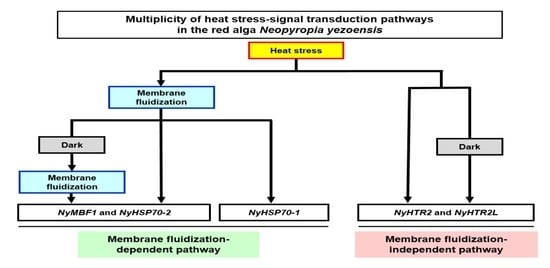
Membrane-Fluidization-Dependent and -Independent Pathways Are Involved in Heat-Stress-Inducible Gene Expression in the Marine Red Alga Neopyropia yezoensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Materials and Culture Conditions
2.2. Identification and Phylogenetic Analysis of HTR2 Homologs
2.3. Total RNA Extraction, DNA Removal, and cDNA Synthesis
2.4. Quantitative Gene Expression Analysis
2.5. Statistical Analysis
3. Results
3.1. Discovery of a Novel Protein Family Specifically Conserved in Bangiales
3.2. Determination of a Reference Gene for Quantitative Analysis of Heat-Inducible Gene Expression in Different Life Cycle Generations of N. yezoensis
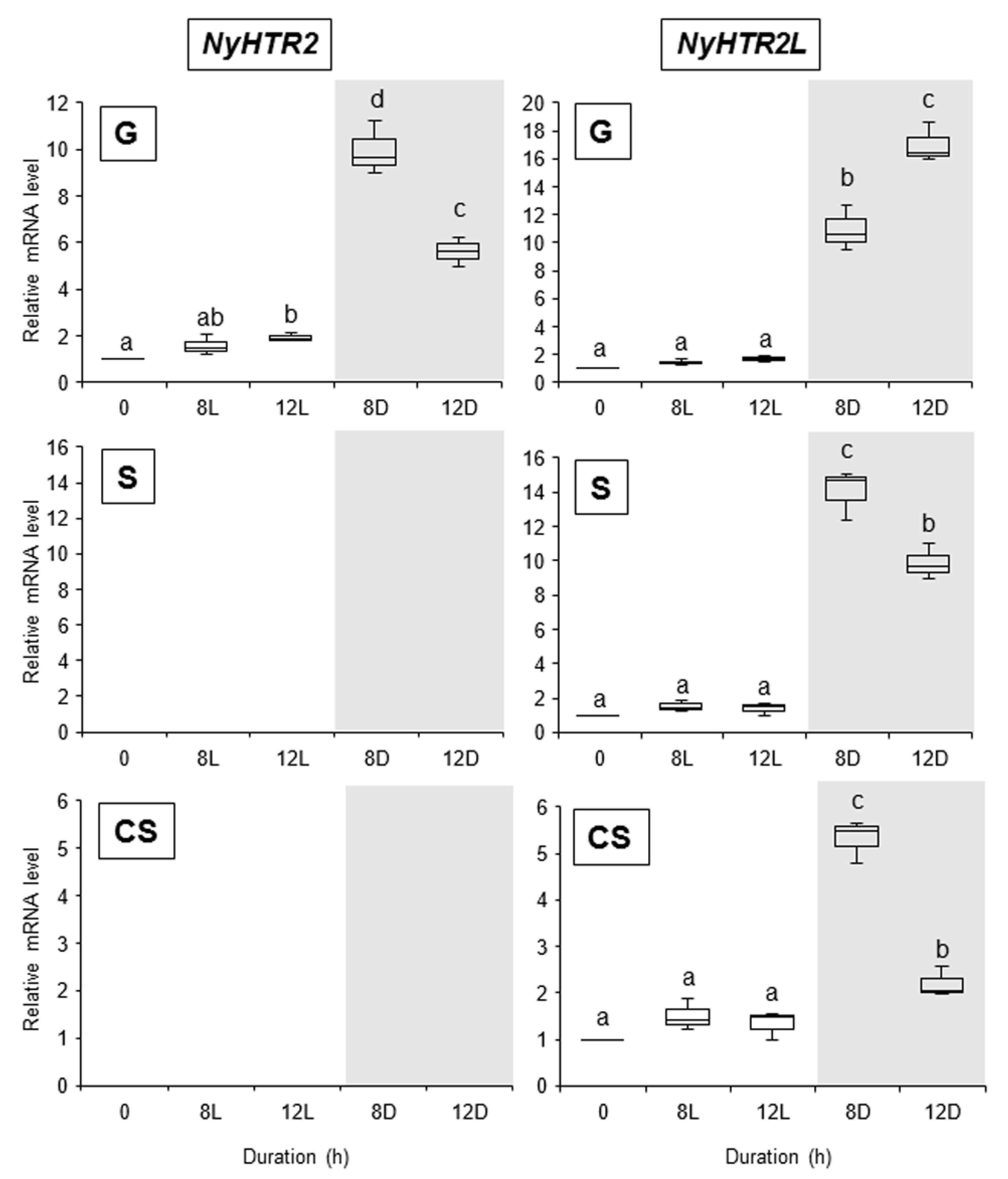
3.3. Heat-Inducible and Dark-Stimulated Expression of NyHTR2 and NyHTR2L
3.4. Relationship between Heat-Inducible Expression and Membrane Fluidization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yoshida, G.; Shimabukuro, H.; Kiyomoto, S.; Kadota, T.; Yoshimura, T.; Murase, N.; Noda, M.; Takenaka, S.; Kono, Y.; Tamura, T.; et al. Assessment and future prediction of climate change impacts on the macroalgal bed ecosystem and cultivation in the Seto Inland Sea. Bull. Jap. Fish. Res. Edu. Agen. 2019, 49, 27–34. [Google Scholar]
- Hwang, E.K.; Park, C.S. Seaweed cultivation and utilization of Korea. Algae 2020, 35, 107–121. [Google Scholar] [CrossRef]
- Sun, P.; Mao, Y.; Li, G.; Cao, M.; Kong, F.; Wang, L.; Bi, G. Comparative transcriptome profiling of Pyropia yezoensis (Ueda) M.S. Hwang & H.G. Choi in response to temperature stresses. BMC Genom. 2015, 16, 463. [Google Scholar] [CrossRef]
- Wang, W.; Teng, F.; Lin, Y.; Ji, D.; Xu, Y.; Chen, C.; Xie, C. Transcriptomic study to understand thermal adaptation in a high temperature-tolerant strain of Pyropia haitanensis. PLoS ONE 2018, 13, e0195842. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, P.; Yan, B.; Li, X.; Yi, L. Characterization and expression patterns of two 70-kDa heat shock protein genes in the intertidal red alga Porphyra yezoensis. Bot. Mar. 2011, 54, 447–455. [Google Scholar] [CrossRef]
- Kim, E.; Park, H.S.; Jung, Y.; Choi, D.W.; Jeong, W.J.; Park, H.S. Identification of the high-temperature response genes from Porphyra seriata (Rhodophyta) expression sequence tags and enhancement of heat tolerance of Chlamydomonas (Chlorophyta) by expression of the Porphyra HTR2 gene1: High-temperature stress in Porphyra. J. Phycol. 2011, 47, 821–828. [Google Scholar] [CrossRef]
- Ji, D.; Li, B.; Xu, Y.; Chen, C.; Xie, C. Cloning and quantitative analysis of five heat shock protein 70 genes from Pyropia haitanensis. J. Appl. Phycol. 2015, 27, 499–509. [Google Scholar] [CrossRef]
- Yu, X.; Mo, Z.; Tang, X. Genome-wide analysis of HSP70 gene superfamily in Pyropia yezoensis (Bangiales, Rhodophyta): Identification, characterization and expression profiles in response to dehydration stress. BMC Plant Biol. 2021, 21, 435. [Google Scholar] [CrossRef]
- Kishimoto, I.; Ariga, I.; Itabashi, Y.; Mikami, K. Heat-stress memory is responsible for acquired thermotolerance in Bangia fuscopurpurea. J. Phycol. 2019, 55, 971–975. [Google Scholar] [CrossRef]
- Khoa, H.V.; Kumari, P.; Uchida, H.; Murakami, A.; Shimada, S.; Mikami, K. Heat-stress responses differ among species from different ‘Bangia’ clades of Bangiales (Rhodophyta). Plants 2021, 22, 1733. [Google Scholar] [CrossRef]
- Mueller, S.P.; Krause, D.M.; Mueller, M.J.; Fekete, A. Accumulation of extra-chloroplastic triacylglycerols in Arabidopsis seedlings during heat acclimation. J. Exp. Bot. 2015, 66, 4517–4526. [Google Scholar] [CrossRef] [PubMed]
- de Vries, J.; de Vries, S.; Curtis, B.A.; Zhou, H.; Penny, S.; Feussner, K.; Pinto, D.M.; Steinert, M.; Cohen, A.M.; von Schwartzenberg, K.; et al. Heat stress response in the closest algal relatives of land plants reveals conserved stress signaling circuits. Plant J. Cell Mol. Biol. 2020, 103, 1025–1048. [Google Scholar] [CrossRef] [PubMed]
- de Vries, J.; Ischebeck, T. Ties between Stress and Lipid Droplets Pre-date Seeds. Trends Plant Sci. 2020, 25, 1203–1214. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, W.; Hayes, S.; Trenner, J.; Delker, C.; Quint, M. On the evolution of plant thermomorphogenesis. J. Exp. Bot. 2021, 72, 7345–7358. [Google Scholar] [CrossRef]
- Rieseberg, T.P.; Dadras, A.; Fürst-Jansen, J.; Dhabalia Ashok, A.; Darienko, T.; de Vries, S.; Irisarri, I.; de Vries, J. Crossroads in the evolution of plant specialized metabolism. Semin. Cell Dev. Biol. 2022. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Döring, P.A. Cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar] [CrossRef]
- Yoshida, T.; Sakuma, Y.; Todaka, D.; Maruyama, K.; Qin, F.; Mizoi, J.; Kidokoro, S.; Fujita, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem. Biophys. Res. Commun. 2008, 368, 515–521. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant heat stress response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Yoshida, T.; Ohama, N.; Nakajima, J.; Kidokoro, S.; Mizoi, J.; Nakashima, K.; Maruyama, K.; Kim, J.-M.; Seki, M.; Todaka, D.; et al. Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol. Genet. Genom. 2011, 286, 321–332. [Google Scholar] [CrossRef]
- Petroll, R.; Schreiber, M.; Finke, H.; Cock, J.M.; Gould, S.B.; Rensing, S.A. Signatures of transcription factor evolution and the secondary gain of red algae complexity. Genes 2021, 12, 1055. [Google Scholar] [CrossRef]
- Thiriet-Rupert, S.; Carrier, G.; Chénais, B.; Trottier, C.; Bougaran, G.; Cadoret, J.P.; Schoefs, B.; Saint-Jean, B. Transcription factors in microalgae: Genome-wide prediction and comparative analysis. BMC Genom. 2016, 17, 282. [Google Scholar] [CrossRef] [PubMed]
- Lawal, G.; Mohd, S.M.A.; Siti, N.O.; Raja, N.Z. Review on fatty acid desaturases and their roles in temperature acclimatisation. J. Appl. Sci. 2017, 17, 282–295. [Google Scholar] [CrossRef][Green Version]
- Rawat, N.; Singla-Pareek, S.L.; Pareek, A. Membrane dynamics during individual and combined abiotic stresses in plants and tools to study the same. Physiol. Plant. 2021, 171, 653–676. [Google Scholar] [CrossRef]
- Falcone, D.L.; Ogas, J.P.; Somerville, C.R. Regulation of membrane fatty acid composition by temperature in mutants of Arabidopsis with alterations in membrane lipid composition. BMC Plant Biol. 2004, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Higashi, Y.; Saito, K. Lipidomic studies of membrane glycerolipids in plant leaves under heat stress. Prog. Lipid Res. 2019, 75, 100990. [Google Scholar] [CrossRef] [PubMed]
- Zoong, L.Z.; Sah, S.; Persaud, L.; Li, J.; Gao, W.; Raja Reddy, K.; Narayanan, S. Alterations in the leaf lipidome of Brassica carinata under high-temperature stress. BMC Plant Biol. 2021, 21, 404. [Google Scholar] [CrossRef]
- Narayanan, S. Comparative lipidomic analysis reveals heat stress responses of two soybean genotypes differing in temperature sensitivity. Plants 2020, 9, 57. [Google Scholar] [CrossRef]
- Shiva, S. Leaf lipid alterations in response to heat stress of Arabidopsis thaliana. Plants 2020, 9, 845. [Google Scholar] [CrossRef]
- Saidi, Y.; Finka, A.; Goloubinoff, P. Heat perception and signaling in plants: A tortuous path to thermotolerance. New Phytol. 2011, 190, 556–565. [Google Scholar] [CrossRef]
- Abdelrahman, M.L.; Ishii, T.; El-Sayed, M.; Tran, L.S.P. Heat sensing and lipid reprograming as a signaling switch for heat stress responses in wheat. Plant Cell Physiol. 2020, 61, 1399–1407. [Google Scholar] [CrossRef]
- Bourgine, B.; Guihur, A. Heat shock signaling in land plants: From plasma membrane sensing to the transcription of small heat shock proteins. Front. Plant Sci. 2021, 12, 710801. [Google Scholar] [CrossRef] [PubMed]
- Cano-Ramirez, D.L.; Carmona-Salazar, L.; Morales-Cedillo, F.; Ramírez-Salcedo, J.; Cahoon, E.B.; Gavilanes-Ruíz, M. Plasma membrane fluidity: An environment thermal detector in Plants. Cells 2021, 10, 2778. [Google Scholar] [CrossRef] [PubMed]
- Hayes, S.; Schachtschabel, J.; Mishkind, M.; Munnik, T.; Arisz, S.A. Hot Topic: Thermosensing in Plants. Plant Cell Environ. 2020, 44, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2010, 31, 629–638. [Google Scholar] [CrossRef]
- Königshofer, H.; Tromballa, H.W.; Löppert, H.G. Early events in signaling high-temperature stress in tobacco BY2 cells involve alterations in membrane fluidity and enhanced hydrogen peroxide production. Plant Cell Environ. 2008, 31, 1771–1780. [Google Scholar] [CrossRef]
- Finka, A.; Goloubinoff, P. The CNGCb and CNGCd genes from Physcomitrella patens moss encode for thermosensory calcium channels responding to fluidity changes in the plasma membrane. Cell Stress Chaperones 2014, 19, 83–90. [Google Scholar] [CrossRef]
- Murakami, Y.; Tsuyama, M.; Kobayashi, Y.; Kodama, H.; Iba, K. Trienoic fatty acids and plant tolerance of high temperature. Science 2000, 287, 476–479. [Google Scholar] [CrossRef]
- Sui, N.; Li, M.; Shu, D.F.; Zhao, S.J.; Meng, Q.W. Antisense-mediated depletion of tomato chloroplast glycerol-3-phosphate acyltransferase affects male fertility and increases thermal tolerance. Physiol. Plant. 2007, 130, 301–314. [Google Scholar] [CrossRef]
- Yan, K.; Chen, N.; Qu, Y.Y.; Dong, X.C.; Meng, Q.W.; Zhao, S.J. Overexpression of sweet pepper glycerol-3-phosphate acyltransferase gene enhanced thermotolerance of photosynthetic apparatus in transgenic tobacco. J. Integr. Plant Biol. 2008, 50, 613–621. [Google Scholar] [CrossRef]
- Uji, T.; Takahashi, M.; Saga, N.; Mikami, K. Visualization of nuclear localization of transcription factors with cyan and green fluorescent proteins in the red alga Porphyra yezoensis. Mar. Biotechnol. 2010, 12, 150–159. [Google Scholar] [CrossRef]
- Li, C.; Ariga, I.; Mikami, K. Difference in nitrogen starvation-inducible expression patterns among phylogenetically diverse ammonium transporter genes in the red seaweed Pyropia yezoensis. Am. J. Plant Sci. 2019, 10, 1325–1349. [Google Scholar] [CrossRef]
- Mikami, K.; Li, C.; Irie, R. A unique life cycle transition in the red seaweed Pyropia yezoensis depends on apospory. Commun. Biol. 2019, 2, 299. [Google Scholar] [CrossRef] [PubMed]
- Brawley, S.H.; Blouin, N.A.; Ficko-Blean, E.; Wheeler, G.L.; Lohr, M.; Goodson, H.V.; Jenkins, J.W.; Blaby-Haas, C.E.; Helliwell, K.E.; Chan, C.X.; et al. Insights into the red algae and eukaryotic evolution from the genome of Porphyra umbilicalis (Bangiophyceae, Rhodophyta). Proc. Natl. Acad. Sci. USA 2017, 114, E6361–E6370. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Uechi, A.; Saito, K. The zinc finger domain of RING finger protein 141 reveals a unique RING fold. Protein Sci. 2017, 26, 1681–1686. [Google Scholar] [CrossRef]
- Miyamoto, K.; Fujiwara, Y.; Saito, K. Zinc finger domain of the human DTX protein adopts a unique RING fold. Protein Sci. 2019, 28, 1151–1156. [Google Scholar] [CrossRef]
- Wu, X.; Huang, A.; Xu, M.; Wang, C.; Jia, Z.; Wang, G.; Niu, J. Variation of expression levels of seven housekeeping genes at different life-history stages in Porphyra yezoensis. PLoS ONE 2013, 8, e60740. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Cao, M.; Sun, P.; Liu, W.; Mao, Y. Selection of reference genes for gene expression normalization in Pyropia yezoensis, using quantitative real-time PCR. J. Appl. Phycol. 2015, 27, 1003–1010. [Google Scholar] [CrossRef]
- Jaimes-Miranda, F.; Chávez Montes, R.A. The plant MBF1 protein family: A bridge between stress and transcription. J. Exp. Bo. 2020, 71, 1782–1791. [Google Scholar] [CrossRef]
- Suri, S.S.; Dhindsa, R.S. A heat-activated MAP kinase (HAMK) as a mediator of heat shock response in tobacco cells. Plant Cell Environ. 2008, 31, 218–226. [Google Scholar] [CrossRef]
- Schmollinger, S.; Schulz-Raffelt, M.; Strenkert, D.; Veyel, D.; Vallon, O.; Schroda, M. Dissecting the heat stress response in Chlamydomonas by pharmaceutical and RNAi approaches reveals conserved and novel aspects. Mol. Plant 2013, 6, 1795–1813. [Google Scholar] [CrossRef]
- Rütgers, M.; Muranaka, L.S.; Schulz-Raffelt, M.; Thoms, S.; Schurig, J.; Willmund, F.; Schroda, M. Not changes in membrane fluidity but proteotoxic stress triggers heat shock protein expression in Chlamydomonas reinhardtii. Plant Cell Environ. 2017, 40, 2987–3001. [Google Scholar] [CrossRef] [PubMed]
- Saidi, Y.; Finka, A.; Muriset, M.; Bromberg, Z.; Weiss, Y.G.; Maathuis, F.J.; Goloubinoff, P. The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 2009, 21, 2829–2843. [Google Scholar] [CrossRef]
- Anderson, J.V.; Li, Q.B.; Haskell, D.W.; Guy, C.L. Structural organization of the spinach endoplasmic reticulum-luminal 70-kilodalton heat-shock cognate gene and expression of 70-kilodalton heat-shock genes during cold acclimation. Plant Physiol. 1994, 104, 1359. [Google Scholar] [CrossRef] [PubMed]
- Sabehat, A.; Lurie, S.; Weiss, D. Expression of small heat-shock proteins at low temperatures. Plant Physiol. 1998, 117, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Caitar, V.S.; de Carvalho, M.C.; Darben, L.M.; Kuwahara, M.K.; Nepomuceno, A.L.; Dias, W.P. Genome-wide analysis of the Hsp 20 gene family in soybean: Comprehensive sequence, genomic organization and expression profile analysis under abiotic and biotic stresses. BMC Genom. 2013, 14, 577. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Wang, J.; Li, Y.; Wu, S.; Yang, S.; Chao, J.; Chen, Y.; Zhang, S.; Shi, M.; Tian, W.M. Comparative transcriptome analysis reveals phytohormone signalings, heat shock module and ROS scavenger mediate the cold-tolerance of rubber tree. Sci. Rep. 2018, 8, 4931. [Google Scholar] [CrossRef]
- Hunter, D.A.; Napier, N.J.; Erridge, Z.A.; Saei, A.; Chen, R.K.Y.; McKenzie, M.J.; O’Donoghue, E.M.; Hunt, M.; Favre, L.; Lill, R.E.; et al. Transcriptome responses of ripe cherry tomato fruit exposed to chilling and rewarming identify reversible and irreversible gene expression changes. Front. Plant Sci. 2021, 12, 685416. [Google Scholar] [CrossRef]
- Li, Q.B.; Haskell, D.W.; Guy, C.L. Coordinate and non-coordinate expression of the stress 70 family and other molecular chaperones at high and low temperature in spinach and tomato. Plant Mol. Biol. 1999, 39, 21–34. [Google Scholar] [CrossRef]
- Volkov, R.A.; Panchuk, I.I.; Mullineaux, P.M.; Schoffl, F. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol. Biol. 2006, 61, 733–746. [Google Scholar] [CrossRef]
- Fedyaeva, A.V.; Stepanov, A.V.; Lyubushkina, I.V.; Pobezhimova, T.P.; Rikhvanov, E.G. Heat shock induces production of reactive oxygen species and increases inner mitochondrial membrane potential in winter wheat cells. Biochemistry 2014, 79, 1202–1210. [Google Scholar] [CrossRef]
- Awasthi, R.; Bhandari, K.; Nayyar, H. Temperature stress and redox homeostasis in agricultural crops. Front. Environ. Sci. 2015, 3, 11. [Google Scholar] [CrossRef]
- Sun, A.Z.; Guo, F.Q. Chloroplast Retrograde Regulation of Heat Stress Responses in Plants. Front. Plant Sci. 2016, 7, 398. [Google Scholar] [CrossRef] [PubMed]
- Katano, K.; Kataoka, R.; Fujii, M.; Suzuki, N. Differences between seedlings and flowers in anti-ROS based heat responses of Arabidopsis plants deficient in cyclic nucleotide gated channel 2. Plant Physiol. Biochem. 2018, 123, 288–296. [Google Scholar] [CrossRef] [PubMed]
- Jajic, I.; Sarna, T.; Szewczyk, G.; Strzalka, K. Changes in production of reactive oxygen species in illuminated thylakoids isolated during development and senescence of barley. J. Plant Physiol. 2015, 184, 49–56. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khoa, H.V.; Mikami, K. Membrane-Fluidization-Dependent and -Independent Pathways Are Involved in Heat-Stress-Inducible Gene Expression in the Marine Red Alga Neopyropia yezoensis. Cells 2022, 11, 1486. https://doi.org/10.3390/cells11091486
Khoa HV, Mikami K. Membrane-Fluidization-Dependent and -Independent Pathways Are Involved in Heat-Stress-Inducible Gene Expression in the Marine Red Alga Neopyropia yezoensis. Cells. 2022; 11(9):1486. https://doi.org/10.3390/cells11091486
Chicago/Turabian StyleKhoa, Ho Viet, and Koji Mikami. 2022. "Membrane-Fluidization-Dependent and -Independent Pathways Are Involved in Heat-Stress-Inducible Gene Expression in the Marine Red Alga Neopyropia yezoensis" Cells 11, no. 9: 1486. https://doi.org/10.3390/cells11091486
APA StyleKhoa, H. V., & Mikami, K. (2022). Membrane-Fluidization-Dependent and -Independent Pathways Are Involved in Heat-Stress-Inducible Gene Expression in the Marine Red Alga Neopyropia yezoensis. Cells, 11(9), 1486. https://doi.org/10.3390/cells11091486