PBAF Subunit Pbrm1 Selectively Influences the Transition from Progenitors to Pre-Myelinating Cells during Oligodendrocyte Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Husbandry and Breeding of Mice
2.2. Immunohistochemical Analysis and In Situ Hybridization
2.3. Statistical Analysis
3. Results
3.1. Pbrm1 Is Present in Oligodendroglial Cells throughout Their Development
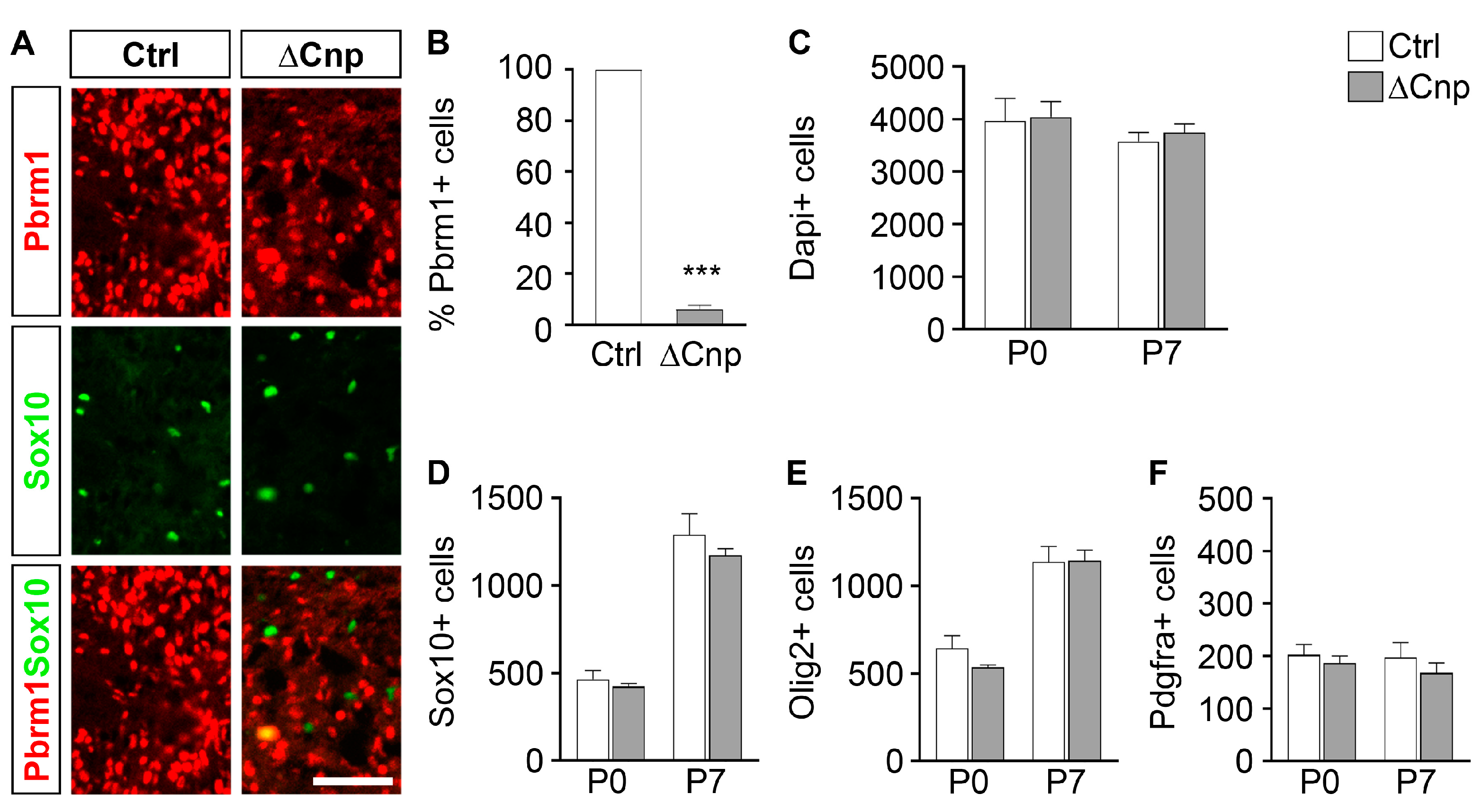
3.2. Pbrm1 Deletion in Late OPCs Fails to Impact Subsequent Oligodendrocyte Development
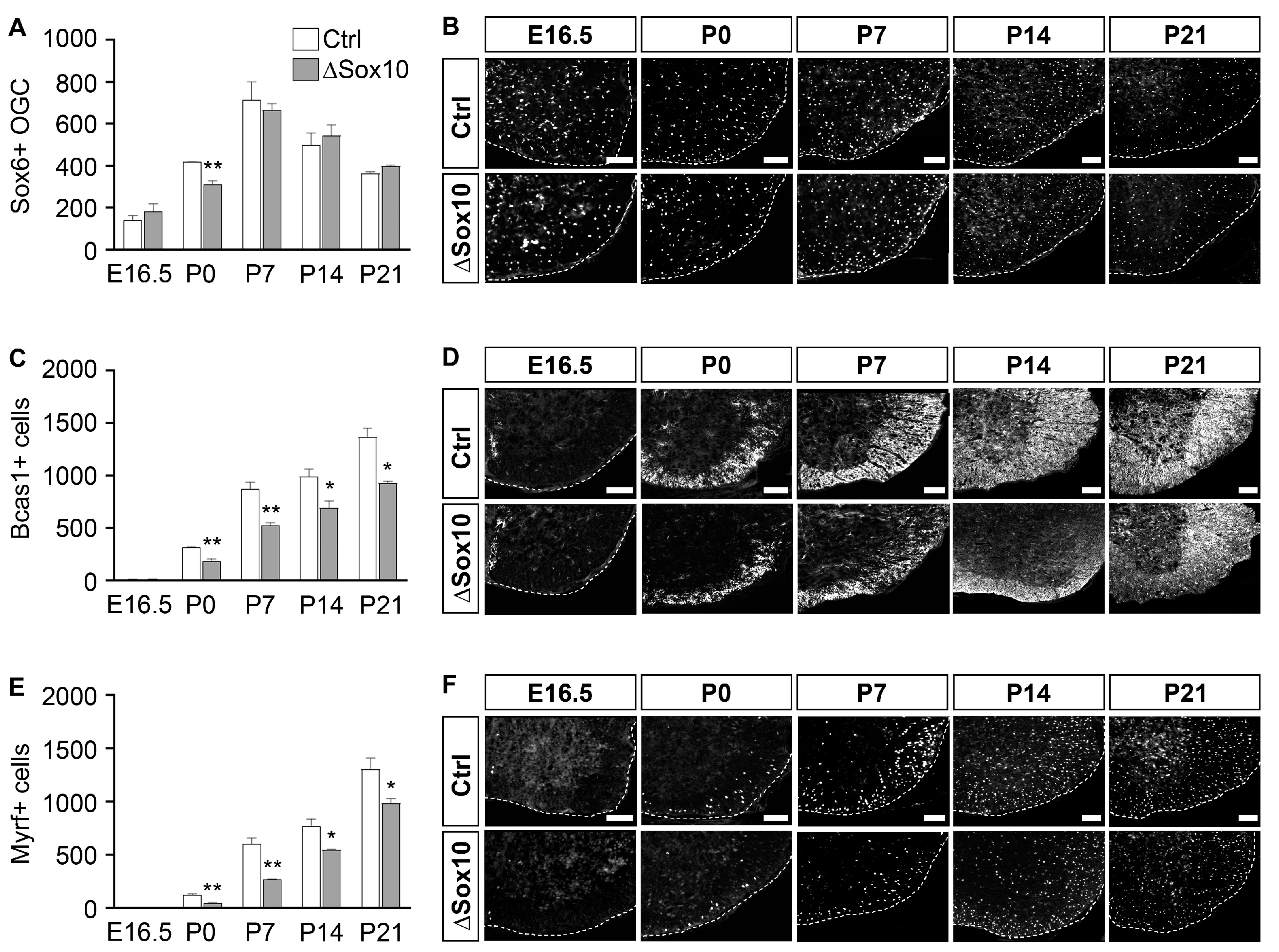
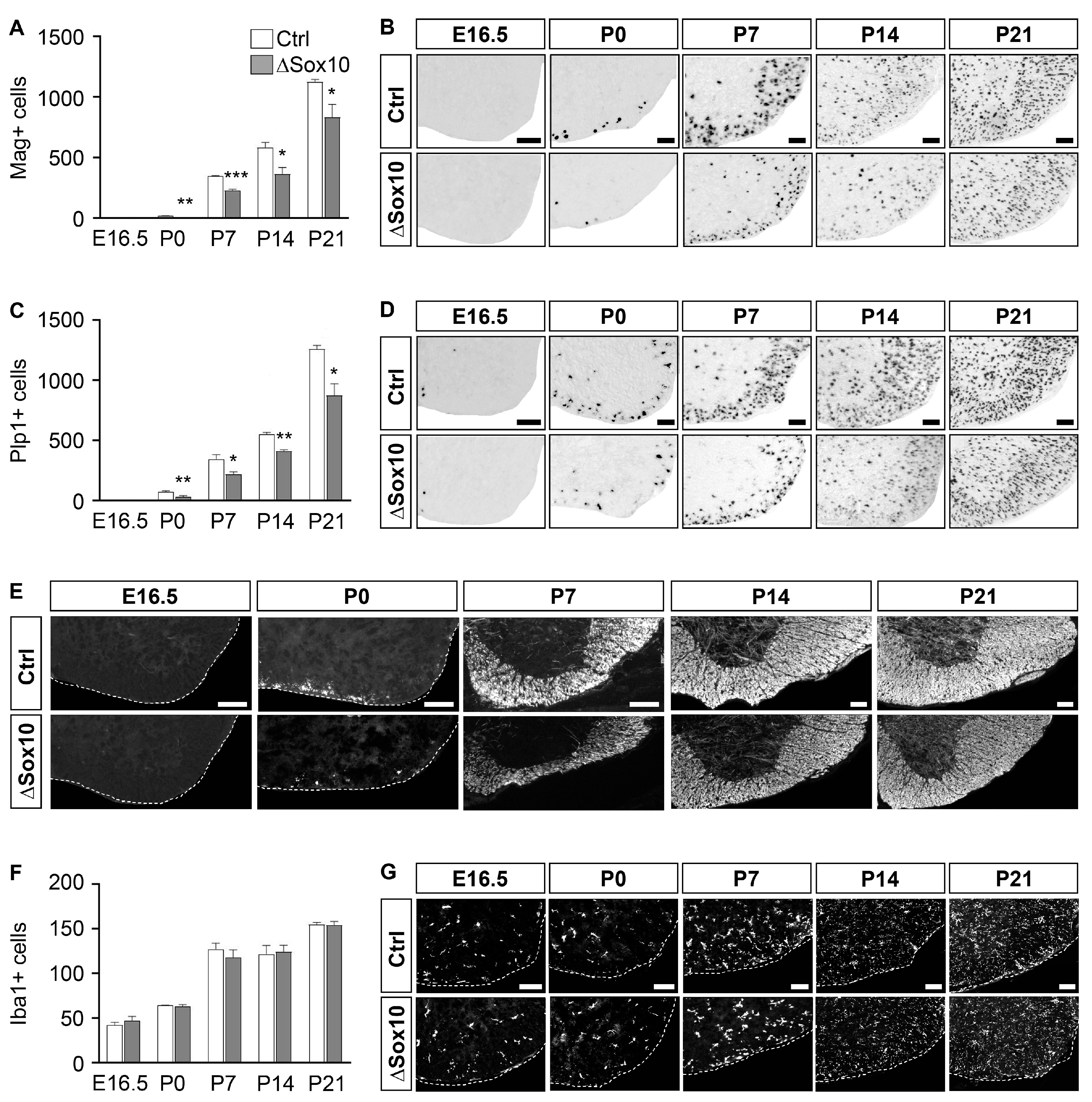
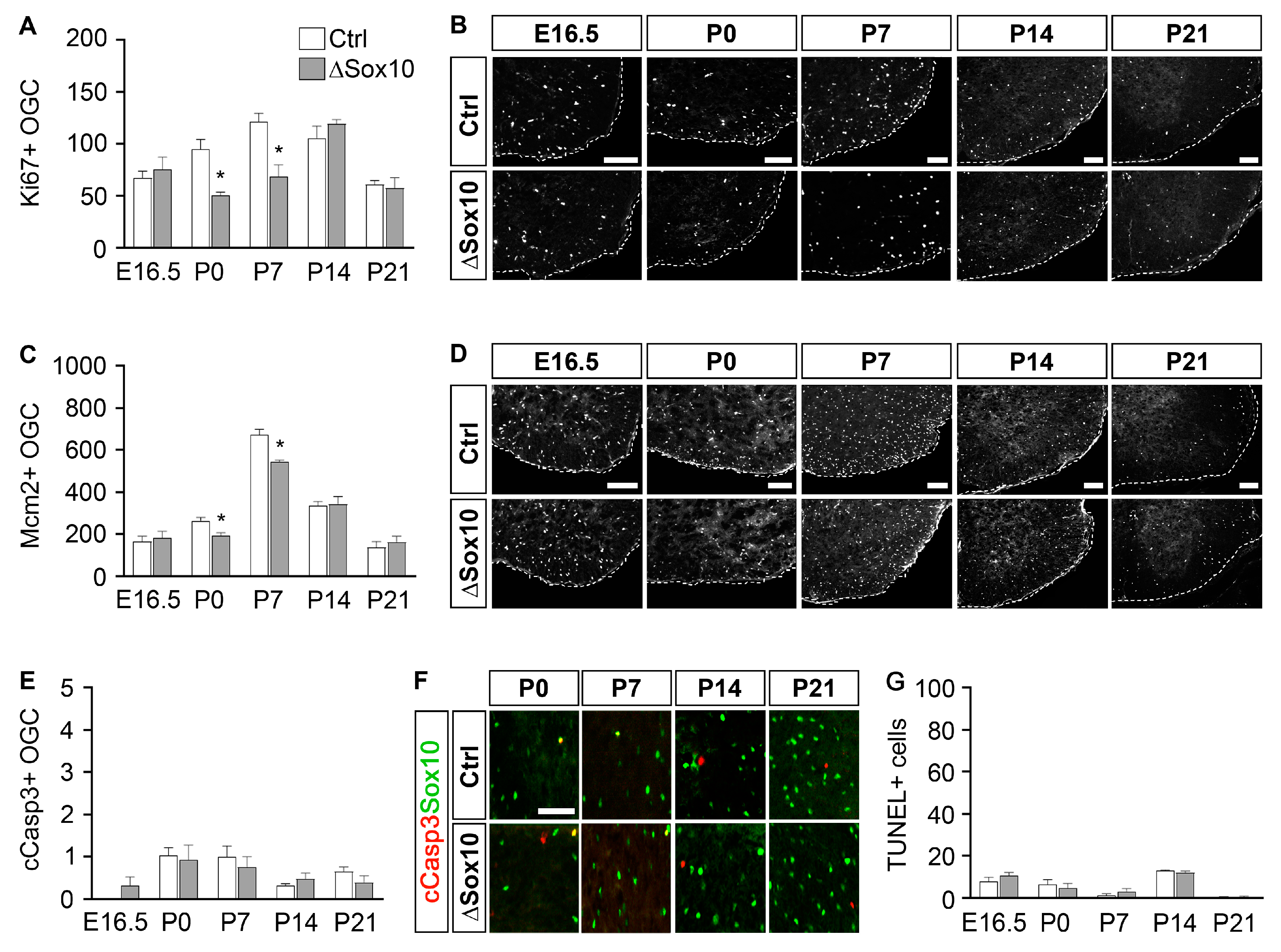
3.3. Pbrm1 Deletion in OPCs Soon after Specification Reduces the Numbers of Myelinating and Total Oligodendroglial Cells
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rowitch, D.H. Glial specification in the vertebrate neural tube. Nat. Rev. Neurosci. 2004, 5, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Sock, E.; Wegner, M. Using the lineage determinants Olig2 and Sox10 to explore transcriptional regulation of oligodendrocyte development. Dev. Neurobiol. 2021, 81, 892–901. [Google Scholar] [CrossRef] [PubMed]
- Parras, C.; Marie, C.; Zhao, C.; Lu, Q.R. Chromatin remodelers in oligodendroglia. Glia 2020, 68, 1604–1618. [Google Scholar] [CrossRef] [PubMed]
- Bischof, M.; Weider, M.; Küspert, M.; Nave, K.A.; Wegner, M. Brg1-Dependent Chromatin Remodelling Is Not Essentially Required during Oligodendroglial Differentiation. J. Neurosci. 2015, 35, 21–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsesser, O.; Fröb, F.; Küspert, M.; Tamm, E.R.; Fujii, T.; Fukunaga, R.; Wegner, M. Chromatin remodeler Ep400 ensures oligodendrocyte survival and is required for myelination in the vertebrate central nervous system. Nucleic Acids Res. 2019, 47, 6208–6224. [Google Scholar] [CrossRef]
- He, D.; Marie, C.; Zhao, C.; Kim, B.; Wang, J.; Deng, Y.; Clavairoly, A.; Frah, M.; Wang, H.; He, X.; et al. Chd7 cooperates with Sox10 and regulates the onset of CNS myelination and remyelination. Nat. Neurosci. 2016, 19, 678–689. [Google Scholar] [CrossRef] [Green Version]
- Marie, C.; Clavairoly, A.; Frah, M.; Hmidan, H.; Yan, J.; Zhao, C.; Van Steenwinckel, J.; Daveau, R.; Zalc, B.; Hassan, B.; et al. Oligodendrocyte progenitor survival and differentiation requires chromatin remodeling by Chd7 and Chd8. Proc. Natl. Acad. Sci. USA 2018, 115, E8246–E8255. [Google Scholar] [CrossRef] [Green Version]
- Zhao, C.; Dong, C.; Frah, M.; Deng, Y.; Marie, C.; Zhang, F.; Xu, L.; Ma, Z.; Dong, X.; Lin, Y.; et al. Dual Requirement of CHD8 for Chromatin Landscape Establishment and Histone Methyltransferase Recruitment to Promote CNS Myelination and Repair. Dev. Cell 2018, 45, 753–768. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Chen, Y.; Kim, B.; Wang, H.; Zhao, C.; He, X.; Liu, L.; Liu, W.; Wu, L.M.; Mao, M.; et al. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell 2013, 152, 248–261. [Google Scholar] [CrossRef] [Green Version]
- Bajpai, R.; Chen, D.A.; Rada-Iglesias, A.; Zhang, J.; Xiong, Y.; Helms, J.; Chang, C.P.; Zhao, Y.; Swigut, T.; Wysocka, J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010, 463, 958–962. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Flowers, S.; Moran, E. Essential role of ARID2 protein-containing SWI/SNF complex in tissue-specific gene expression. J. Biol. Chem. 2012, 287, 5033–5041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Zhai, W.; Richardson, J.A.; Olson, E.N.; Meneses, J.J.; Firpo, M.T.; Kang, C.; Skarnes, W.C.; Tjian, R. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004, 18, 3106–3116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, X.; Enane, F.; Tohme, R.; Schuerger, C.; Radivoyevitch, T.; Parker, Y.; Zuberi, E.; Przychodzen, B.; Jha, B.K.; Lindner, D.; et al. PBRM1 loss in kidney cancer unbalances the proximal tubule master transcription factor hub to repress proximal tubule differentiation. Cell Rep. 2021, 36, 109747. [Google Scholar] [CrossRef] [PubMed]
- Hodges, C.; Kirkland, J.G.; Crabtree, G.R. The Many Roles of BAF (mSWI/SNF) and PBAF Complexes in Cancer. Cold Spring Harb. Perspect. Med. 2016, 6, a026930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemon, B.; Inouye, C.; King, D.S.; Tjian, R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature 2001, 414, 924–928. [Google Scholar] [CrossRef]
- Askree, S.H.; Yehuda, T.; Smolikov, S.; Gurevich, R.; Hawk, J.; Coker, C.; Krauskopf, A.; Kupiec, M.; McEachern, M.J. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 2004, 101, 8658–8663. [Google Scholar] [CrossRef] [Green Version]
- Brownlee, P.M.; Chambers, A.L.; Cloney, R.; Bianchi, A.; Downs, J.A. BAF180 promotes cohesion and prevents genome instability and aneuploidy. Cell Rep. 2014, 6, 973–981. [Google Scholar] [CrossRef] [Green Version]
- Wurster, A.L.; Precht, P.; Becker, K.G.; Wood, W.H., 3rd; Zhang, Y.; Wang, Z.; Pazin, M.J. IL-10 transcription is negatively regulated by BAF180, a component of the SWI/SNF chromatin remodeling enzyme. BMC Immunol. 2012, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- Lappe-Siefke, C.; Goebbels, S.; Gravel, M.; Nicksch, E.; Lee, J.; Braun, P.E.; Griffiths, I.R.; Nave, K.A. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat. Genet. 2003, 33, 366–374. [Google Scholar] [CrossRef]
- Matsuoka, T.; Ahlberg, P.E.; Kessaris, N.; Iannarelli, P.; Dennehy, U.; Richardson, W.D.; McMahon, A.P.; Koentges, G. Neural crest origins of the neck and shoulder. Nature 2005, 436, 347–355. [Google Scholar] [CrossRef] [Green Version]
- Polanetzki, V.; Frob, F.; Baroti, T.; Schimmel, M.; Tamm, E.R.; Wegner, M. Role of the Pbrm1 subunit and the PBAF complex in Schwann cell development. Sci. Rep. 2022, 12, 2651. [Google Scholar] [CrossRef] [PubMed]
- Finzsch, M.; Schreiner, S.; Kichko, T.; Reeh, P.; Tamm, E.R.; Bösl, M.R.; Meijer, D.; Wegner, M. Sox10 is required for Schwann cell identity and progression beyond the immature Schwann cell stage. J. Cell Biol. 2010, 189, 701–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornig, J.; Fröb, F.; Vogl, M.R.; Hermans-Borgmeyer, I.; Tamm, E.R.; Wegner, M. The Transcription Factors Sox10 and Myrf Define an Essential Regulatory Network Module in Differentiating Oligodendrocytes. PLoS Genet. 2013, 9, e1003644. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wüst, H.M.; Wegener, A.; Frob, F.; Hartwig, A.C.; Wegwitz, F.; Kari, V.; Schimmel, M.; Tamm, E.R.; Johnsen, S.A.; Wegner, M.; et al. Egr2-guided histone H2B monoubiquitination is required for peripheral nervous system myelination. Nucleic Acids Res. 2020, 48, 8959–8976. [Google Scholar] [CrossRef] [PubMed]
- Maka, M.; Stolt, C.C.; Wegner, M. Identification of Sox8 as a modifier gene in a mouse model of Hirschsprung disease reveals underlying molecular defect. Dev. Biol. 2005, 277, 155–169. [Google Scholar] [CrossRef] [Green Version]
- Stolt, C.C.; Schlierf, A.; Lommes, P.; Hillgärtner, S.; Werner, T.; Kosian, T.; Sock, E.; Kessaris, N.; Richardson, W.D.; Lefebvre, V.; et al. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev. Cell 2006, 11, 697–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stolt, C.C.; Rehberg, S.; Ader, M.; Lommes, P.; Riethmacher, D.; Schachner, M.; Bartsch, U.; Wegner, M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002, 16, 165–170. [Google Scholar] [CrossRef] [Green Version]
- Baroti, T.; Zimmermann, Y.; Schillinger, A.; Liu, L.; Lommes, P.; Wegner, M.; Stolt, C.C. Transcription factors Sox5 and Sox6 exert direct and indirect influences on oligodendroglial migration in spinal cord and forebrain. Glia 2016, 64, 122–138. [Google Scholar] [CrossRef]
- Emery, B.; Agalliu, D.; Cahoy, J.D.; Watkins, T.A.; Dugas, J.C.; Mulinyawe, S.B.; Ibrahim, A.; Ligon, K.L.; Rowitch, D.H.; Barres, B.A. Myelin gene regulatory factor is a critical transcriptional regulator required for CNS myelination. Cell 2009, 138, 172–185. [Google Scholar] [CrossRef] [Green Version]
- Fard, M.K.; van der Meer, F.; Sanchez, P.; Cantuti-Castelvetri, L.; Mandad, S.; Jakel, S.; Fornasiero, E.F.; Schmitt, S.; Ehrlich, M.; Starost, L.; et al. BCAS1 expression defines a population of early myelinating oligodendrocytes in multiple sclerosis lesions. Sci. Transl. Med. 2017, 9, 419. [Google Scholar] [CrossRef] [Green Version]
- Sock, E.; Wegner, M. Transcriptional control of myelination and remyelination. Glia 2019, 67, 2153–2165. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Canman, J.C.; Lee, C.S.; Nie, Z.; Yang, D.; Moreno, G.T.; Young, M.K.; Salmon, E.D.; Wang, W. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc. Natl. Acad. Sci. USA 2000, 97, 13015–13020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barres, B.A.; Lazar, M.A.; Raff, M.C. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development 1994, 120, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Noll, E.; Miller, R.H. Regulation of oligodendrocyte differentiation: A role for retinoic acid in the spinal cord. Development 1994, 120, 649–660. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waldhauser, V.; Baroti, T.; Fröb, F.; Wegner, M. PBAF Subunit Pbrm1 Selectively Influences the Transition from Progenitors to Pre-Myelinating Cells during Oligodendrocyte Development. Cells 2023, 12, 1556. https://doi.org/10.3390/cells12121556
Waldhauser V, Baroti T, Fröb F, Wegner M. PBAF Subunit Pbrm1 Selectively Influences the Transition from Progenitors to Pre-Myelinating Cells during Oligodendrocyte Development. Cells. 2023; 12(12):1556. https://doi.org/10.3390/cells12121556
Chicago/Turabian StyleWaldhauser, Vanessa, Tina Baroti, Franziska Fröb, and Michael Wegner. 2023. "PBAF Subunit Pbrm1 Selectively Influences the Transition from Progenitors to Pre-Myelinating Cells during Oligodendrocyte Development" Cells 12, no. 12: 1556. https://doi.org/10.3390/cells12121556
APA StyleWaldhauser, V., Baroti, T., Fröb, F., & Wegner, M. (2023). PBAF Subunit Pbrm1 Selectively Influences the Transition from Progenitors to Pre-Myelinating Cells during Oligodendrocyte Development. Cells, 12(12), 1556. https://doi.org/10.3390/cells12121556






