, S.; Zhang, J. Microarray based analysis of microRNA expression in rat cerebral cortex after traumatic brain injury. Brain Res. 2009, 1284, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, D.L.; Zuluaga-Ramirez, V.; Gajghate, S.; Reichenbach, N.L.; Polyak, B.; Persidsky, Y.; Rom, S. miR-98 reduces endothelial dysfunction by protecting blood-brain barrier (BBB) and improves neurological outcomes in mouse ischemia/reperfusion stroke model. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2020, 40, 1953–1965. [Google Scholar] [CrossRef]
- Rom, S.; Dykstra, H.; Zuluaga-Ramirez, V.; Reichenbach, N.L.; Persidsky, Y. miR-98 and let-7g* protect the blood-brain barrier under neuroinflammatory conditions. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2015, 35, 1957–1965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldeira, C.; Oliveira, A.F.; Cunha, C.; Vaz, A.R.; Falcão, A.S.; Fernandes, A.; Brites, D. Microglia change from a reactive to an age-like phenotype with the time in culture. Front. Cell. Neurosci. 2014, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.L.; King, V.R.; Curran, O.E.; Dyall, S.C.; Ward, R.E.; Lal, N.; Priestley, J.V.; Michael-Titus, A.T. A combination of intravenous and dietary docosahexaenoic acid significantly improves outcome after spinal cord injury. Brain A J. Neurol. 2007, 130, 3004–3019. [Google Scholar] [CrossRef] [Green Version]
- Keating, G.M. Dexmedetomidine: A Review of Its Use for Sedation in the Intensive Care Setting. Drugs 2015, 75, 1119–1130. [Google Scholar] [CrossRef]
- Wang, W.; Ferguson, D.J.; Quinn, J.M.; Simpson, A.H.; Athanasou, N.A. Osteoclasts are capable of particle phagocytosis and bone resorption. J. Pathol. 1997, 182, 92–98. [Google Scholar] [CrossRef]
- Nijweide, P.J.; Burger, E.H.; Feyen, J.H. Cells of bone: Proliferation, differentiation, and hormonal regulation. Physiol. Rev. 1986, 66, 855–886. [Google Scholar] [CrossRef]
- Teitelbaum, S.L. Bone resorption by osteoclasts. Science 2000, 289, 1504–1508. [Google Scholar] [CrossRef]
- Hu, C.H.; Sui, B.D.; Du, F.Y.; Shuai, Y.; Zheng, C.X.; Zhao, P.; Yu, X.R.; Jin, Y. miR-21 deficiency inhibits osteoclast function and prevents bone loss in mice. Sci. Rep. 2017, 7, 43191. [Google Scholar] [CrossRef] [Green Version]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.A.; Leupold, J.H.; Colburn, N.H.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liew, P.X.; Kubes, P. The Neutrophil’s Role During Health and Disease. Physiol. Rev. 2019, 99, 1223–1248. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Kim, S.E.; Yano, H.; Matsumoto, G.; Ohuchida, R.; Ishikura, Y.; Araki, M.; Araki, K.; Park, S.; Komatsu, T.; et al. MiR-142 Is Required for Staphylococcus aureus Clearance at Skin Wound Sites via Small GTPase-Mediated Regulation of the Neutrophil Actin Cytoskeleton. J. Investig. Dermatol. 2017, 137, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Du, S.W.; Palczewski, K. MicroRNA regulation of critical retinal pigment epithelial functions. Trends Neurosci. 2022, 45, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, J.; Zheng, L.; Wang, M.; Lu, Y.; Li, Z.; Lian, C.; Mao, S.; Hou, X.; Li, S.; et al. miR-25 Mediates Retinal Degeneration Via Inhibiting ITGAV and PEDF in Rat. Curr. Mol. Med. 2017, 17, 359–374. [Google Scholar] [CrossRef]
- Murad, N.; Kokkinaki, M.; Gunawardena, N.; Gunawan, M.S.; Hathout, Y.; Janczura, K.J.; Theos, A.C.; Golestaneh, N. miR-184 regulates ezrin, LAMP-1 expression, affects phagocytosis in human retinal pigment epithelium and is downregulated in age-related macular degeneration. FEBS J. 2014, 281, 5251–5264. [Google Scholar] [CrossRef]
- Cui, L.; Lyu, Y.; Jin, X.; Wang, Y.; Li, X.; Wang, J.; Zhang, J.; Deng, Z.; Yang, N.; Zheng, Z.; et al. miR-194 suppresses epithelial-mesenchymal transition of retinal pigment epithelial cells by directly targeting ZEB1. Ann. Transl. Med. 2019, 7, 751. [Google Scholar] [CrossRef]
- Zhang, C.; Miyagishima, K.J.; Dong, L.; Rising, A.; Nimmagadda, M.; Liang, G.; Sharma, R.; Dejene, R.; Wang, Y.; Abu-Asab, M.; et al. Regulation of phagolysosomal activity by miR-204 critically influences structure and function of retinal pigment epithelium/retina. Hum. Mol. Genet. 2019, 28, 3355–3368. [Google Scholar] [CrossRef] [Green Version]
- Xia, Z.; Liu, F.; Zhang, J.; Liu, L. Decreased Expression of MiRNA-204-5p Contributes to Glioma Progression and Promotes Glioma Cell Growth, Migration and Invasion. PLoS ONE 2015, 10, e0132399. [Google Scholar] [CrossRef] [Green Version]
- Naso, F.; Intartaglia, D.; Falanga, D.; Soldati, C.; Polishchuk, E.; Giamundo, G.; Tiberi, P.; Marrocco, E.; Scudieri, P.; Di Malta, C.; et al. Light-responsive microRNA miR-211 targets Ezrin to modulate lysosomal biogenesis and retinal cell clearance. EMBO J. 2020, 39, e102468. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, H.; Zhang, D.; Xie, S.; Wang, W.; Li, Q.; Lin, Z.; Wang, Y. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018, 9, 742. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Xie, P.; Sun, R.; Sun, X.; Liu, G.; Ding, S.; Zhu, M.; Yan, B.; Liu, Q.; Chen, X.; et al. c-Jun-mediated microRNA-302d-3p induces RPE dedifferentiation by targeting p21(Waf1/Cip1). Cell Death Dis. 2018, 9, 451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Jiang, C.; Sun, R.; Yang, D.; Liu, Q. Circular Noncoding RNA NR3C1 Acts as a miR-382-5p Sponge to Protect RPE Functions via Regulating PTEN/AKT/mTOR Signaling Pathway. Mol. Ther. J. Am. Soc. Gene Ther. 2020, 28, 929–945. [Google Scholar] [CrossRef]
- Choi, S.W.; Kim, J.J.; Seo, M.S.; Park, S.B.; Shin, T.H.; Shin, J.H.; Seo, Y.; Kim, H.S.; Kang, K.S. Inhibition by miR-410 facilitates direct retinal pigment epithelium differentiation of umbilical cord blood-derived mesenchymal stem cells. J. Vet. Sci. 2017, 18, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.; Lu, Q.; Wei, Y.; Han, L.; Ji, R.; Li, Q.; Lu, Q. Mertk deficiency alters expression of micrornas in the retinal pigment epithelium cells. Metab. Brain Dis. 2015, 30, 943–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordes, K.R.; Sheehy, N.T.; White, M.P.; Berry, E.C.; Morton, S.U.; Muth, A.N.; Lee, T.H.; Miano, J.M.; Ivey, K.N.; Srivastava, D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 2009, 460, 705–710. [Google Scholar] [CrossRef] [Green Version]
- Vengrenyuk, Y.; Nishi, H.; Long, X.; Ouimet, M.; Savji, N.; Martinez, F.O.; Cassella, C.P.; Moore, K.J.; Ramsey, S.A.; Miano, J.M.; et al. Cholesterol loading reprograms the microRNA-143/145-myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 535–546. [Google Scholar] [CrossRef] [Green Version]

| MicroRNA | Organism | Cell | Setting | Target | Effect | Ref. |
|---|---|---|---|---|---|---|
| let-7a-5p | human | monocytes | downregulated in macrophages compared to monocytes | WASL and VASP | enhanced phagocytosis | [107] |
| let-7b-5p | human | THP-1 | M. tuberculosis infection | FAS | inhibition of let-7b-5p augments apoptosis and pathogen clearance | [108] |
| let-7b-5p | human | monocytes | S. aureus infection | SOCS1/STAT | regulates M2 polarization | [109] |
| let-7c | mice | bone marrow-derived macrophages (M1 and M2) | bleomycin-induced pulmonary fibrosis | C/EBP-δ | let-7c promotes M2 polarization and stimulates phagocytosis of apoptotic cells, whereas its knock-out leads to M1 polarization | [110] |
| let-7e | mice | RAW264.7 | LPS stimulation | TLR4 | let-7e is upregulated upon LPS stimulation and targets TLR4 to modulate inflammatory response | [57] |
| let-7i-5p | human | monocytes | downregulated in macrophages compared to monocytes | WASL and VASP | enhanced phagocytosis | [107] |
| miR-1 | mice | RAW264.7 | experimental overexpression | clathrin heavy chain 1 (CLTC1) | decrease of E. coli uptake | [111] |
| miR-9-1 | human | blood monocyte | LPS stimulation | NFKB1 | negative feedback on pro-inflammatory response | [112] |
| miR-15a/16 | miR-15a/16 knocked-out mice | bone marrow-derived macrophages | exposure to E. coli | derepression of PU.1 after miR-15a/16 knock-out | increased E. coli uptake and generation of mitochondrial reactive oxygen species in miR-15a/16 knocked-out mice | [113] |
| miR-17 | human | HL-60, U937, THP-1 | LPS-induced upregulation of miR-17, miR-20a, and miR-106a | SIRPα | decreased migration, zymosan particles uptake, and secretion of pro-inflammatory cytokines upon simultaneous microRNAs inhibition | [114] |
| miR-20a-5p | human | HL-60, U937, THP-1 | LPS-induced upregulation of miR-17, miR-20a, and miR-106a | SIRPα | decreased migration, zymosan particles uptake, and secretion of pro-inflammatory cytokines upon simultaneous microRNAs inhibition | [114] |
| miR-20a-5p | human | monocytes, THP-1 | M. tuberculosis infection | JNK2 | expression of miR-20a-5p is reduced upon infection, which enhance pathogen clearance | [115] |
| miR-20b-5p | mice | RAW264.7 | M. tuberculosis infection | Mcl-1 (direct interaction not confirmed) | expression of miR-20b-5p is reduced upon infection, which enhance pathogen survival | [116] |
| miR-21 | mice | RAW264.7 | miR-21 transfection, induction by miR-21-rich exosomes | not specified | polarization towards M1 phenotype | [117] |
| miR-21 | miR-21 knock-out mice | peritoneal macrophages | miR-21 is downregulated through PGE2 | STAT3 (suppressed by miR-21) | promoting M2 over M1 polarization upon miR-21 knock-out | [118] |
| miR-21 | miR-21 knock-out mice | bone marrow-derived macrophages | miR-21-deficient mice exposed to L. monocytogenes | myristoylated alanine-rich C-kinase substrate (MARCKS) and Ras homolog gene family, member B (RhoB)—upregulated in miR-21 knock-out mice; lack of experimental confirmation of direct binding of microRNA with 3′UTRs | increased uptake of L. monocytogenes, E. coli and dextran | [119] |
| miR-21 | human, mice | bone marrow-derived macrophages, RAW264.7, PDCD4 knock-out mice | LPS stimulation | PDCD4 | induction of miR-21 protects from LPS-mediated overstimulation | [120] |
| miR-21 | human | THP-1, bone marrow-derived macrophages | wound healing | PTEN, PDCD4 | expression of miR-21 upon LPS stimulation is higher in macrophages performing efferocytosis; miR-21 promotes resolving of inflammation through suppression of NF-κB and induction of IL-10 | [121] |
| miR-23a-3p | human | bone marrow-derived macrophages | M. tuberculosis infection | IRF1/SP1 | reduction of reactive oxygen species generation and inhibition of TLR4/TNF-α/TGF-β1/IL-10 signaling pathway | [122] |
| miR-24 | human | monocyte | E. coli infection, IgG-opsonized beads infection | PKC-α | reduced secretion of TNF-α and IL-8, suppressed superoxide generation and reduction in expression of FcRs including FCGR2A, FcɛR1G and FCER2 | [123] |
| miR-24 | human | monocyte | LPS stimulation | p110δ | reduced secretion of cytokines, and promotion of anti-inflammatory phenotype | [124] |
| miR-24 | human | monocytes | E. coli and S. aureus infection | PKCα | modulation of phagocytosis and cytokine production | [125] |
| miR-26a | human, mice | bone marrow-derived macrophages, RAW264.7 | M. tuberculosis infection | KLF4 | downregulation of miR-26a promotes M2 polarization and intracellular pathogen survival due to decreased trafficking to lysosomes | [126]. |
| miR-26a | rat | bone marrow-derived macrophages | co-culture with dying cells | C1qa | promotion of M1 phenotype | [127] |
| miR-27a | human | monocytes, THP-1 | alcohol-exposed monocytes | not specified | monocytes polarize into M2 macrophages as indicated by increased surface expression of CD68 (macrophage marker), M2 markers (CD206 (mannose receptor) and CD163 (scavenger receptor)), secretion of IL-10, and TGFβ and increased phagocytic activity | [128] |
| miR-30b | human | monocyte | E. coli infection, IgG-opsonized beads infection | PKCα | reduced secretion of TNF-α and IL-8, suppressed superoxide generation and reduction in expression of FcRs including FCGR2A, FcɛR1G and FCER2 | [123] |
| miR-30b | human | monocytes | E. coli and S. aureus infection | PKCα | modulation of phagocytosis and cytokine production | [125] |
| miR-30b | human | monocytes | experimental overexpression | Vinculin, Dab2 and Skap2 directly associated with cytoskeletal rearrangement | regulation of cytoskeletal rearrangement and cell movement | [129] |
| miR-30b/30c | mice | RAW264.7 | B. pseudomallei infection | Rab32 | enhanced phagosome maturation | [130] |
| miR-30e-5p | mice | BALB/c macrophages | L. amazonensis infection | increased nitric oxide synthase 2 (Nos2) mRNA expression levels and nitric oxide (NO) production | nitric oxide is secreted as free radicals in an immune response and is toxic to intracellular parasites | [131] |
| miR-33 | mice macrophages’ specific miR-33 inhibition | bone marrow-derived macrophages | inflammation in atherosclerotic plaque | AMP-activated protein kinase and retinoic acid-producing enzyme aldehyde dehydrogenase family 1, subfamily A2 (direct or indirect) | increased oxidative respiration, promoted M2 polarization and reduction of atherosclerotic plaque upon miR-33 knock-out (partially due to Treg lymphocyte-mediated effects) | [132] |
| miR-34a | mice | C57BL/6 | induction of apoptosis by dexamethasone treatment | SIRT1 | negatively regulates efferocytosis | [133] |
| miR-92a | mice | RAW264.7, MyD88 knock-out mice | stimulation of multiple TLRs (mainly TLR4 by LPS), which leads to downregulation of miR-92a | mitogen-activated protein kinase kinase 4 (MKK4) | TLR-mediated miR-92a derepresses production of pro-inflammatory cytokines and impedes resolution of inflammation | [134] |
| miR-99b | human | monocytes | experimental differentiation into macrophages or dendritic cells | TLR4 | reduced differentiation into dendritic cells | [135] |
| miR-106a | human | HL-60, U937, THP-1 | LPS-induced upregulation of miR-17, miR-20a, and miR-106a | SIRPα | decreased migration, zymosan particles uptake, and secretion of pro-inflammatory cytokines upon simultaneous microRNAs inhibition | [114] |
| miR-106b-5p | human | monocytes | M. tuberculosis infection | cathepsin S (CtsS) | decreased host lysosomal enzymatic activity | [136] |
| mir-124-5p | human | monocytes | experimental overexpression | ARP2/3 complex | rearrangement of actin cytoskeleton | [137] |
| miR-125a-5p | mice | bone marrow-derived macrophages, KLF13 knock-out mice | TLR2 and TLR4-dependent upregulation of miR-125a-5p | Kruppel-like Factor 13 (KLF13) | miR-125a-5p upregulation decrease of bactericidal activity, promote switch from M1 to M2 polarization and resolution of inflammation | [138] |
| miR-125a-5p | mice | bone marrow-derived macrophages, BALB/c mice | stimulation with LPS and T. crassiceps-excreted/secreted antigens | not specified | miR-125a-5p upregulation promotes of M2 phenotype | [139] |
| miR-125b-5p | mice | bone marrow-derived macrophages, C57bl/6 mice | exposition on biomaterials | not specified | miR-125b-5p downregulation promotes of M1 phenotype | [140] |
| miR-125b-5p | mice | RAW264.7, C57BL/6 mice | LPS stimulation | TNF-α | LPS-induced miR-125b-5p downregulation promotes M1 phenotype via TNF-α production | [141] |
| miR-125b-5p | human | bone marrow-derived macrophages | M. tuberculosis and M. smegmatis lipomannan stimulation | TNF-α | Mtb-induced miR-125b-5p upregulation promote M2 phenotype, and repression of TNF-α production, whereas M. smegmatis promotes miR-125b-5p downregulation and thus M1 phenotype and enhanced phagocytic capacity | [142] |
| miR-125b-5p | mice, C57Bl/6 mice | RAW264.7, bone marrow-derived macrophages | experimental overexpression or silencing | IFN regulatory factor 4 (IRF4) | suppression of IRF4 and induction of CD80, what enhances macrophages’ antigen presenting cells capacities | [143] |
| miR-128 | C57BL/6 mice | bone marrow-derived macrophages | co-culturing with Panc02 cells | not specified | increased phagocytosis | [144] |
| miR-139-5p | human | monocytes | experimental differentiation into macrophages or dendritic cells | TLR4 | reduced differentiation into dendritic cells | [135] |
| hsa-miR-142-3p | human, mice | monocytes, J774A | M. tuberculosis infection | N-Wasp, an actin-binding protein involved in actin dynamics during bacterial uptake | modulation of bacteria uptake, decreased internalization | [145] |
| miR-142-3p | human | monocyte | E. coli infection, IgG-opsonized beads infection | PKC-α | reduced secretion of TNF-α and IL-8, suppressed superoxide generation and reduction in expression of FcRs including FCGR2A, FcɛR1G and FCER2 | [123] |
| miR-142-3p | human | monocytes | experimental overexpression | Vinculin, Dab2 and Skap2 directly associated with cytoskeletal rearrangement | regulation of cytoskeletal rearrangement and cell movement | [129] |
| miR-142-3p | human | monocytes | E. coli and S. aureus infection | PKCα | modulation of phagocytosis and cytokine production | [125] |
| miR-143-3p | mice | bone marrow-derived macrophages, C57bl/6 mice | exposition on biomaterials | not specified | promotion of M1 phenotype | [140] |
| miR-144 | rat | macrophages | HIV infection | Nrf2 | impaired bacterial phagocytic capacity and H2O2 scavenging ability | [146] |
| miR-145-5p | mice | bone marrow-derived macrophages, C57bl/6 mice | exposition on biomaterials | not specified | promotion of M1 phenotype | [140] |
| miR-145-3p | human | THP-1 | LPS stimulation | not specified | promotion of M2 polarization | [147] |
| miR-146a | mice | RAW264.7 | experimental overexpression | TLR2 | SNP in miR-146a affects regulation of expression of TLR2, which regulates amyloid uptake, and may contribute to the risk of Alzheimer’s disease | [148] |
| miR-146a | human | THP-1 | macrophages of atherosclerotic plaque | TLR4 | overexpression of miR-146a reduces intracellular LDL cholesterol content and secretion of interleukin 6, interleukin 8, chemokine (C-C motif) ligand 2 and matrix metallopeptidase 9, thus may suppress atherosclerosis | [149] |
| miR-146a | miR-146a knock-out mice | macrophages | B. burgdorferi infection | overactivation of NF-κB (due to miR-146a knock-out) | increased pathogen uptake, impaired resolving of inflammation and more severe Lyme arthritis | [150] |
| miR-146a | IL-10 and Rag1 double knock-out mice | bone marrow-derived macrophages | colitis model | interferon regulatory factor 5 (IRF5) | miR-146a knock-out leads to M1 polarization and intestinal inflammation, whereas treatment with miR-146a mimic ameliorates colitis | [151] |
| miR-146a | mice | RAW264.7 | experimental overexpression or silencing | Notch1, Peroxisome proliferator-activated receptor γ (PPARγ) (directly or indirectly) | M2 polarization | [152] |
| miR-146a | mice | bone marrow derived macrophages | L. donovani infection | TRAF6, IRAK1 | M2 polarization, decreased phagocytosis of L. donovani upon miR-146a inhibition | [153] |
| miR-146a | human, mice | THP-1, C57BL/6.NOD-Aec1Aec2 (mouse model of Sjögren’s syndrome) | verification of the role of miR-146a, which is upregulated in Sjögren’s syndrome patients (and mice model) | not specified | increased uptake of E. coli and suppression of pro-inflammatory cytokine production upon miR-146a overexpression | [154] |
| miR-155 | human | THP-1 | experimental overexpression or silencing | SCG2 | overexpression of miR-155 decrease lipid uptake, potentially affecting atherosclerosis | [155] |
| miR-155 | human | monocytes | experimental overexpression or silencing | not specified | increased ROS production, and M1 phenotype promotion | [156] |
| miR-155 | human | monocytes | Vibrio anguillarum infection | not specified | M1 phenotype promotion | [157] |
| miR-155 | mice | RAW264.7, C57BL/6 mice | LPS stimulation | Fas-associated death domain protein (FADD), IkappaB kinase epsilon (IKKepsilon), and the receptor (TNFR superfamily)-interacting serine-threonine kinase 1 (Ripk1) | LPS stimulation upregulates miR-155, thus increases TNF-α production | [141] |
| miR-155 | mice | RAW264.7 | LPS stimulation | Suppressor of Cytokine Signaling 1 (SOCS1) | miR-155 is downregulated upon LPS stimulation, which derepresses SOCS1 to modulate inflammatory response | [57] |
| miR-155 | mice | RAW264.7 | experimental overexpression or silencing | Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) | increased activation of Akt upon LPS stimulation | [158] |
| miR-155 | human | bone marrow-derived macrophages | M. tuberculosis lipomannan stimulation | SH-2 containing inositol 5′ polyphosphatase 1 (SHIP1) | lipomannan stimulation downregulates miR-155, thus derepressing SHIP1, consequently downregulating TNF-α | [142] |
| miR-155 | mice | C57 or TLR2KO mice | S. aureus or S. pneumoniae infection | SH-2 containing inositol 5′ polyphosphatase 1 (SHIP1) | enhanced bacteria uptake | [159] |
| miR-155 | mice | miR-155 knock-out mice | macrophages of atherosclerotic plaque | BCL6 | mildly oxidized LDL increases expression of miR-155, which downregulates BCL6, thus attenuating NF-κB signaling | [160] |
| miR-155 | human | monocytes | co-culture with abnormal red blood cell | BACH1 | enhanced phagocytic activity | [161] |
| miR-155 | mice | RAW264.7, bone marrow-derived macrophages | induction of miR-155 by M. tuberculosis infection | Ras homologue enriched in brain (Rheb) | promotion of maturation of mycobacterium-containing phagosomes and decreasing the survival rate of intracellular mycobacteria | [162] |
| miR-155 | human, mice | corneas, RAW264.7, bone marrow-derived macrophages | induction of miR-155 in P. aeruginosa–induced keratitis | Ras homologue enriched in brain (Rheb) | suppression of phagocytosis and intracellular killing of P. aeruginosa | [163] |
| miR-181a | human, mice | THP-1, RAW264.7 | experimental overexpression or silencing | KLF6, C/EBPα | M2 polarization | [164] |
| miR-181b | human | monocytes | zymosan stimulation | ALX/FPR2 | stimulated phagocytic activity | [165] |
| miR-181b | human | monocytes | zymosan stimulation or P. aeruginosa infection | not specified | modulation of receptor-dependent LXA4-induced phagocytosis | [166] |
| miR-183 | human | monocytes | M. tuberculosis infection | NF-κB | increased phagocytosis | [167] |
| miR-183/96/182 cluster | mice | miR-183/96/182 knockout mice, RAW264.7 | P. aeruginosa-induced keratitis | not specified | knock-out and experimental silencing increases phagocytic capacity, knock-out decreases inflammatory response and severity of keratitis | [168] |
| miR-185-5p | human, mice | THP-1, RAW264.7 | phagocytosis of intrarenal CaOx crystals | not specified | stimulation of M2 phenotype | [169] |
| miR-200a | human | monocytes | co-culturing with nasopharyngeal carcinoma cells | CD47 | increased phagocytosis | [170] |
| miR-212 | human | monocytes | experimental differentiation into macrophages or dendritic cells | TLR4 | reduced differentiation into dendritic cells | [135] |
| miR-218 | human | monocytes | experimental differentiation into macrophages or dendritic cells | TLR4 | reduced differentiation into dendritic cells | [135] |
| miR-223 | miR-223 knock-out mice | bone marrow derived macrophages | obesity-associated adipose tissue inflammation | Pknox1 | preferential M1 polarization and exacerbation of insulin resistance and adipose tissue inflammation in miR-223-deficient mice | [171] |
| miR-223 | human | monocytes | M tuberculosis infection | FOXO3 | suppressed apoptosis of macrophages | [172] |
| miR-302d-3p | mice | BALB/c mice macrophages | Leishmania amazonensis infection | increased nitric oxide synthase 2 (Nos2) mRNA expression levels and nitric oxide (NO) production | nitric oxide is secreted as free radicals in an immune response and is toxic to intracellular parasites | [131] |
| miR-328 | mice, human | monocytes | H. influenzae infection | not specified | miR-328 inhibition augments phagocytosis and production of reactive oxygen species | [173] |
| miR-340 | mice | bone marrow derived macrophages of C57BL/6 mice | co-culture macrophages with pancreatic cancer cells | not specified | stimulation of M1 phenotype | [174] |
| miR-378a | mice | ApoE−/− mice macrophages | zymosan stimulation, or co-culture with Ishikawa cells | SIRPα | modulation of phagocytosis and differentiation | [175] |
| miR-466 | human | monocytes | P. aeruginosa infection | TIRAP | miR-466 delivery in extracellular vesicles promotes M2 polarization, increases pathogen phagocytosis, suppresses pro-inflammatory factors, decreases neutrophil efflux, and reduces infected mice mortality | [176] |
| miR-484 | mice | bone marrow-derived macrophages of BALB/c mice | stimulation with LPS and T. crassiceps-excreted/secreted antigens | not specified | promotion of M2 phenotype | [139] |
| miR-511 | human | monocytes | experimental differentiation into macrophages or dendritic cells | TLR4 | reduced differentiation into dendritic cells | [135] |
| miR-582-5p | human | monocytes, THP-1 | M. tuberculosis infection | FOXO1 | suppressed apoptosis of macrophages | [177] |
| miR-590 | apoE−/− mice | macrophages | experimental overexpression and silencing | lipoprotein lipase | miR-590 decreases concentration of proinflammatory cytokines and atherosclerotic plaque | [178] |
| miR-615-3p | human | THP-1, splenic macrophages | hypersplenism (resulting in overexpression of miR-615-3p | ligand-dependent nuclear receptor corepressor (LCoR), which is a derepressor of peroxisome proliferator-activated receptor gamma (PPARγ) | inhibition on miR-615-3p reduces uptake of E. coli | [179] |
| miR-708-5p | BALB/c mice | macrophages | co-incubation with CFSE-labelled breast cancer cells | CD47 | enhanced phagocytosis | [180] |
| miR-708-5p | human | THP-1, U937 | M. tuberculosis infection | TLR4 | miR-708-5p expression increases upon infection and enhances pathogen survival | [181] |
| miR-762 | BALB/c mice | bone marrow-derived macrophages | stimulation with LPS and T. crassiceps-excreted/secreted antigens | not specified | promotion of M2 phenotype | [139] |
| miR-1246 | human | monocytes | co-culture with glioma-derived exosomes (GDEs) | TERF2IP | induced M2 polarization | [182] |
| miR-4270 | human | monocytes | H. pylori infection | CD300E | induced M1 polarization | [183] |
| MicroRNA | Organism | Cell | Setting | Target | Effect | Ref. |
|---|---|---|---|---|---|---|
| miR-21-5p | murine/rat cell lines | PC12 (murine neuronal cell line) | experimental overexpression | BV2 (rat microglia cell line) | M1 polarization | [228] |
| miR-34a | human/mice | C8B4-microglial cells | age-related macular degeneration (AMD) | triggering receptor expressed in myeloid/microglial cells-2 (TREM2) | decreased uptake of Aβ42-peptides | [229] |
| miR-34a | p53-deficient mice | RAW cell line | experimental overexpression | Twist2 | p53-dependent miR-34a upregulation represses Twist2, and consequently anti-inflammatory c-Maf | [230] |
| miR-98 | mice | extracellular vesicles secreted by neurons | murine model of ischemic stroke | platelet activating factor receptor in microglia | prevention of stress-but-viable neurons from microglial phagocytosis | [231] |
| miR-124 | C57BL/6 mice | microglia | experimental autoimmune encephalomyelitis | CCAAT/enhancer-binding protein-α (C/EBP-α) | suppression of inflammation | [232] |
| miR-124 | primary adult rat spinal microglia cultures and in the murine microglial cell line BV2 | microglia | spinal cord injury | not specified | reduced myelin phagocytosis | [233] |
| miR-124 | Danio rerio | microglia | experimental silencing and overexpression | not specified | overexpression of miR-124 reduces microglia motility and phagocytosis | [234] |
| miR-142-5p | human brain autopsy samples, C57BL/6 mice | murine splenocytes | multiple sclerosis, experimental autoimmune encephalomyelitis | SOCS1 | promotion of differentiation towards Th1 subtype | [235] |
| miR-142-3p | TGFBR1 | |||||
| miR-145 | p53-deficient mice | RAW cell line | experimental overexpression | Twist2 | p53-dependent miR-34a upregulation represses Twist2, and consequently anti-inflammatory c-Maf | [230] |
| miR-146 | SPF KM mice | U251 human astrocyte cell line | E. coli strain PCN033 infection | IRAK1 and TRAF6 | fine-tuning of inflammation through negative feedback loop with NF-κB | [236] |
| miR-155 | SPF KM mice | U251 human astrocyte cell line | E. coli strain PCN033 infection | TAB2 | fine-tuning of inflammation through negative feedback loop with NF-κB | [236] |
| miR-155 | cell line | N9 microglia cells | LPS stimulation | SOCS1 | downregulation of inflammatory cytokines and inducible nitric oxide synthase, decreased production of nitric oxide; decreased neurons phagocytosis by activated microglia | [237] |
| miR-155 | p53- or miR-155 deficient mice | microglia | model of neuroinflammation | c-Maf | p53-dependent miR-155 upregulation represses anti-inflammatory c-Maf | [230] |
| miR-155 | SOD1 mice | microglia | miR-155 knock-out | not specified | increased phagocytic function and disease amelioration upon miR-155 knock-out | [238] |
| miR-155 | C57/BL6 wild-type mice | primary microglia | miR-155 knock-out or overexpression | not specified | increased amyloid uptake and catabolism upon miR-155 knock-out | [239] |
| miR-181c | mice, rat | BV-2, primary microglia | oxygen-glucose deprivation, experimental overexpression and silencing | TLR4 | oxygen-glucose deprivation leads to miR-181c downregulation and subsequent derepression of TLR4, promoting inflammatory response | [205] |
| miR-223 | SOD1 mice | macrophages, microglia | miR-155 knock-out | not specified | miR-223 takes part in M2 polarization and stimulates myelin debris clearance | [240] |
| miR-340 | rat cell line | BV2 (microglia cell line) | LPS-induced inflammation attenuated by dexmedetomidine | NF-κB | suppression of pro-inflammatory cytokines (TNF-α, IL-6, IL-1β, IL-2 and IL-12), induction of anti-inflammatory IL-10, and inhibition of phagocytosis | [241] |
| MicroRNA | Organism | Cell | Setting | Target | Effect | Refs. |
|---|---|---|---|---|---|---|
| miR-21 | mice | osteoclasts | experimental knock-out | PDCD4 | decreased bone loss in miR-21 knock-out mice | [260,261] |
| MicroRNA | Organism | Cell | Setting | Target | Effect | Ref. |
|---|---|---|---|---|---|---|
| miR-142-5p and miR-142-3p | mir-142 knock-out mice | neutrophils | S. aureus skin wound infection | small GTPases Rho, Rac, and Cdc42 | increased bacteria load, impaired abscess formation, and decreased phagocytic activity due to impaired cytoskeleton remodeling in neutrophils of mir-142 knock-out mice | [263] |
| miR-183/96/182 cluster | miR-183/96/182 knockout mice | neutrophils | P. aeruginosa-induced keratitis | not specified | knock-out increases phagocytic capacity, knock-out decreases inflammatory response and severity of keratitis | [168] |
| miR-328 | mice, human | neutrophils | experimental inhibition, H. influenzae infection | not specified | miR-328 inhibition augments phagocytosis of H. influenzae and increases ROS production | [173] |
| MicroRNA | Organism | Cell | Setting | Target | Effect | Refs. |
|---|---|---|---|---|---|---|
| miR-25 | rat | retinal pigment epithelium | model of age-related macular degeneration | IGTAV and PEDF | decreased phagocytosis, retina degeneration and visual impairment | [265] |
| miR-184 | human | primary retinal pigment epithelium | age-related macular degeneration | ezrin | impaired phagocytosis and visual impairment due to low expression of miR-184 | [266] |
| miR-194 | ARPE-19 cell line, rat | primary retinal pigment epithelium | model of proliferative vitreoretinopathy | zinc finger E-box binding homeobox 1 (ZEB1) | miR-194 administration in vivo suppressed proliferative vitreoretinopathy in the rat model | [267] |
| miR-204 | mice, human | retinal pigment epithelium | miR-204 knock-out | Rab22a | impaired phagocytosis and visual impairment due to low expression of miR-204 | [268,269] |
| miR-211 | mice, ARPE-19 cell line | retinal pigment epithelium | miR-211 knock-out and overexpression | ezrin | overexpression of miR-211 stimulates lysosomal biogenesis and increase autophagosome–lysosome fusion | [270,271] |
| miR-302d-3p | HiPSC-RPE, ARPE-19 cell lines | retinal pigment epithelium | miR-302d-3p silencing and overexpression | p21Waf1/Cip1 | miR-302d-3p induces RPE dedifferentiation, cell cycle progression, proliferation, migration, inhibits phagocytosis | [272] |
| miR-382-5p | ARPE-19 cell line | retinal pigment epithelium | miR-382-5p silencing and overexpression, direct or mediated via manipulation of circNR3C1 | PTEN | miR-382-5p induces RPE dedifferentiation, proliferation, migration, inhibits phagocytosis | [273] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gierlikowski, W.; Gierlikowska, B. MicroRNAs as Regulators of Phagocytosis. Cells 2022, 11, 1380. https://doi.org/10.3390/cells11091380
Gierlikowski W, Gierlikowska B. MicroRNAs as Regulators of Phagocytosis. Cells. 2022; 11(9):1380. https://doi.org/10.3390/cells11091380
Chicago/Turabian StyleGierlikowski, Wojciech, and Barbara Gierlikowska. 2022. "MicroRNAs as Regulators of Phagocytosis" Cells 11, no. 9: 1380. https://doi.org/10.3390/cells11091380
APA StyleGierlikowski, W., & Gierlikowska, B. (2022). MicroRNAs as Regulators of Phagocytosis. Cells, 11(9), 1380. https://doi.org/10.3390/cells11091380






