Proteasomes of Autophagy-Deficient Cells Exhibit Alterations in Regulatory Proteins and a Marked Reduction in Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dictyostelium Strains
2.2. Vector Construction and Transformation
2.3. Native Gel Immunoblotting of the Proteasome
2.4. SDS PAGE and Western Blotting
2.5. Co-Immunoprecipitation (IP) Assays
2.6. Mass Spectrometry
2.7. Miscellaneous Methods
3. Results
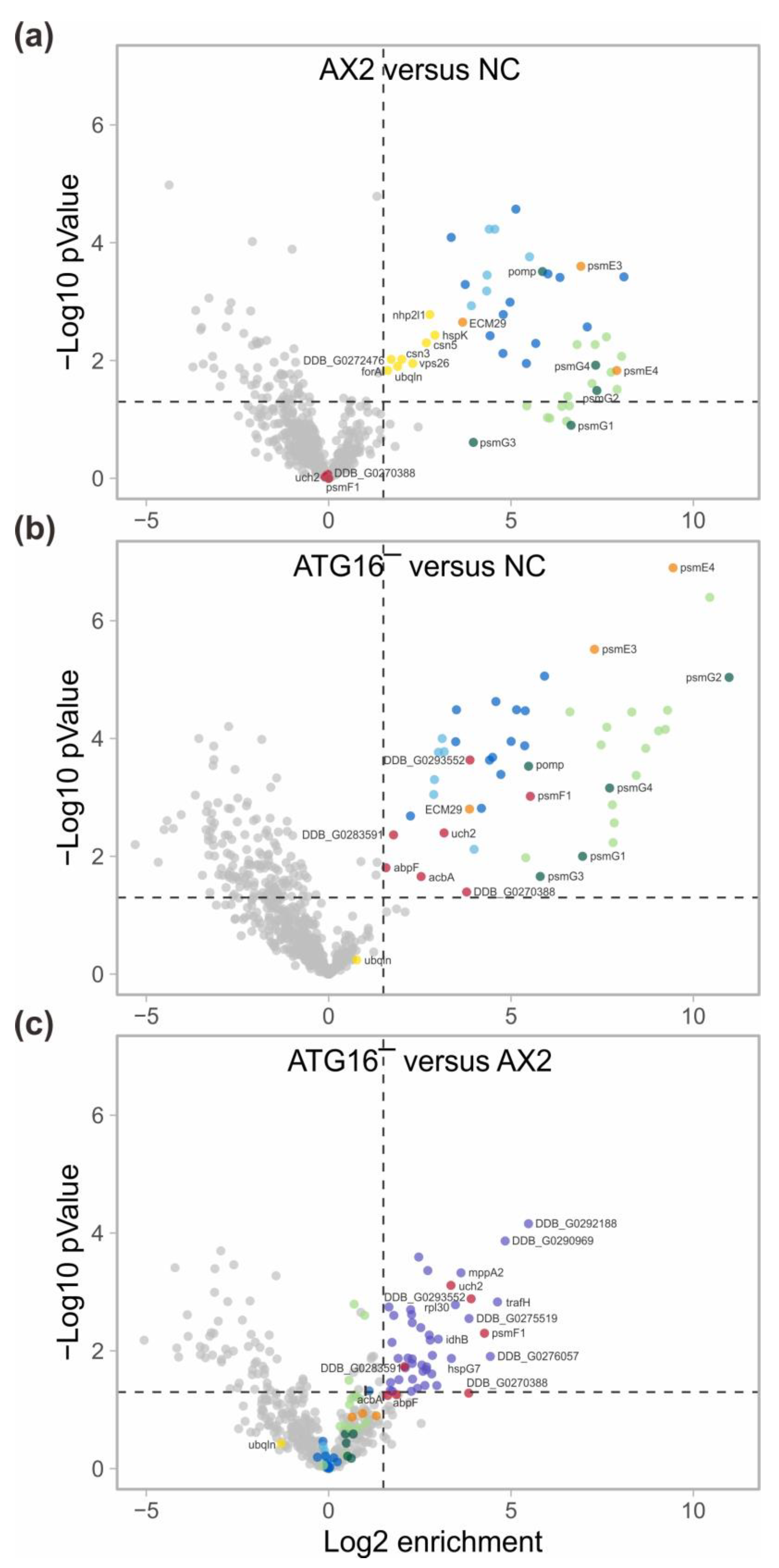
3.1. Autophagy Mutants Exhibit a Strong Decrease in Proteasomal Activity but No Quantitative Change in Proteasomal Proteins
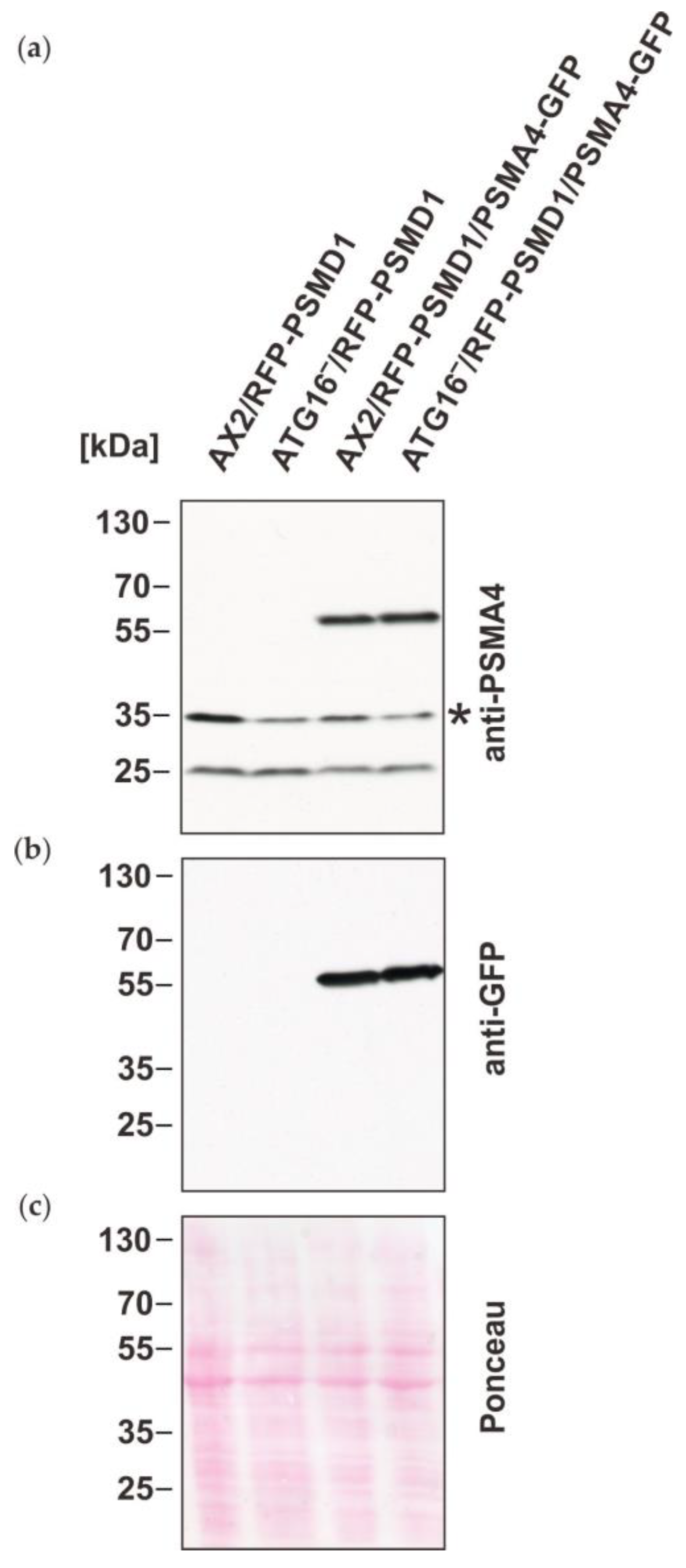
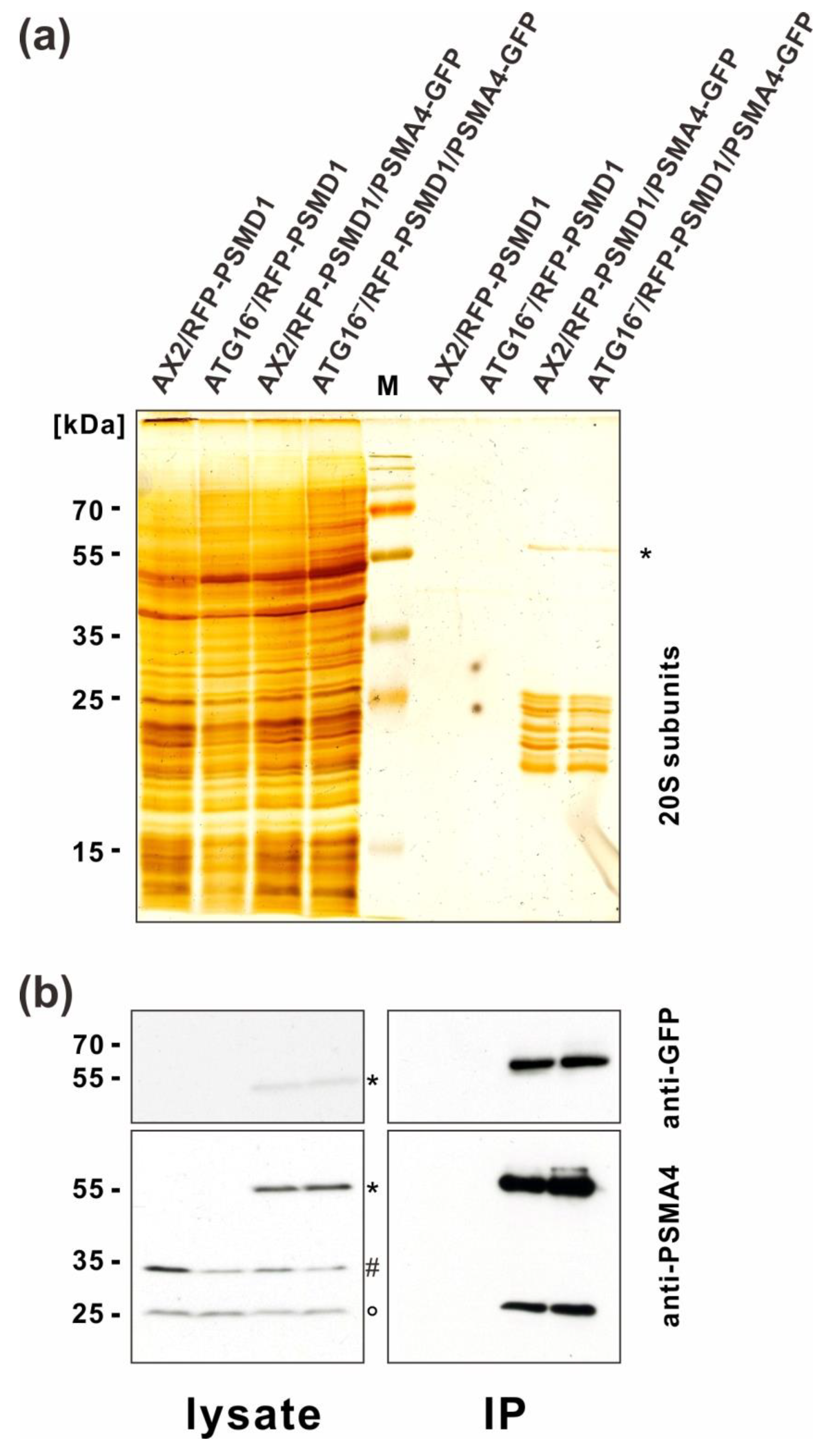
3.2. Immunoprecipitation of Tagged Proteasomes and Mass Spectrometry Support the Finding of Similar Amounts of Proteasomes in AX2 and ATG16− Cells
3.3. Some Proteasome-Associated Proteins Were Ubiquitinated
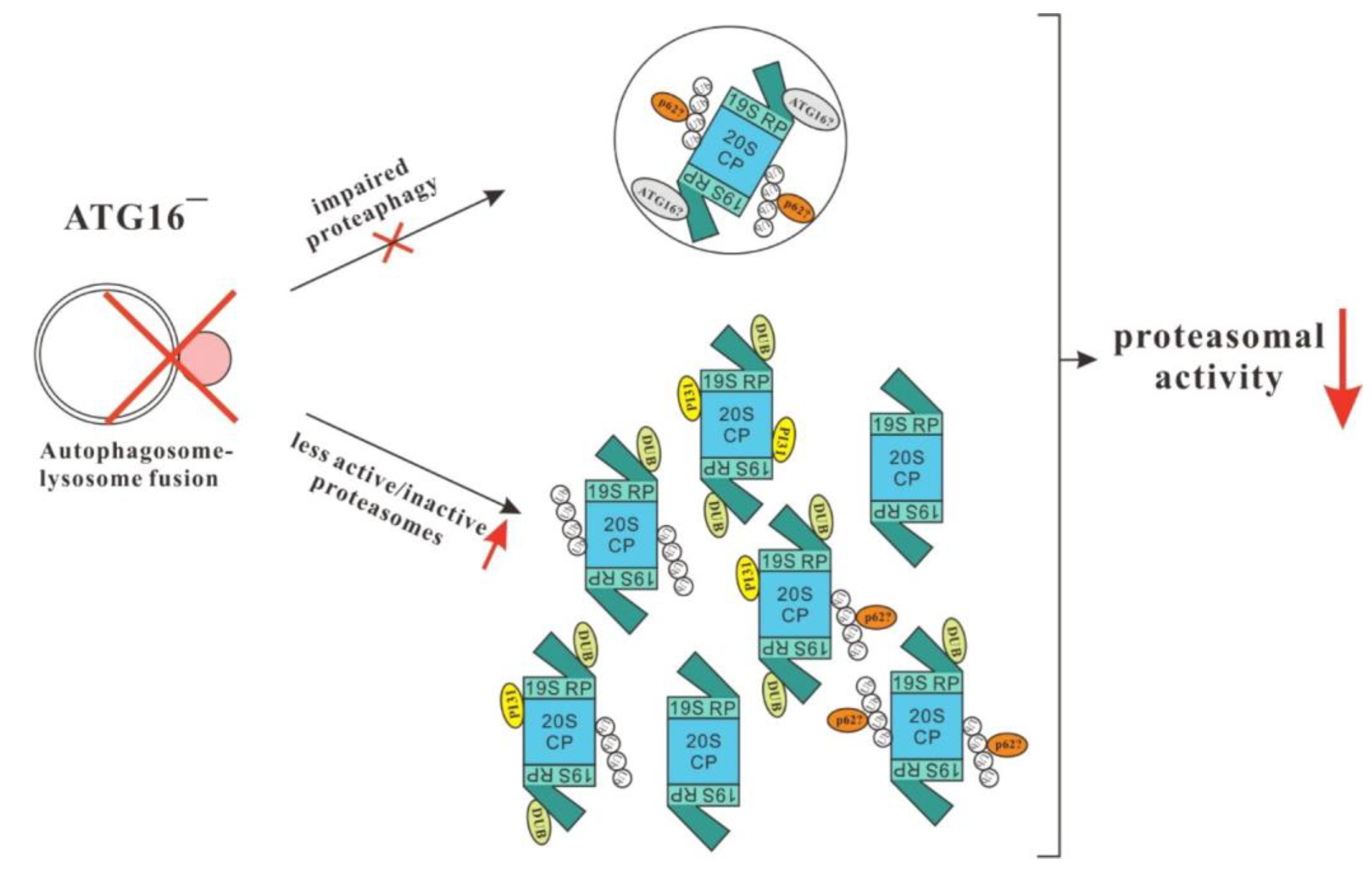
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, H.M.; Yu, Y.; Cheng, Y. Structure characterization of the 26S proteasome. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2011, 1809, 67–79. [Google Scholar] [CrossRef]
- Schreiber, A.; Peter, M. Substrate recognition in selective autophagy and the ubiquitin–proteasome system. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2014, 1843, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2009, 584, 1393–1398. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Mansilla, A.; Menzies, F.M.; Rubinsztein, D.C. Autophagy Inhibition Compromises Degradation of Ubiquitin-Proteasome Pathway Substrates. Mol. Cell 2009, 33, 517–527. [Google Scholar] [CrossRef]
- Driscoll, J.J.; Chowdhury, R.D. Molecular crosstalk between the proteasome, aggresomes and autophagy: Translational potential and clinical implications. Cancer Lett. 2012, 325, 147–154. [Google Scholar] [CrossRef]
- Kraft, C.; Peter, M.; Hofmann, K. Selective autophagy: Ubiquitin-mediated recognition and beyond. Nature 2010, 12, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Gammoh, N.; Wong, P.-M.; Erdjument-Bromage, H.; Tempst, P.; Jiang, X. Processing of autophagic protein LC3 by the 20S proteasome. Autophagy 2010, 6, 126–137. [Google Scholar] [CrossRef]
- Hu, G.; Rios, L.; Yan, Z.; Jasper, A.M.; Luera, D.; Luo, S.; Rao, H. Autophagy regulator Atg9 is degraded by the proteasome. Biochem. Biophys. Res. Commun. 2020, 522, 254–258. [Google Scholar] [CrossRef]
- Haller, M.; Hock, A.K.; Giampazolias, E.; Oberst, A.; Green, D.R.; Debnath, J.; Ryan, K.M.; Vousden, K.H.; Tait, S.W. Ubiquitination and proteasomal degradation of ATG12 regulates its proapoptotic activity. Autophagy 2014, 10, 2269–2278. [Google Scholar] [CrossRef]
- Liu, C.-C.; Lin, Y.-C.; Chen, Y.-H.; Chen, C.-M.; Pang, L.-Y.; Chen, H.-A.; Wu, P.-R.; Lin, M.-Y.; Jiang, S.-T.; Tsai, T.-F.; et al. Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination. Mol. Cell 2016, 61, 84–97. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Saitoh, T.; Kageyama, S.; Akira, S.; Noda, T.; Yoshimori, T. Differential involvement of atg16l1 in crohn disease and canonical autophagy: Analysis of the organization of the atg16l1 complex in fibroblasts. J. Biol. Chem. 2009, 284, 32602–32609. [Google Scholar] [CrossRef] [PubMed]
- Cuervo, A.M.; Palmer, A.; Rivett, A.J.; Knecht, E. Degradation of proteasomes by lysosomes in rat liver. Eur. J. Biochem. 1995, 227, 792–800. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26s proteasome is mediated by the dual atg8/ubiquitin receptor rpn10 in arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Cohen-Kaplan, V.; Livneh, I.; Avni, N.; Fabre, B.; Ziv, T.; Kwon, Y.T.; Ciechanover, A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, E7490–E7499. [Google Scholar] [CrossRef]
- Marshall, R.S.; McLoughlin, F.; Vierstra, R.D. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell Rep. 2016, 16, 1717–1732. [Google Scholar] [CrossRef]
- Xiong, Q.; Fischer, S.; Karow, M.; Müller, R.; Meßling, S.; Eichinger, L. ATG16 mediates the autophagic degradation of the 19S proteasomal subunits PSMD1 and PSMD2. Eur. J. Cell Biol. 2018, 97, 523–532. [Google Scholar] [CrossRef]
- Xiong, Q.; Li, W.; Li, P.; Yang, M.; Wu, C.; Eichinger, L. The Role of ATG16 in Autophagy and The Ubiquitin Proteasome System. Cells 2018, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; Vierstra, R.D. Dynamic Regulation of the 26S Proteasome: From Synthesis to Degradation. Front. Mol. Biosci. 2019, 6, 40. [Google Scholar] [CrossRef]
- Ding, W.-X.; Ni, H.-M.; Gao, W.; Yoshimori, T.; Stolz, D.B.; Ron, D.; Yin, X.-M. Linking of Autophagy to Ubiquitin-Proteasome System Is Important for the Regulation of Endoplasmic Reticulum Stress and Cell Viability. Am. J. Pathol. 2007, 171, 513–524. [Google Scholar] [CrossRef]
- Pandey, U.B.; Nie, Z.; Batlevi, Y.; McCray, B.A.; Ritson, G.P.; Nedelsky, N.B.; Schwartz, S.L.; DiProspero, N.A.; Knight, M.A.; Schuldiner, O.; et al. Hdac6 rescues neurodegeneration and provides an essential link between autophagy and the ups. Nature 2007, 447, 859–863. [Google Scholar] [CrossRef] [PubMed]
- Rideout, H.J.; Lang-Rollin, I.; Stefanis, L. Involvement of macroautophagy in the dissolution of neuronal inclusions. Int. J. Biochem. Cell Biol. 2004, 36, 2551–2562. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.J.; Yu, J.; Wong, S.H.; Cheng, A.S.; Chan, F.K.; Ng, S.S.; Cho, C.H.; Sung, J.J.; Wu, W.K. A novel crosstalk between two major protein degradation systems: Regulation of proteasomal activity by autophagy. Autophagy 2013, 9, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.-I.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, J. Inhibition of lysosomal functions reduces proteasomal activity. Neurosci. Lett. 2009, 456, 15–19. [Google Scholar] [CrossRef]
- Qiao, L.; Hamamichi, S.; A Caldwell, K.; A Caldwell, G.; A Yacoubian, T.; Wilson, S.; Xie, Z.-L.; Speake, L.D.; Parks, R.; Crabtree, D.; et al. Lysosomal enzyme cathepsin D protects against alpha-synuclein aggregation and toxicity. Mol. Brain 2008, 1, 17–18. [Google Scholar] [CrossRef]
- Xiong, Q.; Unal, C.; Matthias, J.; Steinert, M.; Eichinger, L. The phenotypes of atg9, atg16 and atg9/16 knock-out mutants imply autophagy-dependent and -independent functions. Open Biol. 2015, 5, 150008. [Google Scholar] [CrossRef]
- Arhzaouy, K.; Strucksberg, K.H.; Tung, S.M.; Tangavelou, K.; Stumpf, M.; Faix, J.; Schroder, R.; Clemen, C.S.; Eichinger, L. Heteromeric p97/p97r155c complexes induce dominant negative changes in wild-type and autophagy 9-deficient dictyostelium strains. PLoS ONE 2012, 7, e46879. [Google Scholar] [CrossRef]
- Meßling, S.; Matthias, J.; Xiong, Q.; Fischer, S.; Eichinger, L. The two Dictyostelium discoideum autophagy 8 proteins have distinct autophagic functions. Eur. J. Cell Biol. 2017, 96, 312–324. [Google Scholar] [CrossRef]
- Fischer, S.; Rijal, R.; Frommolt, P.; Wagle, P.; Konertz, R.; Faix, J.; Meßling, S.; Eichinger, L. Functional Characterization of Ubiquitin-Like Core Autophagy Protein ATG12 in Dictyostelium discoideum. Cells 2019, 8, 72. [Google Scholar] [CrossRef]
- Karow, M.; Fischer, S.; Messling, S.; Konertz, R.; Riehl, J.; Xiong, Q.; Rijal, R.; Wagle, P.; Clemen, C.S.; Eichinger, L. Functional characterisation of the autophagy atg12~5/16 complex in dictyostelium discoideum. Cells 2020, 9, 1179. [Google Scholar] [CrossRef] [PubMed]
- Mesquita, A.; Cardenal Muñoz, E.; Dominguez, E.; Muñoz-Braceras, S.; Nuñez-Corcuera, B.; Phillips, B.A.; Tábara, L.C.; Xiong, Q.; Coria, R.; Eichinger, L.; et al. Autophagy in Dictyostelium: Mechanisms, regulation and disease in a simple biomedical model. Autophagy 2017, 13, 24–40. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Otto, G.P.; Wu, M.Y.; Kazgan, N.; Anderson, O.R.; Kessin, R.H. Dictyostelium Macroautophagy Mutants Vary in the Severity of Their Developmental Defects. J. Biol. Chem. 2004, 279, 15621–15629. [Google Scholar] [CrossRef] [PubMed]
- Calvo-Garrido, J.; Escalante, R. Autophagy dysfunction and ubiquitin-positive protein aggregates in Dictyostelium cells lacking Vmp1. Autophagy 2010, 6, 100–109. [Google Scholar] [CrossRef]
- Calvo-Garrido, J.; King, J.S.; Muñoz-Braceras, S.; Escalante, R. Vmp1 Regulates PtdIns3P Signaling During Autophagosome Formation inDictyostelium discoideum. Traffic 2014, 15, 1235–1246. [Google Scholar] [CrossRef]
- Tung, S.M.; Ünal, C.; Ley, A.; Peña, C.; Tunggal, B.; Noegel, A.A.; Krut, O.; Steinert, M.; Eichinger, L. Loss of Dictyostelium ATG9 results in a pleiotropic phenotype affecting growth, development, phagocytosis and clearance and replication of Legionella pneumophila. Cell. Microbiol. 2010, 12, 765–780. [Google Scholar] [CrossRef]
- Muñoz-Braceras, S.; Calvo, R.; Escalante, R. TipC and the chorea-acanthocytosis protein VPS13A regulate autophagy in Dictyostelium and human HeLa cells. Autophagy 2015, 11, 918–927. [Google Scholar] [CrossRef]
- Xiong, Q.; Song, N.; Li, P.; Fischer, S.; Konertz, R.; Wagle, P.; Glockner, G.; Wu, C.; Eichinger, L. Rna(seq) and quantitative proteomic analysis of dictyostelium knock-out cells lacking the core autophagy proteins atg9 and/or atg16. BMC Genom. 2021, 22, 444. [Google Scholar]
- Brink, M.; Gerisch, G.; Isenberg, G.; Noegel, A.A.; Segall, J.E.; Wallraff, E.; Schleicher, M. A Dictyostelium mutant lacking an F-actin cross-linking protein, the 120-kD gelation factor. J. Cell Biol. 1990, 111, 1477–1489. [Google Scholar] [CrossRef]
- Watts, D.J.; Ashworth, J.M. Growth of myxameobae of the cellular slime mould dictyostelium discoideum in axenic culture. Biochem. J. 1970, 119, 171–174. [Google Scholar] [CrossRef]
- Williams, K.L.; Newell, P.C. A genetic study of aggregation in the cellular slime mould dictyostelium discoideum using complementation analysis. Genetics 1976, 82, 287–307. [Google Scholar] [CrossRef]
- Raper, K.B. Dictyostelium discoideum, a new species of slime mold from decaying forest leaves. J. Agr. Res. 1935, 50, 135–147. [Google Scholar]
- Myeku, N.; Metcalfe, M.J.; Huang, Q.; Figueiredo-Pereira, M. Assessment of Proteasome Impairment and Accumulation/Aggregation of Ubiquitinated Proteins in Neuronal Cultures. Neurodegener. Methods Protoc. 2011, 793, 273–296. [Google Scholar] [CrossRef]
- Elsasser, S.; Schmidt, M.; Finley, D. Characterization of the Proteasome Using Native Gel Electrophoresis. Methods Enzymol. 2005, 398, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Noegel, A.A.; Blau-Wasser, R.; Sultana, H.; Müller, R.; Israel, L.; Schleicher, M.; Patel, H.; Weijer, C.J. The Cyclase-associated Protein CAP as Regulator of Cell Polarity and cAMP Signaling in Dictyostelium. Mol. Biol. Cell 2004, 15, 934–945. [Google Scholar] [CrossRef]
- Schauer, T.M.; Nesper, M.; Kehl, M.; Lottspeich, F.; Müller-Taubenberger, A.; Gerisch, G.; Baumeister, W. Proteasomes from Dictyostelium discoideum: Characterization of Structure and Function. J. Struct. Biol. 1993, 111, 135–147. [Google Scholar] [CrossRef]
- Simpson, P.A.; Spudich, J.A.; Parham, P. Monoclonal antibodies prepared against Dictyostelium actin: Characterization and interactions with actin. J. Cell Biol. 1984, 99, 287–295. [Google Scholar] [CrossRef]
- Farbrother, P.; Wagner, C.; Na, J.; Tunggal, B.; Morio, T.; Urushihara, H.; Tanaka, Y.; Schleicher, M.; Steinert, M.; Eichinger, L. Dictyostelium transcriptional host cell response upon infection with Legionella. Cell. Microbiol. 2006, 8, 438–456. [Google Scholar] [CrossRef]
- Wang, Y.; Le, W.D. Autophagy and ubiquitin-proteasome system. Adv. Exp. Med. Biol. 2019, 1206, 527–550. [Google Scholar]
- Bartel, B. Proteaphagy—Selective Autophagy of Inactive Proteasomes. Mol. Cell 2015, 58, 970–971. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.C.; Higgins, N.R.; Phung, T.H.; Monteiro, M.J. UBQLN proteins in health and disease with a focus on UBQLN2 in ALS/FTD. FEBS J. 2022, 289, 6132–6153. [Google Scholar] [CrossRef]
- Chu-Ping, M.; Slaughter, C.A.; DeMartino, G.N. Purification and characterization of a protein inhibitor of the 20S proteasome (macropain). Biochim. Biophys. Acta (BBA) Protein Struct. Mol. Enzym. 1992, 1119, 303–311. [Google Scholar] [CrossRef]
- Rawson, S.; Walsh, R.M.; Velez, B.; Schnell, H.M.; Jiao, F.; Blickling, M.; Ang, J.; Bhanu, M.K.; Huang, L.; Hanna, J. Yeast PI31 inhibits the proteasome by a direct multisite mechanism. Nat. Struct. Mol. Biol. 2022, 29, 791–800. [Google Scholar] [CrossRef]
- Clemen, C.S.; Marko, M.; Strucksberg, K.-H.; Behrens, J.; Wittig, I.; Gärtner, L.; Winter, L.; Chevessier, F.; Matthias, J.; Türk, M.; et al. VCP and PSMF1: Antagonistic regulators of proteasome activity. Biochem. Biophys. Res. Commun. 2015, 463, 1210–1217. [Google Scholar] [CrossRef]
- Ge, J.; Hu, W.; Zhou, H.; Yu, J.; Sun, C.; Chen, W. Ubiquitin carboxyl-terminal hydrolase isozyme L5 inhibits human glioma cell migration and invasion via downregulating SNRPF. Oncotarget 2017, 8, 113635–113649. [Google Scholar] [CrossRef] [PubMed]
- Hameed, D.S.; Ovaa, H.; Noort, G.J.V.D.H.V.; Sapmaz, A. Inhibiting UCH-L5: Rational Design of a Cyclic Ubiquitin-Based Peptide Inhibitor. Front. Mol. Biosci. 2022, 9, 866467. [Google Scholar] [CrossRef] [PubMed]
- Santonico, E. Old and New Concepts in Ubiquitin and NEDD8 Recognition. Biomolecules 2020, 10, 566. [Google Scholar] [CrossRef]
- Kim, D.-Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced Proteomic Analyses Yield a Deep Catalog of Ubiquitylation Targets in Arabidopsis. Plant Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef]
- Finley, D.; Ulrich, H.D.; Sommer, T.; Kaiser, P. The ubiquitin-proteasome system of saccharomyces cerevisiae. Genetics 2012, 192, 319–360. [Google Scholar] [CrossRef] [PubMed]
- Yip, M.C.J.; Bodnar, N.O.; Rapoport, T.A. Ddi1 is a ubiquitin-dependent protease. Proc. Natl. Acad. Sci. USA 2020, 117, 7776–7781. [Google Scholar] [CrossRef] [PubMed]
- Fey, P.; Dodson, R.J.; Basu, S.; Hartline, E.C.; Chisholm, R.L. Dictybase and the dicty stock center (version 2.0)—A progress report. Int. J. Dev. Biol. 2019, 63, 563–572. [Google Scholar] [CrossRef] [PubMed]

| Strains | Summary | Reference |
|---|---|---|
| AX2 | Axenically growing derivate of wild isolate NC-4 | [43] |
| ATG9− | ATG9 knock-out mutant | [37] |
| ATG16− | ATG16 knock-out mutant | [27] |
| ATG9−/16− | ATG9 and ATG16 double knock-out mutant | [27] |
| AX2/RFP-PSMD1 | AX2 cells expressing RFP-tagged PSMD1 | [17] |
| AX2/RFP-PSMD1/PSMA4-GFP | AX2 cells expressing RFP-tagged PSMD1 and GFP-tagged PSMA4 | This work |
| ATG16−/RFP-PSMD1 | ATG16 null cells expressing RFP-tagged PSMD1 | [17] |
| ATG16−/RFP-PSMD1/PSMA4-GFP | ATG16 null cells expressing RFP-tagged PSMD1 and GFP-tagged PSMA4 | This work |
| UniProt ID | DDB_G ID | Description | AA | Position | Mean Intensities (log2) | ||
|---|---|---|---|---|---|---|---|
| AX2 | ATG16− | ||||||
| Exclusive in AX2 | Q54SZ7 | DDB_G0282111 | Uncharacterized protein | K | 700 | 24.51 | 0.00 |
| Q75JX9 | DDB_G0272214 | Uncharacterized protein | K | 303 | 24.20 | 0.00 | |
| Q54LV8 | DDB_G0286389 | 60S ribosomal protein L34 | K | 106 | 21.81 | 0.00 | |
| Ubiquitinated in AX2 and ATG16− cells | Q54VN6 | DDB_G0280229 | 60S ribosomal protein L24 | K | 70 | 26.22 | 27.42 |
| Q55B20 | DDB_G0271470 | Uncharacterized protein | K | 3 | 26.21 | 28.01 | |
| Q86HT3 | DDB_G0274439 | BolA-like protein | K | 19 | 25.16 | 27.28 | |
| Q54KW5 | DDB_G0287061 | Uncharacterized protein | K | 927 | 24.20 | 26.19 | |
| Q54QR2 | DDB_G0283679 | Proteasome subunit beta type-7 | K | 63 | 23.78 | 25.52 | |
| P14794 | DDB_G0280755 | Ubiquitin-60S ribosomal protein L40 | K | 48 | 23.61 | 25.40 | |
| Exclusive in ATG16− | B0G0Z1 | DDB_G0270910 | Eukaryotic translation initiation factor 4E | K | 870 | 0.00 | 30.46 |
| Q54T94 | DDB_G0281913 | Uncharacterized protein | K | 146 | 0.00 | 28.58 | |
| Q55BM4 | DDB_G0269190 | T-complex protein 1 subunit alpha | K | 468 | 0.00 | 26.14 | |
| Q55GK4 | DDB_G0267634 | Uncharacterized protein | K | 827 | 0.00 | 26.05 | |
| Q54ED9 | DDB_G0291538 | Uncharacterized protein | K | 3 | 0.00 | 25.71 | |
| Q54HN5 | DDB_G0289337 | Uncharacterized protein | K | 3 | 0.00 | 25.64 | |
| Q55CG3 | DDB_G0271028 | Uncharacterized protein | K | 18 | 0.00 | 25.50 | |
| Q54JB0 | DDB_G0288187 | DDI1 homolog | K | 55 | 0.00 | 24.82 | |
| Q54QE4 | DDB_G0283987 | Bifunctional purine synth. protein purC/E | K | 234 | 0.00 | 24.71 | |
| Q8MMQ2 | DDB_G0275539 | Uncharacterized protein | K | 86 | 0.00 | 24.64 | |
| Q54GS9 | DDB_G0289933 | Uncharacterized protein | K | 346 | 0.00 | 24.30 | |
| Q54YL1 | DDB_G0278621 | Uncharacterized protein | K | 604 | 0.00 | 23.89 | |
| Q559T1 | DDB_G0272438 | Uncharacterized protein | K | 231 | 0.00 | 23.73 | |
| Q55C03 | DDB_G0270284 | Uncharacterized protein | K | 373 | 0.00 | 23.67 | |
| Q54WT5 | DDB_G0279449 | Uncharacterized protein | K | 115 | 0.00 | 23.24 | |
| Q54GX0 | DDB_G0289871 | Uncharacterized protein | K | 2 | 0.00 | 22.15 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, Q.; Feng, R.; Fischer, S.; Karow, M.; Stumpf, M.; Meßling, S.; Nitz, L.; Müller, S.; Clemen, C.S.; Song, N.; et al. Proteasomes of Autophagy-Deficient Cells Exhibit Alterations in Regulatory Proteins and a Marked Reduction in Activity. Cells 2023, 12, 1514. https://doi.org/10.3390/cells12111514
Xiong Q, Feng R, Fischer S, Karow M, Stumpf M, Meßling S, Nitz L, Müller S, Clemen CS, Song N, et al. Proteasomes of Autophagy-Deficient Cells Exhibit Alterations in Regulatory Proteins and a Marked Reduction in Activity. Cells. 2023; 12(11):1514. https://doi.org/10.3390/cells12111514
Chicago/Turabian StyleXiong, Qiuhong, Rong Feng, Sarah Fischer, Malte Karow, Maria Stumpf, Susanne Meßling, Leonie Nitz, Stefan Müller, Christoph S. Clemen, Ning Song, and et al. 2023. "Proteasomes of Autophagy-Deficient Cells Exhibit Alterations in Regulatory Proteins and a Marked Reduction in Activity" Cells 12, no. 11: 1514. https://doi.org/10.3390/cells12111514
APA StyleXiong, Q., Feng, R., Fischer, S., Karow, M., Stumpf, M., Meßling, S., Nitz, L., Müller, S., Clemen, C. S., Song, N., Li, P., Wu, C., & Eichinger, L. (2023). Proteasomes of Autophagy-Deficient Cells Exhibit Alterations in Regulatory Proteins and a Marked Reduction in Activity. Cells, 12(11), 1514. https://doi.org/10.3390/cells12111514









