Genome Editing for Cystic Fibrosis
Abstract
1. Introduction
2. CF Gene Mutations and CF Clinical Diseases
2.1. CF Genomic Mutations
2.2. CF Clinical Diseases
3. General Considerations in Gene Editing Design for CF
4. Gene Editing Tools for Selection
4.1. Cas Nuclease Editors
4.2. Base Editors
4.3. Prime Editors
5. CRISPR-Based CFTR Gene Editing
5.1. In Vitro Correction of CFTR Mutations
5.2. Creation of CF Models
5.3. In Vivo Correction of CFTR Mutations
6. Non-CRISPR-Based CFTR Gene Editing
7. Comparison of CRISPR-Editors and Non-CRISPR PNA Editors for CF Gene Editing
8. Barriers to Overcome to Improve In Vivo Gene Editing Efficiency for CF
8.1. Gene Editor Delivery through Epithelial Lumens
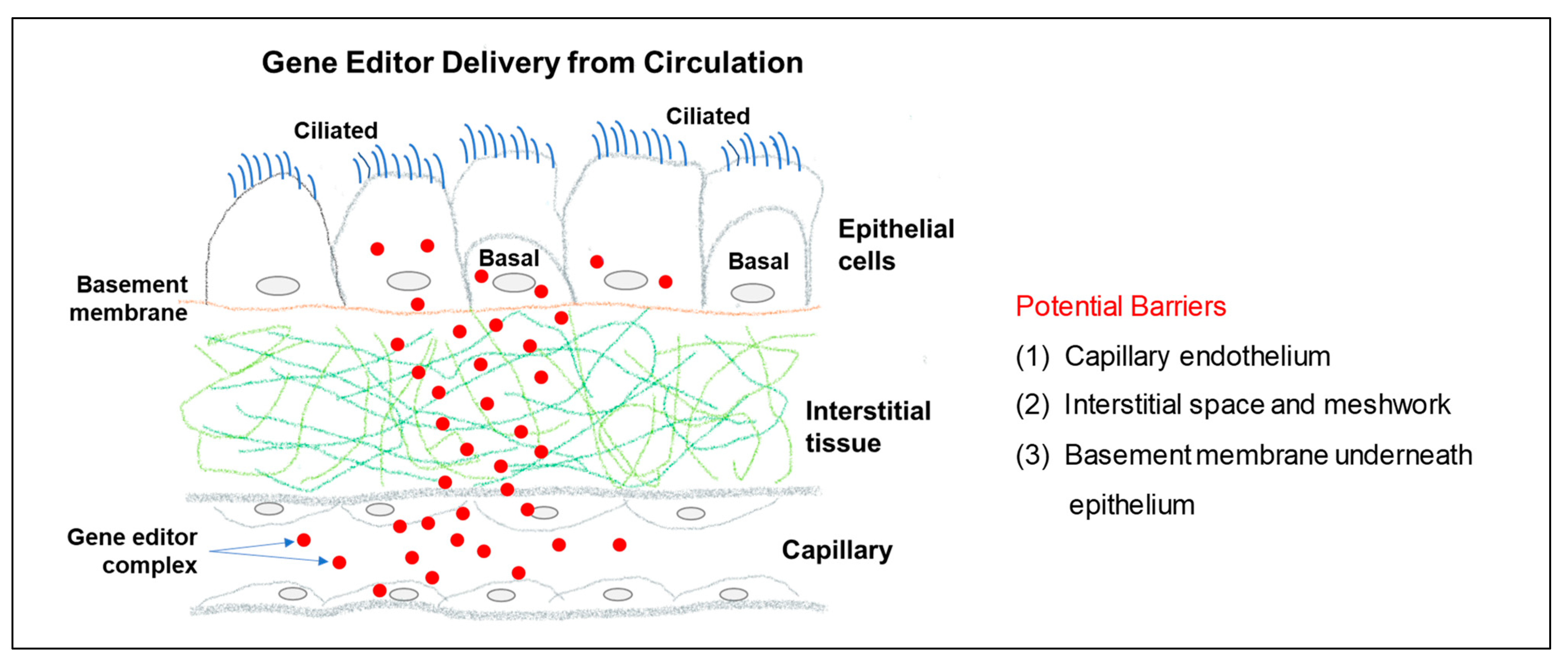
8.2. Gene Editor Delivery through the Circulatio
9. Prospects for a Cure in CF
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Garcia-Martinez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Masepohl, B.; Gorlitz, K.; Bohme, H. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim. Biophys. Acta 1996, 1307, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Diez-Villasenor, C.; Soria, E.; Juez, G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol. Microbiol. 2000, 36, 244–246. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Gostimskaya, I. CRISPR-Cas9: A History of Its Discovery and Ethical Considerations of Its Use in Genome Editing. Biochem. (Mosc.) 2022, 87, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Garneau, J.E.; Dupuis, M.E.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadan, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.S.; Domm, J.; Eustace, B.K.; Foell, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Welsh, M.J.; Boat, T.F.; Tsui, L.C.; Beaudet, A.L. Cystic Fibrosis. In The Metabolic Basis of Inherited Disease; Scriver, C.R., Beaudet, A.L., Sly, W.S., Valle, D., Eds.; McGraw-Hill Inc.: New York, NY, USA, 1995; pp. 3799–3876. [Google Scholar]
- Shteinberg, M.; Haq, I.J.; Polineni, D.; Davies, J.C. Cystic fibrosis. Lancet 2021, 397, 2195–2211. [Google Scholar] [CrossRef] [PubMed]
- Porteous, D.J.; Dorin, J.R. Cystic fibrosis. 3. Cloning the cystic fibrosis gene: Implications for diagnosis and treatment. Thorax 1991, 46, 46–55. [Google Scholar] [CrossRef]
- Rommens, J.M.; Iannuzzi, M.C.; Kerem, B.; Drumm, M.L.; Melmer, G.; Dean, M.; Rozmahel, R.; Cole, J.L.; Kennedy, D.; Hidaka, N.; et al. Identification of the cystic fibrosis gene: Chromosome walking and jumping. Science 1989, 245, 1059–1065. [Google Scholar] [CrossRef]
- Anderson, M.P.; Gregory, R.J.; Thompson, S.; Souza, D.W.; Paul, S.; Mulligan, R.C.; Smith, A.E.; Welsh, M.J. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 1991, 253, 202–205. [Google Scholar] [CrossRef]
- Kerem, B.; Rommens, J.M.; Buchanan, J.A.; Markiewicz, D.; Cox, T.K.; Chakravarti, A.; Buchwald, M.; Tsui, L.C. Identification of the cystic fibrosis gene: Genetic analysis. Science 1989, 245, 1073–1080. [Google Scholar] [CrossRef]
- Cutting, G.R. Cystic fibrosis genetics: From molecular understanding to clinical application. Nat. Rev. Genet. 2015, 16, 45–56. [Google Scholar] [CrossRef]
- Sosnay, P.R.; Raraigh, K.S.; Gibson, R.L. Molecular Genetics of Cystic Fibrosis Transmembrane Conductance Regulator: Genotype and Phenotype. Pediatr. Clin. N. Am. 2016, 63, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Solomon, G.M.; Marshall, S.G.; Ramsey, B.W.; Rowe, S.M. Breakthrough therapies: Cystic fibrosis (CF) potentiators and correctors. Pediatr. Pulmonol. 2015, 50 (Suppl. 40), S3–S13. [Google Scholar] [CrossRef] [PubMed]
- Veit, G.; Avramescu, R.G.; Chiang, A.N.; Houck, S.A.; Cai, Z.; Peters, K.W.; Hong, J.S.; Pollard, H.B.; Guggino, W.B.; Balch, W.E.; et al. From CFTR biology toward combinatorial pharmacotherapy: Expanded classification of cystic fibrosis mutations. Mol. Biol. Cell. 2016, 27, 424–433. [Google Scholar] [CrossRef]
- Foundation, C.F. Cystic Fibrosis Foundation Patient Rigestry: 2021 Annual Data Report; Cystic Fibrosis Foundation: Bethesda, MD, USA, 2021. [Google Scholar]
- Davis, P.B.; Drumm, M.; Konstan, M.W. Cystic fibrosis. Am. J. Respir. Crit. Care Med. 1996, 154, 1229–1256. [Google Scholar] [CrossRef] [PubMed]
- Stoltz, D.A.; Meyerholz, D.K.; Welsh, M.J. Origins of cystic fibrosis lung disease. N. Engl. J. Med. 2015, 372, 1574–1575. [Google Scholar] [CrossRef]
- Boucher, R.C. Cystic fibrosis: A disease of vulnerability to airway surface dehydration. Trends Mol. Med. 2007, 13, 231–240. [Google Scholar] [CrossRef]
- Andersen, D.H. Cystic fibrosis of the pancreas and its relation to celiac disease: A clinical and pathological study. Am. J. Dis. Child. 1938, 56, 344–399. [Google Scholar] [CrossRef]
- Gibson-Corley, K.N.; Meyerholz, D.K.; Engelhardt, J.F. Pancreatic pathophysiology in cystic fibrosis. J. Pathol. 2016, 238, 311–320. [Google Scholar] [CrossRef]
- Moran, A.; Dunitz, J.; Nathan, B.; Saeed, A.; Holme, B.; Thomas, W. Cystic fibrosis-related diabetes: Current trends in prevalence, incidence, and mortality. Diabetes Care 2009, 32, 1626–1631. [Google Scholar] [CrossRef]
- Sathe, M.; Houwen, R. Meconium ileus in Cystic Fibrosis. J. Cyst. Fibros. 2017, 16 (Suppl. 2), S32–S39. [Google Scholar] [CrossRef]
- Sankararaman, S.; Schindler, T.; Sferra, T.J. Management of Exocrine Pancreatic Insufficiency in Children. Nutr. Clin. Pract. 2019, 34 (Suppl. 1), S27–S42. [Google Scholar] [CrossRef] [PubMed]
- Houwen, R.H.; van der Doef, H.P.; Sermet, I.; Munck, A.; Hauser, B.; Walkowiak, J.; Robberecht, E.; Colombo, C.; Sinaasappel, M.; Wilschanski, M.; et al. Defining DIOS and constipation in cystic fibrosis with a multicentre study on the incidence, characteristics, and treatment of DIOS. J. Pediatr. Gastroenterol. Nutr. 2010, 50, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Kelly, T.; Buxbaum, J. Gastrointestinal Manifestations of Cystic Fibrosis. Dig. Dis. Sci. 2015, 60, 1903–1913. [Google Scholar] [CrossRef] [PubMed]
- Dorsey, J.; Gonska, T. Bacterial overgrowth, dysbiosis, inflammation, and dysmotility in the Cystic Fibrosis intestine. J. Cyst. Fibros. 2017, 16 (Suppl. 2), S14–S23. [Google Scholar] [CrossRef]
- Rogers, G.B.; Narkewicz, M.R.; Hoffman, L.R. The CF gastrointestinal microbiome: Structure and clinical impact. Pediatr. Pulmonol. 2016, 51, S35–S44. [Google Scholar] [CrossRef]
- Haber, H.P.; Benda, N.; Fitzke, G.; Lang, A.; Langenberg, M.; Riethmuller, J.; Stern, M. Colonic wall thickness measured by ultrasound: Striking differences in patients with cystic fibrosis versus healthy controls. Gut 1997, 40, 406–411. [Google Scholar] [CrossRef]
- Taussig, L.M.; Saldino, R.M.; Di Sant’Agnese, P.A. Radiographic abnormalities of the duodenum and small bowel in cystic fibrosis of the pancreas (mucoviscidosis). Radiology 1973, 106, 369–376. [Google Scholar] [CrossRef]
- Littlewood, J.M. Update on intestinal strictures. J. R. Soc. Med. 1999, 92 (Suppl. 37), 41–49. [Google Scholar] [CrossRef]
- Dodge, J.A.; Macpherson, C. Colonic strictures in cystic fibrosis. J. R. Soc. Med. 1995, 88 (Suppl. 25), 3–8. [Google Scholar]
- Phelan, M.S.; Fine, D.R.; Zentler-Munro, P.L.; Hodson, M.E.; Batten, J.C.; Kerr, I.H. Radiographic abnormalities of the duodenum in cystic fibrosis. Clin. Radiol. 1983, 34, 573–577. [Google Scholar] [CrossRef]
- Lloyd-Still, J.D.; Beno, D.W.; Kimura, R.M. Cystic fibrosis colonopathy. Curr. Gastroenterol. Rep. 1999, 1, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Norkina, O.; Kaur, S.; Ziemer, D.; De Lisle, R.C. Inflammation of the cystic fibrosis mouse small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 286, G1032–G1041. [Google Scholar] [CrossRef]
- Kobelska-Dubiel, N.; Klincewicz, B.; Cichy, W. Liver disease in cystic fibrosis. Prz. Gastroenterol. 2014, 9, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Cohn, J.A.; Strong, T.V.; Picciotto, M.R.; Nairn, A.C.; Collins, F.S.; Fitz, J.G. Localization of the cystic fibrosis transmembrane conductance regulator in human bile duct epithelial cells. Gastroenterology 1993, 105, 1857–1864. [Google Scholar] [CrossRef] [PubMed]
- Beuers, U.; Hohenester, S.; de Buy Wenniger, L.J.; Kremer, A.E.; Jansen, P.L.; Elferink, R.P. The biliary HCO3− umbrella: A unifying hypothesis on pathogenetic and therapeutic aspects of fibrosing cholangiopathies. Hepatology 2010, 52, 1489–1496. [Google Scholar] [CrossRef]
- Jarzabek, K.; Zbucka, M.; Pepinski, W.; Szamatowicz, J.; Domitrz, J.; Janica, J.; Wolczynski, S.; Szamatowicz, M. Cystic fibrosis as a cause of infertility. Reprod. Biol. 2004, 4, 119–129. [Google Scholar]
- Phillipson, G. Cystic fibrosis and reproduction. Reprod. Fertil. Dev. 1998, 10, 113–119. [Google Scholar] [CrossRef]
- Tousson, A.; Van Tine, B.A.; Naren, A.P.; Shaw, G.M.; Schwiebert, L.M. Characterization of CFTR expression and chloride channel activity in human endothelia. Am. J. Physiol. 1998, 275, C1555–C1564. [Google Scholar] [CrossRef]
- Lamhonwah, A.M.; Bear, C.E.; Huan, L.J.; Kim Chiaw, P.; Ackerley, C.A.; Tein, I. Cystic fibrosis transmembrane conductance regulator in human muscle: Dysfunction causes abnormal metabolic recovery in exercise. Ann. Neurol. 2010, 67, 802–808. [Google Scholar] [CrossRef]
- Robert, R.; Norez, C.; Becq, F. Disruption of CFTR chloride channel alters mechanical properties and cAMP-dependent Cl- transport of mouse aortic smooth muscle cells. J. Physiol. 2005, 568, 483–495. [Google Scholar] [CrossRef]
- Risse, P.A.; Kachmar, L.; Matusovsky, O.S.; Novali, M.; Gil, F.R.; Javeshghani, S.; Keary, R.; Haston, C.K.; Michoud, M.C.; Martin, J.G.; et al. Ileal smooth muscle dysfunction and remodeling in cystic fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G1–G8. [Google Scholar] [CrossRef]
- Hume, J.R.; Hart, P.; Levesque, P.C.; Collier, M.L.; Geary, Y.; Warth, J.; Chapman, T.; Horowitz, B. Molecular physiology of CFTR Cl- channels in heart. Jpn. J. Physiol. 1994, 44 (Suppl. 2), S177–S182. [Google Scholar] [PubMed]
- Sterling, K.M., Jr.; Shah, S.; Kim, R.J.; Johnston, N.I.; Salikhova, A.Y.; Abraham, E.H. Cystic fibrosis transmembrane conductance regulator in human and mouse red blood cell membranes and its interaction with ecto-apyrase. J. Cell. Biochem. 2004, 91, 1174–1182. [Google Scholar] [CrossRef] [PubMed]
- Mattoscio, D.; Evangelista, V.; De Cristofaro, R.; Recchiuti, A.; Pandolfi, A.; Di Silvestre, S.; Manarini, S.; Martelli, N.; Rocca, B.; Petrucci, G.; et al. Cystic fibrosis transmembrane conductance regulator (CFTR) expression in human platelets: Impact on mediators and mechanisms of the inflammatory response. FASEB J. 2010, 24, 3970–3980. [Google Scholar] [CrossRef] [PubMed]
- Reznikov, L.R.; Dong, Q.; Chen, J.H.; Moninger, T.O.; Park, J.M.; Zhang, Y.; Du, J.; Hildebrand, M.S.; Smith, R.J.; Randak, C.O.; et al. CFTR-deficient pigs display peripheral nervous system defects at birth. Proc. Natl. Acad. Sci. USA 2013, 110, 3083–3088. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.D.; Li, J.C.; Cheung, K.H.; Leung, G.P.; Chew, S.B.; Wong, P.Y. Expression of the cystic fibrosis transmembrane conductance regulator in rat spermatids: Implication for the site of action of antispermatogenic agents. Mol. Hum. Reprod. 2001, 7, 705–713. [Google Scholar] [CrossRef]
- Painter, R.G.; Valentine, V.G.; Lanson, N.A., Jr.; Leidal, K.; Zhang, Q.; Lombard, G.; Thompson, C.; Viswanathan, A.; Nauseef, W.M.; Wang, G. CFTR Expression in human neutrophils and the phagolysosomal chlorination defect in cystic fibrosis. Biochemistry 2006, 45, 10260–10269. [Google Scholar] [CrossRef]
- Di, A.; Brown, M.E.; Deriy, L.V.; Li, C.; Szeto, F.L.; Chen, Y.; Huang, P.; Tong, J.; Naren, A.P.; Bindokas, V.; et al. CFTR regulates phagosome acidification in macrophages and alters bactericidal activity. Nat. Cell Biol. 2006, 8, 933–944. [Google Scholar] [CrossRef]
- Yoshimura, K.; Nakamura, H.; Trapnell, B.C.; Chu, C.S.; Dalemans, W.; Pavirani, A.; Lecocq, J.P.; Crystal, R.G. Expression of the cystic fibrosis transmembrane conductance regulator gene in cells of non-epithelial origin. Nucleic Acids Res. 1991, 19, 5417–5423. [Google Scholar] [CrossRef]
- Zhou, Y.; Song, K.; Painter, R.G.; Aiken, M.; Reiser, J.; Stanton, B.A.; Nauseef, W.M.; Wang, G. Cystic fibrosis transmembrane conductance regulator recruitment to phagosomes in neutrophils. J. Innate Immun. 2013, 5, 219–230. [Google Scholar] [CrossRef]
- Aiken, M.L.; Painter, R.G.; Zhou, Y.; Wang, G. Chloride transport in functionally active phagosomes isolated from Human neutrophils. Free. Radic. Biol. Med. 2012, 53, 2308–2317. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. Chloride flux in phagocytes. Immunol. Rev. 2016, 273, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Nauseef, W.M. Salt, chloride, bleach, and innate host defense. J. Leukoc. Biol. 2015, 98, 163–172. [Google Scholar] [CrossRef]
- Ng, H.P.; Valentine, V.G.; Wang, G. CFTR targeting during activation of human neutrophils. J. Leukoc. Biol. 2016, 100, 1413–1424. [Google Scholar] [CrossRef] [PubMed]
- Painter, R.G.; Marrero, L.; Lombard, G.A.; Valentine, V.G.; Nauseef, W.M.; Wang, G. CFTR-mediated halide transport in phagosomes of human neutrophils. J. Leukoc. Biol. 2010, 87, 933–942. [Google Scholar] [CrossRef]
- Painter, R.G.; Bonvillain, R.W.; Valentine, V.G.; Lombard, G.A.; LaPlace, S.G.; Nauseef, W.M.; Wang, G. The role of chloride anion and CFTR in killing of Pseudomonas aeruginosa by normal and CF neutrophils. J. Leukoc. Biol. 2008, 83, 1345–1353. [Google Scholar] [CrossRef]
- Ng, H.P.; Zhou, Y.; Song, K.; Hodges, C.A.; Drumm, M.L.; Wang, G. Neutrophil-mediated phagocytic host defense defect in myeloid cftr-inactivated mice. PLoS ONE 2014, 9, e106813. [Google Scholar] [CrossRef]
- Ng, H.P.; Jennings, S.; Wellems, D.; Sun, F.; Xu, J.; Nauseef, W.M.; Wang, G. Myeloid CFTR loss-of-function causes persistent neutrophilic inflammation in cystic fibrosis. J. Leukoc. Biol. 2020, 108, 1777–1785. [Google Scholar] [CrossRef]
- Sajjan, U.; Corey, M.; Humar, A.; Tullis, E.; Cutz, E.; Ackerley, C.; Forstner, J. Immunolocalisation of Burkholderia cepacia in the lungs of cystic fibrosis patients. J. Med. Microbiol. 2001, 50, 535–546. [Google Scholar] [CrossRef]
- Hartl, D.; Latzin, P.; Hordijk, P.; Marcos, V.; Rudolph, C.; Woischnik, M.; Krauss-Etschmann, S.; Koller, B.; Reinhardt, D.; Roscher, A.A.; et al. Cleavage of CXCR1 on neutrophils disables bacterial killing in cystic fibrosis lung disease. Nat. Med. 2007, 13, 1423–1430. [Google Scholar] [CrossRef]
- Su, X.; Looney, M.R.; Su, H.E.; Lee, J.W.; Song, Y.; Matthay, M.A. Role of CFTR expressed by neutrophils in modulating acute lung inflammation and injury in mice. Inflamm. Res. 2011, 60, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Witko-Sarsat, V.; Allen, R.C.; Paulais, M.; Nguyen, A.T.; Bessou, G.; Lenoir, G.; Descamps-Latscha, B. Disturbed myeloperoxidase-dependent activity of neutrophils in cystic fibrosis homozygotes and heterozygotes, and its correction by amiloride. J. Immunol. 1996, 157, 2728–2735. [Google Scholar] [CrossRef] [PubMed]
- Adib-Conquy, M.; Pedron, T.; Petit-Bertron, A.F.; Tabary, O.; Corvol, H.; Jacquot, J.; Clement, A.; Cavaillon, J.M. Neutrophils in cystic fibrosis display a distinct gene expression pattern. Mol. Med. 2008, 14, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Tirouvanziam, R.; Gernez, Y.; Conrad, C.K.; Moss, R.B.; Schrijver, I.; Dunn, C.E.; Davies, Z.A.; Herzenberg, L.A. Profound functional and signaling changes in viable inflammatory neutrophils homing to cystic fibrosis airways. Proc. Natl. Acad. Sci. USA 2008, 105, 4335–4339. [Google Scholar] [CrossRef]
- Reeves, E.P.; Williamson, M.; O’Neill, S.J.; Greally, P.; McElvaney, N.G. Nebulized hypertonic saline decreases IL-8 in sputum of patients with cystic fibrosis. Am. J. Respir. Crit. Care Med. 2011, 183, 1517–1523. [Google Scholar] [CrossRef]
- Corvol, H.; Fitting, C.; Chadelat, K.; Jacquot, J.; Tabary, O.; Boule, M.; Cavaillon, J.M.; Clement, A. Distinct cytokine production by lung and blood neutrophils from children with cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 2003, 284, L997–L1003. [Google Scholar] [CrossRef]
- Moriceau, S.; Lenoir, G.; Witko-Sarsat, V. In cystic fibrosis homozygotes and heterozygotes, neutrophil apoptosis is delayed and modulated by diamide or roscovitine: Evidence for an innate neutrophil disturbance. J. Innate Immun. 2010, 2, 260–266. [Google Scholar] [CrossRef]
- Marcos, V.; Zhou, Z.; Yildirim, A.O.; Bohla, A.; Hector, A.; Vitkov, L.; Wiedenbauer, E.M.; Krautgartner, W.D.; Stoiber, W.; Belohradsky, B.H.; et al. CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat. Med. 2010, 16, 1018–1023. [Google Scholar] [CrossRef]
- Kettle, A.J.; Turner, R.; Gangell, C.L.; Harwood, D.T.; Khalilova, I.S.; Chapman, A.L.; Winterbourn, C.C.; Sly, P.D. Oxidation contributes to low glutathione in the airways of children with cystic fibrosis. Eur. Respir. J. 2014, 44, 122–129. [Google Scholar] [CrossRef]
- Pohl, K.; Hayes, E.; Keenan, J.; Henry, M.; Meleady, P.; Molloy, K.; Jundi, B.; Bergin, D.A.; McCarthy, C.; McElvaney, O.J.; et al. A neutrophil intrinsic impairment affecting Rab27a and degranulation in cystic fibrosis is corrected by CFTR potentiator therapy. Blood 2014, 24, 999–1009. [Google Scholar] [CrossRef]
- Bruscia, E.M.; Zhang, P.X.; Satoh, A.; Caputo, C.; Medzhitov, R.; Shenoy, A.; Egan, M.E.; Krause, D.S. Abnormal trafficking and degradation of TLR4 underlie the elevated inflammatory response in cystic fibrosis. J. Immunol. 2011, 186, 6990–6998. [Google Scholar] [CrossRef] [PubMed]
- Bruscia, E.M.; Zhang, P.X.; Ferreira, E.; Caputo, C.; Emerson, J.W.; Tuck, D.; Krause, D.S.; Egan, M.E. Macrophages directly contribute to the exaggerated inflammatory response in cystic fibrosis transmembrane conductance regulator−/− mice. Am. J. Respir. Cell Mol. Biol. 2009, 40, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Hartl, D.; Gaggar, A.; Bruscia, E.; Hector, A.; Marcos, V.; Jung, A.; Greene, C.; McElvaney, G.; Mall, M.; Doring, G. Innate immunity in cystic fibrosis lung disease. J. Cyst. Fibros. 2012, 11, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Deriy, L.V.; Gomez, E.A.; Zhang, G.; Beacham, D.W.; Hopson, J.A.; Gallan, A.J.; Shevchenko, P.D.; Bindokas, V.P.; Nelson, D.J. Disease-causing mutations in the cystic fibrosis transmembrane conductance regulator determine the functional responses of alveolar macrophages. J. Biol. Chem. 2009, 284, 35926–35938. [Google Scholar] [CrossRef]
- Del Porto, P.; Cifani, N.; Guarnieri, S.; Di Domenico, E.G.; Mariggio, M.A.; Spadaro, F.; Guglietta, S.; Anile, M.; Venuta, F.; Quattrucci, S.; et al. Dysfunctional CFTR alters the bactericidal activity of human macrophages against Pseudomonas aeruginosa. PLoS ONE 2011, 6, e19970. [Google Scholar] [CrossRef]
- Soleti, R.; Porro, C.; Martinez, M.C. Apoptotic process in cystic fibrosis cells. Apoptosis 2013, 18, 1029–1038. [Google Scholar] [CrossRef]
- Turner, M.A.; Baildam, E.; Patel, L.; David, T.J. Joint disorders in cystic fibrosis. J. R. Soc. Med. 1997, 90 (Suppl. 31), 13–20. [Google Scholar] [CrossRef]
- Botton, E.; Saraux, A.; Laselve, H.; Jousse, S.; Le Goff, P. Musculoskeletal manifestations in cystic fibrosis. Jt. Bone Spine 2003, 70, 327–335. [Google Scholar] [CrossRef]
- Thornton, J.; Rangaraj, S. Disease Modifying Anti-Rheumatic Drugs in People with Cystic Fibrosis-Related Arthritis; Cochrane: London, UK, 2012. [Google Scholar] [CrossRef]
- Rush, P.J.; Shore, A.; Coblentz, C.; Wilmot, D.; Corey, M.; Levison, H. The musculoskeletal manifestations of cystic fibrosis. Semin. Arthritis Rheum. 1986, 15, 213–225. [Google Scholar] [CrossRef]
- Dixey, J.; Redington, A.N.; Butler, R.C.; Smith, M.J.; Batchelor, J.R.; Woodrow, D.F.; Hodson, M.E.; Batten, J.C.; Brewerton, D.A. The arthropathy of cystic fibrosis. Ann. Rheum. Dis. 1988, 47, 218–223. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S. Origins and evolution of CRISPR-Cas systems. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [PubMed]
- Deltcheva, E.; Chylinski, K.; Sharma, C.M.; Gonzales, K.; Chao, Y.; Pirzada, Z.A.; Eckert, M.R.; Vogel, J.; Charpentier, E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011, 471, 602–607. [Google Scholar] [CrossRef]
- Chen, H.; Choi, J.; Bailey, S. Cut site selection by the two nuclease domains of the Cas9 RNA-guided endonuclease. J. Biol. Chem. 2014, 289, 13284–13294. [Google Scholar] [CrossRef]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, E2579–E2586. [Google Scholar] [CrossRef] [PubMed]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef]
- Chen, M.; Qi, L.S. Repurposing CRISPR System for Transcriptional Activation. Adv. Exp. Med. Biol. 2017, 983, 147–157. [Google Scholar] [CrossRef]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR-Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016, 17, 5–15. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Karvelis, T.; Gasiunas, G.; Siksnys, V. Harnessing the natural diversity and in vitro evolution of Cas9 to expand the genome editing toolbox. Curr. Opin. Microbiol. 2017, 37, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Wang, W.; Zhang, J. CRISPR-Cas12 and Cas13: The lesser known siblings of CRISPR-Cas9. Cell Biol. Toxicol. 2019, 35, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Ghosh, A.; Chakravarti, R.; Singh, R.; Ravichandiran, V.; Swarnakar, S.; Ghosh, D. Cas13d: A New Molecular Scissor for Transcriptome Engineering. Front. Cell Dev. Biol. 2022, 10, 866800. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.P.; Newby, G.A.; Liu, D.R. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat. Protoc. 2021, 16, 1089–1128. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Chen, P.J.; Liu, D.R. Prime editing for precise and highly versatile genome manipulation. Nat. Rev. Genet. 2023, 24, 161–177. [Google Scholar] [CrossRef]
- Doman, J.L.; Sousa, A.A.; Randolph, P.B.; Chen, P.J.; Liu, D.R. Designing and executing prime editing experiments in mammalian cells. Nat. Protoc. 2022, 17, 2431–2468. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Schwank, G.; Koo, B.K.; Sasselli, V.; Dekkers, J.F.; Heo, I.; Demircan, T.; Sasaki, N.; Boymans, S.; Cuppen, E.; van der Ent, C.K.; et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell 2013, 13, 653–658. [Google Scholar] [CrossRef]
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional Gene Correction for Cystic Fibrosis in Lung Epithelial Cells Generated from Patient iPSCs. Cell Rep. 2015, 12, 1385–1390. [Google Scholar] [CrossRef] [PubMed]
- Sanz, D.J.; Hollywood, J.A.; Scallan, M.F.; Harrison, P.T. Cas9/gRNA targeted excision of cystic fibrosis-causing deep-intronic splicing mutations restores normal splicing of CFTR mRNA. PLoS ONE 2017, 12, e0184009. [Google Scholar] [CrossRef] [PubMed]
- Vaidyanathan, S.; Baik, R.; Chen, L.; Bravo, D.T.; Suarez, C.J.; Abazari, S.M.; Salahudeen, A.A.; Dudek, A.M.; Teran, C.A.; Davis, T.H.; et al. Targeted replacement of full-length CFTR in human airway stem cells by CRISPR-Cas9 for pan-mutation correction in the endogenous locus. Mol. Ther. 2022, 30, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.P.; Yang, L.L.; Cao, H.; Chen, Z.R.; Zhang, Y.; Wen, X.Y.; Hu, J. In Vitro Validation of a CRISPR-Mediated CFTR Correction Strategy for Preclinical Translation in Pigs. Hum. Gene Ther. 2019, 30, 1448–1458. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.; Traore, S.; Cooney, A.L.; Brommel, C.M.; Kulhankova, K.; Sinn, P.L.; Newby, G.A.; Liu, D.R.; McCray, P.B. Functional correction of CFTR mutations in human airway epithelial cells using adenine base editors. Nucleic Acids Res. 2021, 49, 10558–10572. [Google Scholar] [CrossRef]
- Geurts, M.H.; de Poel, E.; Pleguezuelos-Manzano, C.; Oka, R.; Carrillo, L.; Andersson-Rolf, A.; Boretto, M.; Brunsveld, J.E.; van Boxtel, R.; Beekman, J.M.; et al. Evaluating CRISPR-Based Prime Editing for Cancer Modeling and CFTR Repair in Organoids; Life Science Alliance LLC: Baltimore, MD, USA, 2021. [Google Scholar] [CrossRef]
- Bellec, J.; Bacchetta, M.; Losa, D.; Anegon, I.; Chanson, M.; Nguyen, T.H. CFTR inactivation by lentiviral vector-mediated RNA interference and CRISPR-Cas9 genome editing in human airway epithelial cells. Curr. Gene Ther. 2015, 15, 447–459. [Google Scholar] [CrossRef]
- Jennings, S.; Ng, H.P.; Wang, G. Establishment of a DeltaF508-CF promyelocytic cell line for cystic fibrosis research and drug screening. J. Cyst. Fibros. 2019, 18, 44–53. [Google Scholar] [CrossRef]
- Chung, W.Y.; Song, M.; Park, J.; Namkung, W.; Lee, J.; Kim, H.; Lee, M.G.; Kim, J.Y. Generation of DeltaF508-CFTR T84 cell lines by CRISPR/Cas9-mediated genome editing. Biotechnol. Lett. 2016, 38, 2023–2034. [Google Scholar] [CrossRef]
- Valley, H.C.; Bukis, K.M.; Bell, A.; Cheng, Y.; Wong, E.; Jordan, N.J.; Allaire, N.E.; Sivachenko, A.; Liang, F.; Bihler, H.; et al. Isogenic cell models of cystic fibrosis-causing variants in natively expressing pulmonary epithelial cells. J. Cyst. Fibros. 2019, 18, 476–483. [Google Scholar] [CrossRef]
- Hao, S.; Roesch, E.A.; Perez, A.; Weiner, R.L.; Henderson, L.C.; Cummings, L.; Consiglio, P.; Pajka, J.; Eisenberg, A.; Yeh, L.; et al. Inactivation of CFTR by CRISPR/Cas9 alters transcriptional regulation of inflammatory pathways and other networks. J. Cyst. Fibros. 2019, 18, e1–e5. [Google Scholar] [CrossRef]
- McHugh, D.R.; Steele, M.S.; Valerio, D.M.; Miron, A.; Mann, R.J.; LePage, D.F.; Conlon, R.A.; Cotton, C.U.; Drumm, M.L.; Hodges, C.A. A G542X cystic fibrosis mouse model for examining nonsense mutation directed therapies. PLoS ONE 2018, 13, e0199573. [Google Scholar] [CrossRef] [PubMed]
- Sharma, J.; Abbott, J.; Klaskala, L.; Zhao, G.; Birket, S.E.; Rowe, S.M. A Novel G542X CFTR Rat Model of Cystic Fibrosis Is Sensitive to Nonsense Mediated Decay. Front. Physiol. 2020, 11, 611294. [Google Scholar] [CrossRef] [PubMed]
- McCarron, A.; Cmielewski, P.; Reyne, N.; McIntyre, C.; Finnie, J.; Craig, F.; Rout-Pitt, N.; Delhove, J.; Schjenken, J.E.; Chan, H.Y.; et al. Phenotypic Characterization and Comparison of Cystic Fibrosis Rat Models Generated Using CRISPR/Cas9 Gene Editing. Am. J. Pathol. 2020, 190, 977–993. [Google Scholar] [CrossRef]
- Yang, D.; Liang, X.; Pallas, B.; Hoenerhoff, M.; Ren, Z.; Han, R.; Zhang, J.; Chen, Y.E.; Jin, J.P.; Sun, F.; et al. Production of CFTR-DeltaF508 Rabbits. Front. Genet. 2020, 11, 627666. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Vorhies, K.; Feng, Z.; Park, S.Y.; Choi, S.H.; Zhang, Y.; Winter, M.; Sun, X.; Engelhardt, J.F. Recombinant Adeno-Associated Virus-Mediated Editing of the G551D Cystic Fibrosis Transmembrane Conductance Regulator Mutation in Ferret Airway Basal Cells. Hum. Gene Ther. 2022, 33, 1023–1036. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Wohlford-Lenane, C.; Kandimalla, S.; Sartre, G.; Meyerholz, D.K.; Theberge, V.; Hallee, S.; Duperre, A.M.; Del’Guidice, T.; Lepetit-Stoffaes, J.P.; et al. Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia. Nat. Commun. 2019, 10, 4906. [Google Scholar] [CrossRef]
- Kulhankova, K.; Traore, S.; Cheng, X.; Benk-Fortin, H.; Hallee, S.; Harvey, M.; Roberge, J.; Couture, F.; Gross, T.; Newby, G.; et al. Shuttle Peptide Delivers Base Editor RNPs to Rhesus Monkey Airway Epithelial Cells In Vivo; Research Square Company LLC: Durham, NC, USA, 2023. [Google Scholar] [CrossRef]
- Suzuki, S.; Crane, A.M.; Anirudhan, V.; Barilla, C.; Matthias, N.; Randell, S.H.; Rab, A.; Sorscher, E.J.; Kerschner, J.L.; Yin, S.; et al. Highly Efficient Gene Editing of Cystic Fibrosis Patient-Derived Airway Basal Cells Results in Functional CFTR Correction. Mol. Ther. 2020, 28, 1684–1695. [Google Scholar] [CrossRef]
- Xia, E.; Zhang, Y.; Cao, H.; Li, J.; Duan, R.; Hu, J. TALEN-Mediated Gene Targeting for Cystic Fibrosis-Gene Therapy. Genes 2019, 10, 39. [Google Scholar] [CrossRef]
- McNeer, N.A.; Anandalingam, K.; Fields, R.J.; Caputo, C.; Kopic, S.; Gupta, A.; Quijano, E.; Polikoff, L.; Kong, Y.; Bahal, R.; et al. Nanoparticles that deliver triplex-forming peptide nucleic acid molecules correct F508del CFTR in airway epithelium. Nat. Commun. 2015, 6, 6952. [Google Scholar] [CrossRef]
- Egan, M.E. Emerging technologies for cystic fibrosis transmembrane conductance regulator restoration in all people with CF. Pediatr. Pulmonol. 2021, 56 (Suppl. 1), S32–S39. [Google Scholar] [CrossRef]
- Rogers, F.A.; Vasquez, K.M.; Egholm, M.; Glazer, P.M. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc. Natl. Acad. Sci. USA 2002, 99, 16695–16700. [Google Scholar] [CrossRef]
- McNeer, N.A.; Schleifman, E.B.; Cuthbert, A.; Brehm, M.; Jackson, A.; Cheng, C.; Anandalingam, K.; Kumar, P.; Shultz, L.D.; Greiner, D.L.; et al. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther. 2013, 20, 658–669. [Google Scholar] [CrossRef] [PubMed]
- Piotrowski-Daspit, A.S.; Barone, C.; Lin, C.Y.; Deng, Y.; Wu, D.; Binns, T.C.; Xu, E.; Ricciardi, A.S.; Putman, R.; Garrison, A.; et al. In vivo correction of cystic fibrosis mediated by PNA nanoparticles. Sci. Adv. 2022, 8, eabo0522. [Google Scholar] [CrossRef] [PubMed]
- Roesch, E.A.; Drumm, M.L. Powerful tools for genetic modification: Advances in gene editing. Pediatr. Pulmonol. 2017, 52, S15–S20. [Google Scholar] [CrossRef]
- Flotte, T.R.; Ng, P.; Dylla, D.E.; McCray, P.B., Jr.; Wang, G.; Kolls, J.K.; Hu, J. Viral vector-mediated and cell-based therapies for treatment of cystic fibrosis. Mol. Ther. 2007, 15, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; McCray, P.B., Jr.; Engelhardt, J.F. Advances in gene therapy for cystic fibrosis lung disease. Hum. Mol. Genet. 2019, 28, R88–R94. [Google Scholar] [CrossRef] [PubMed]
- Vu, A.; McCray, P., Jr. New Directions in Pulmonary Gene Therapy. Hum. Gene Ther. 2020, 31, 921–939. [Google Scholar] [CrossRef]
- Welsh, M.J.; Zabner, J.; Graham, S.M.; Smith, A.E.; Moscicki, R.; Wadsworth, S. Adenovirus-mediated gene transfer for cystic fibrosis: Part A. Safety of dose and repeat administration in the nasal epithelium. Part B. Clinical efficacy in the maxillary sinus. Hum. Gene Ther. 1995, 6, 205–218. [Google Scholar] [CrossRef]
- Zabner, J.; Couture, L.A.; Gregory, R.J.; Graham, S.M.; Smith, A.E.; Welsh, M.J. Adenovirus-mediated gene transfer transiently corrects the chloride transport defect in nasal epithelia of patients with cystic fibrosis. Cell 1993, 75, 207–216. [Google Scholar] [CrossRef]
- Halbert, C.L.; Allen, J.M.; Miller, A.D. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 2001, 75, 6615–6624. [Google Scholar] [CrossRef]
- Wagner, J.A.; Nepomuceno, I.B.; Messner, A.H.; Moran, M.L.; Batson, E.P.; Dimiceli, S.; Brown, B.W.; Desch, J.K.; Norbash, A.M.; Conrad, C.K.; et al. A phase II, double-blind, randomized, placebo-controlled clinical trial of tgAAVCF using maxillary sinus delivery in patients with cystic fibrosis with antrostomies. Hum. Gene Ther. 2002, 13, 1349–1359. [Google Scholar] [CrossRef]
- Moss, R.B.; Rodman, D.; Spencer, L.T.; Aitken, M.L.; Zeitlin, P.L.; Waltz, D.; Milla, C.; Brody, A.S.; Clancy, J.P.; Ramsey, B.; et al. Repeated adeno-associated virus serotype 2 aerosol-mediated cystic fibrosis transmembrane regulator gene transfer to the lungs of patients with cystic fibrosis: A multicenter, double-blind, placebo-controlled trial. Chest 2004, 125, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Guggino, W.B.; Benson, J.; Seagrave, J.; Yan, Z.; Engelhardt, J.; Gao, G.; Conlon, T.J.; Cebotaru, L. A Preclinical Study in Rhesus Macaques for Cystic Fibrosis to Assess Gene Transfer and Transduction by AAV1 and AAV5 with a Dual-Luciferase Reporter System. Hum. Gene Ther. Clin. Dev. 2017, 28, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Cooney, A.L.; Abou Alaiwa, M.H.; Shah, V.S.; Bouzek, D.C.; Stroik, M.R.; Powers, L.S.; Gansemer, N.D.; Meyerholz, D.K.; Welsh, M.J.; Stoltz, D.A.; et al. Lentiviral-Mediated Phenotypic Correction of Cystic Fibrosis Pigs; American Society for Clinical Investigation (ASCI): Rockville, MD, USA, 2016. [Google Scholar] [CrossRef]
- Wang, G.; Slepushkin, V.; Zabner, J.; Keshavjee, S.; Johnston, J.C.; Sauter, S.L.; Jolly, D.J.; Dubensky, T.W., Jr.; Davidson, B.L.; McCray, P.B., Jr. Feline immunodeficiency virus vectors persistently transduce nondividing airway epithelia and correct the cystic fibrosis defect [see comments]. J. Clin. Investig. 1999, 104, R55–R62. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Sinn, P.L.; McCray, P.B., Jr. Development of retroviral vectors for gene transfer to airway epithelia. Curr. Opin. Mol. Ther. 2000, 2, 497–506. [Google Scholar] [PubMed]
- Sinn, P.L.; Cooney, A.L.; Oakland, M.; Dylla, D.E.; Wallen, T.J.; Pezzulo, A.A.; Chang, E.H.; McCray, P.B., Jr. Lentiviral vector gene transfer to porcine airways. Mol. Ther. Nucleic Acids 2012, 1, e56. [Google Scholar] [CrossRef] [PubMed]
- Alton, E.; Armstrong, D.K.; Ashby, D.; Bayfield, K.J.; Bilton, D.; Bloomfield, E.V.; Boyd, A.C.; Brand, J.; Buchan, R.; Calcedo, R.; et al. Repeated nebulisation of non-viral CFTR gene therapy in patients with cystic fibrosis: A randomised, double-blind, placebo-controlled, phase 2b trial. Lancet Respir. Med. 2015, 3, 684–691. [Google Scholar] [CrossRef]
- Cooney, A.L.; McCray, P.B., Jr.; Sinn, P.L. Cystic Fibrosis Gene Therapy: Looking Back, Looking Forward. Genes 2018, 9, 538. [Google Scholar] [CrossRef]
- Bandara, R.A.; Chen, Z.R.; Hu, J. Potential of helper-dependent Adenoviral vectors in CRISPR-cas9-mediated lung gene therapy. Cell Biosci. 2021, 11, 145. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G. Genome Editing for Cystic Fibrosis. Cells 2023, 12, 1555. https://doi.org/10.3390/cells12121555
Wang G. Genome Editing for Cystic Fibrosis. Cells. 2023; 12(12):1555. https://doi.org/10.3390/cells12121555
Chicago/Turabian StyleWang, Guoshun. 2023. "Genome Editing for Cystic Fibrosis" Cells 12, no. 12: 1555. https://doi.org/10.3390/cells12121555
APA StyleWang, G. (2023). Genome Editing for Cystic Fibrosis. Cells, 12(12), 1555. https://doi.org/10.3390/cells12121555




