Metabolic Pathway Modeling in Muscle of Male Marathon Mice (DUhTP) and Controls (DUC)—A Possible Role of Lactate Dehydrogenase in Metabolic Flexibility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Experimental Design and Sample Collection
2.2. Immunohistochemical Evaluation of Musculus rectus femoris
2.3. RNA Isolation, Next-Generation Sequencing (NGS), and Differential Gene Expression Analysis
2.4. Validation by Fluidigm
2.5. Pathway Analysis
2.6. Determination of Enzyme Activities
2.7. Determination of Plasma Glucose, Insulin, and Lactate
2.8. Untargeted LC-MS/MS Analysis of Lipids and Fatty Acids in Serum
2.9. Determination of Muscular Glycogen Content
2.10. Statistical Analysis
3. Results
3.1. Reduced Body and Muscle Weight in Response to Selection and Training in Marathon Mice
3.2. Analysis of mRNA Indicates Induction of Glycolysis, Beta-Oxidation, TCA-Cycle, and Oxidative Phosphorylation in Muscles from DUhTP Mice
3.3. Plasma Levels of Energy Metabolites in Response to Phenotype Selection and Physical Activity
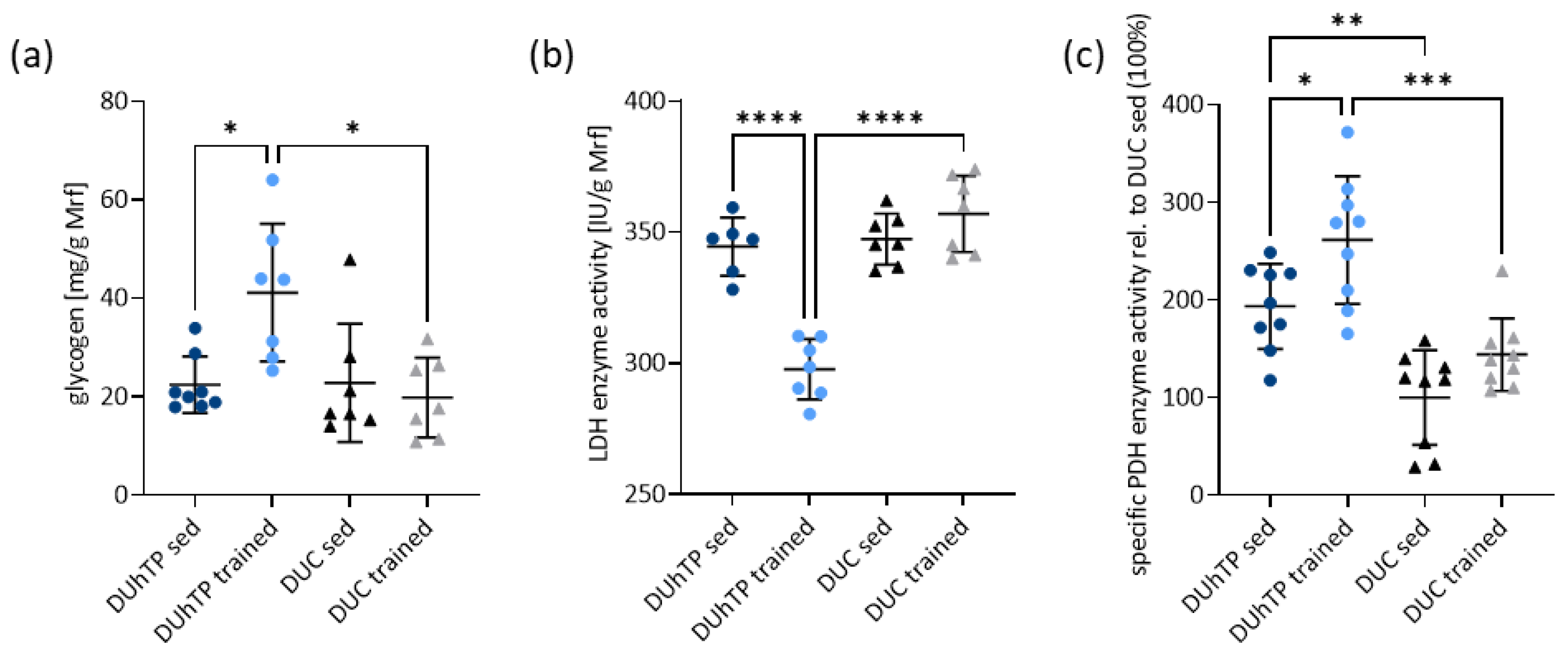
3.4. Treadmill Running Results in Higher Glycogen Levels and Reduced Lactate Dehydrogenase Activity in Muscle from DUhTP Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hargreaves, M.; Spriet, L.L. Skeletal muscle energy metabolism during exercise. Nat. Metab. 2020, 2, 817–828. [Google Scholar] [CrossRef] [PubMed]
- Sahlin, K.; Tonkonogi, M.; Soderlund, K. Energy supply and muscle fatigue in humans. Acta Physiol. Scand. 1998, 162, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Watt, M.J.; Heigenhauser, G.J.; Dyck, D.J.; Spriet, L.L. Intramuscular triacylglycerol, glycogen and acetyl group metabolism during 4 h of moderate exercise in man. J. Physiol. 2002, 541, 969–978. [Google Scholar] [CrossRef] [PubMed]
- Brooks, G.A. Lactate: Glycolytic End Product and Oxidative Substrate During Sustained Exercise in Mammals—The “Lactate Shuttle”. In Proceedings of Circulation, Respiration, and Metabolism; Springer: Berlin/Heidelberg, Germany, 1985; pp. 208–218. [Google Scholar]
- Stanley, W.C.; Gertz, E.W.; Wisneski, J.A.; Neese, R.A.; Morris, D.L.; Brooks, G.A. Lactate extraction during net lactate release in legs of humans during exercise. J. Appl. Physiol. 1986, 60, 1116–1120. [Google Scholar] [CrossRef]
- Brooks, G.A. Lactate shuttles in nature. Biochem. Soc. Trans. 2002, 30, 258–264. [Google Scholar] [CrossRef]
- Kitaoka, Y.; Hoshino, D.; Hatta, H. Monocarboxylate transporter and lactate metabolism. J. Phys. Fit. Sports Med. 2012, 1, 247–252. [Google Scholar] [CrossRef] [Green Version]
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [CrossRef]
- Brooks, G.A. The Science and Translation of Lactate Shuttle Theory. Cell Metab. 2018, 27, 757–785. [Google Scholar] [CrossRef] [Green Version]
- Adijanto, J.; Philp, N.J. The SLC16A family of monocarboxylate transporters (MCTs)—Physiology and function in cellular metabolism, pH homeostasis, and fluid transport. Curr. Top. Membr. 2012, 70, 275–311. [Google Scholar] [CrossRef]
- Fishbein, W.N.; Merezhinskaya, N.; Foellmer, J.W. Relative distribution of three major lactate transporters in frozen human tissues and their localization in unfixed skeletal muscle. Muscle Nerve 2002, 26, 101–112. [Google Scholar] [CrossRef]
- Hashimoto, T.; Masuda, S.; Taguchi, S.; Brooks, G.A. Immunohistochemical analysis of MCT1, MCT2 and MCT4 expression in rat plantaris muscle. J. Physiol. 2005, 567, 121–129. [Google Scholar] [CrossRef]
- Herzig, S.; Raemy, E.; Montessuit, S.; Veuthey, J.L.; Zamboni, N.; Westermann, B.; Kunji, E.R.; Martinou, J.C. Identification and functional expression of the mitochondrial pyruvate carrier. Science 2012, 337, 93–96. [Google Scholar] [CrossRef]
- Dubouchaud, H.; Butterfield, G.E.; Wolfel, E.E.; Bergman, B.C.; Brooks, G.A. Endurance training, expression, and physiology of LDH, MCT1, and MCT4 in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2000, 278, E571–E579. [Google Scholar] [CrossRef] [Green Version]
- Cruz, R.S.; de Aguiar, R.A.; Turnes, T.; Dos Santos, R.P.; de Oliveira, M.F.; Caputo, F. Intracellular shuttle: The lactate aerobic metabolism. ScientificWorldJournal 2012, 2012, 420984. [Google Scholar] [CrossRef] [Green Version]
- Koonen, D.P.; Glatz, J.F.; Bonen, A.; Luiken, J.J. Long-chain fatty acid uptake and FAT/CD36 translocation in heart and skeletal muscle. Biochim. Biophys. Acta 2005, 1736, 163–180. [Google Scholar] [CrossRef]
- Jeukendrup, A.E. Regulation of fat metabolism in skeletal muscle. Ann. N. Y. Acad. Sci. 2002, 967, 217–235. [Google Scholar] [CrossRef]
- Ahlborg, G.; Felig, P.; Hagenfeldt, L.; Hendler, R.; Wahren, J. Substrate Turnover during Prolonged Exercise in Man: Splanchnic and leg metabolism of glucose, free fatty acids, and amino acids. J. Clin. Investig. 1974, 53, 1080–1090. [Google Scholar] [CrossRef]
- O’Brien, M.J.; Viguie, C.A.; Mazzeo, R.S.; Brooks, G.A. Carbohydrate dependence during marathon running. Med. Sci. Sports Exerc. 1993, 25, 1009–1017. [Google Scholar]
- Achten, J.; Gleeson, M.; Jeukendrup, A.E. Determination of the exercise intensity that elicits maximal fat oxidation. Med. Sci. Sports Exerc. 2002, 34, 92–97. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; Sparks, L.M. Metabolic Flexibility in Health and Disease. Cell Metab. 2017, 25, 1027–1036. [Google Scholar] [CrossRef] [Green Version]
- Smith, R.L.; Soeters, M.R.; Wüst, R.C.I.; Houtkooper, R.H. Metabolic Flexibility as an Adaptation to Energy Resources and Requirements in Health and Disease. Endocr. Rev. 2018, 39, 489–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkenberg, H.; Langhammer, M.; Renne, U. Comparison of biochemical blood traits after long-term selection on high or low locomotory activity in mice. Arch. Anim. Breed. 2000, 43, 513–522. [Google Scholar] [CrossRef]
- Brenmoehl, J.; Walz, C.; Renne, U.; Ponsuksili, S.; Wolf, C.; Langhammer, M.; Schwerin, M.; Hoeflich, A. Metabolic adaptations in the liver of born long-distance running mice. Med. Sci. Sports Exerc. 2013, 45, 841–850. [Google Scholar] [CrossRef] [PubMed]
- Ohde, D.; Moeller, M.; Brenmoehl, J.; Walz, C.; Ponsuksili, S.; Schwerin, M.; Fuellen, G.; Hoeflich, A. Advanced Running Performance by Genetic Predisposition in Male Dummerstorf Marathon Mice (DUhTP) Reveals Higher Sterol Regulatory Element-Binding Protein (SREBP) Related mRNA Expression in the Liver and Higher Serum Levels of Progesterone. PLoS ONE 2016, 11, e0146748. [Google Scholar] [CrossRef] [PubMed]
- Brenmoehl, J.; Ohde, D.; Walz, C.; Schultz, J.; Tuchscherer, A.; Rieder, F.; Renne, U.; Hoeflich, A. Dynamics of Fat Mass in DUhTP Mice Selected for Running Performance—Fat Mobilization in a Walk. Obes. Facts 2015, 8, 373–385. [Google Scholar] [CrossRef] [PubMed]
- Brenmoehl, J.; Ohde, D.; Albrecht, E.; Walz, C.; Tuchscherer, A.; Hoeflich, A. Browning of subcutaneous fat and higher surface temperature in response to phenotype selection for advanced endurance exercise performance in male DUhTP mice. J. Comp. Physiol. B 2017, 187, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Dietl, G.; Langhammer, M.; Renne, U. Model simulations for genetic random drift in the outbred strain Fzt: DU. Arch. Fur Tierz. Arch. Anim. Breed. 2004, 47, 595–604. [Google Scholar] [CrossRef]
- Petkov, S.; Brenmoehl, J.; Langhammer, M.; Hoeflich, A.; Röntgen, M. Myogenic Precursor Cells Show Faster Activation and Enhanced Differentiation in a Male Mouse Model Selected for Advanced Endurance Exercise Performance. Cells 2022, 11, 1001. [Google Scholar] [CrossRef]
- Walz, C.; Brenmoehl, J.; Trakooljul, N.; Noce, A.; Caffier, C.; Ohde, D.; Langhammer, M.; Wimmers, K.; Ponsuksili, S.; Hoeflich, A. Control of Protein and Energy Metabolism in the Pituitary Gland in Response to Three-Week Running Training in Adult Male Mice. Cells 2021, 10, 736. [Google Scholar] [CrossRef]
- Brenmoehl, J.; Walz, C.; Spitschak, M.; Wirthgen, E.; Walz, M.; Langhammer, M.; Tuchscherer, A.; Naumann, R.; Hoeflich, A. Partial phenotype conversion and differential trait response to conditions of husbandry in mice. J. Comp. Physiol. B 2018, 188, 527–539. [Google Scholar] [CrossRef] [Green Version]
- Rehfeldt, C.; Ott, G.; Gerrard, D.; Varga, L.; Schlote, W.; Williams, J.L.; Renne, U.; Bunger, L. Effects of the Compact mutant myostatin allele Mstn(Cmpt-dl1Abc) introgressed into a high growth mouse line on skeletal muscle cellularity. J. Muscle Res. Cell Motil. 2005, 26, 103–112. [Google Scholar] [CrossRef]
- Spurgeon, S.L.; Jones, R.C.; Ramakrishnan, R. High Throughput Gene Expression Measurement with Real Time PCR in a Microfluidic Dynamic Array. PLoS ONE 2008, 3, e1662. [Google Scholar] [CrossRef] [Green Version]
- Ballester, M.; Cordón, R.; Folch, J.M. DAG Expression: High-Throughput Gene Expression Analysis of Real-Time PCR Data Using Standard Curves for Relative Quantification. PLoS ONE 2013, 8, e80385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G., Jr.; Brouwer, C. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassid, W.Z.; Abraham, S. [7] Chemical procedures for analysis of polysaccharides. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 1957; Volume 3, pp. 34–50. [Google Scholar]
- Nakatani, T.; Nakashima, T.; Kita, T.; Hirofuji, C.; Itoh, K.; Itoh, M.; Ishihara, A. Cell size and oxidative enzyme activity of different types of fibers in different regions of the rat plantaris and tibialis anterior muscles. Jpn. J. Physiol. 2000, 50, 413–418. [Google Scholar] [CrossRef] [Green Version]
- Soffe, Z.; Radley-Crabb, H.G.; McMahon, C.; Grounds, M.D.; Shavlakadze, T. Effects of loaded voluntary wheel exercise on performance and muscle hypertrophy in young and old male C57Bl/6J mice. Scand. J. Med. Sci. Sports 2016, 26, 172–188. [Google Scholar] [CrossRef]
- Faulkner, J.A.; Maxwell, L.C.; Lieberman, D.A. Histochemical characteristics of muscle fibers from trained and detrained guinea pigs. Am. J. Physiol. 1972, 222, 836–840. [Google Scholar] [CrossRef] [Green Version]
- Storlien, L.; Oakes, N.D.; Kelley, D.E. Metabolic flexibility. Proc. Nutr. Soc. 2004, 63, 363–368. [Google Scholar] [CrossRef]
- Wilson, J.M.; Loenneke, J.P.; Jo, E.; Wilson, G.J.; Zourdos, M.C.; Kim, J.-S. The Effects of Endurance, Strength, and Power Training on Muscle Fiber Type Shifting. J. Strength. Cond. Res. 2012, 26, 1724–1729. [Google Scholar] [CrossRef]
- Yaghoob Nezhad, F.; Verbrugge, S.A.J.; Schonfelder, M.; Becker, L.; de Angelis, M.H.; Wackerhage, H. Genes Whose Gain or Loss-of-Function Increases Endurance Performance in Mice: A Systematic Literature Review. Front. Physiol. 2019, 10, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fueger, P.T.; Shearer, J.; Krueger, T.M.; Posey, K.A.; Bracy, D.P.; Heikkinen, S.; Laakso, M.; Rottman, J.N.; Wasserman, D.H. Hexokinase II protein content is a determinant of exercise endurance capacity in the mouse. J. Physiol. 2005, 566, 533–541. [Google Scholar] [CrossRef]
- Romijn, J.A.; Coyle, E.F.; Sidossis, L.S.; Gastaldelli, A.; Horowitz, J.F.; Endert, E.; Wolfe, R.R. Regulation of endogenous fat and carbohydrate metabolism in relation to exercise intensity and duration. Am. J. Physiol. 1993, 265, E380–E391. [Google Scholar] [CrossRef] [Green Version]
- Dyck, D.J.; Peters, S.J.; Wendling, P.S.; Chesley, A.; Hultman, E.; Spriet, L.L. Regulation of muscle glycogen phosphorylase activity during intense aerobic cycling with elevated FFA. Am. J. Physiol. Endocrinol. Metab. 1996, 270, E116–E125. [Google Scholar] [CrossRef]
- Green, H.J.; Ball-Burnett, M.E.; Morrissey, M.A.; Spalding, M.J.; Hughson, R.L.; Fraser, I.G. Fiber type specific glycogen utilization in rat diaphragm during treadmill exercise. J. Appl. Physiol. 1987, 63, 75–83. [Google Scholar] [CrossRef]
- Murase, T.; Haramizu, S.; Shimotoyodome, A.; Tokimitsu, I.; Hase, T. Green tea extract improves running endurance in mice by stimulating lipid utilization during exercise. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 290, R1550–R1556. [Google Scholar] [CrossRef]
- Gladden, L.B. Muscle as a consumer of lactate. Med. Sci. Sports Exerc. 2000, 32, 764–771. [Google Scholar] [CrossRef]
- Ohkuwa, T.; Kato, Y.; Katsumata, K.; Nakao, T.; Miyamura, M. Blood lactate and glycerol after 400-m and 3,000-m runs in sprint and long distance runners. Eur. J. Appl. Physiol. Occup. Physiol. 1984, 53, 213–218. [Google Scholar] [CrossRef]
- Fujitsuka, N.; Yamamoto, T.; Ohkuwa, T.; Saito, M.; Miyamura, M. Peak blood lactate after short periods of maximal treadmill running. Eur. J. Appl. Physiol. Occup. Physiol. 1982, 48, 289–296. [Google Scholar] [CrossRef]
- Zhang, S.; Hulver, M.W.; McMillan, R.P.; Cline, M.A.; Gilbert, E.R. The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr. Metab. 2014, 11, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arner, P.; Kriegholm, E.; Engfeldt, P.; Bolinder, J. Adrenergic regulation of lipolysis in situ at rest and during exercise. J. Clin. Investig. 1990, 85, 893–898. [Google Scholar] [CrossRef] [Green Version]
- Frayn, K.N. Adipose tissue as a buffer for daily lipid flux. Diabetologia 2002, 45, 1201–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Verdejo, R.; Bajpeyi, S.; Ravussin, E.; Galgani, J.E. Metabolic flexibility to lipid availability during exercise is enhanced in individuals with high insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2018, 315, E715–E722. [Google Scholar] [CrossRef]
- Hagberg, C.E.; Falkevall, A.; Wang, X.; Larsson, E.; Huusko, J.; Nilsson, I.; van Meeteren, L.A.; Samen, E.; Lu, L.; Vanwildemeersch, M.; et al. Vascular endothelial growth factor B controls endothelial fatty acid uptake. Nature 2010, 464, 917–921. [Google Scholar] [CrossRef] [Green Version]
- Muoio, D.M. Metabolism and vascular fatty acid transport. N. Engl. J. Med. 2010, 363, 291–293. [Google Scholar] [CrossRef]
- Bruce, C.R.; Brolin, C.; Turner, N.; Cleasby, M.E.; van der Leij, F.R.; Cooney, G.J.; Kraegen, E.W. Overexpression of carnitine palmitoyltransferase I in skeletal muscle in vivo increases fatty acid oxidation and reduces triacylglycerol esterification. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E1231–E1237. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, K.; Tunaru, S.; Tang, C.; Muller, M.; Gille, A.; Sassmann, A.; Hanson, J.; Offermanns, S. An autocrine lactate loop mediates insulin-dependent inhibition of lipolysis through GPR81. Cell Metab. 2010, 11, 311–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Wu, J.; Zhu, J.; Kuei, C.; Yu, J.; Shelton, J.; Sutton, S.W.; Li, X.; Yun, S.J.; Mirzadegan, T.; et al. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 2009, 284, 2811–2822. [Google Scholar] [CrossRef] [Green Version]




| Pathway | Involved Genes | Gene Name | DUhTP Sed vs. DUC Sed | DUhTP Trained vs. DUC Trained | DUhTP Trained vs. DUhTP Sed | DUC Trained vs. DUC Sed | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| logFC | FDR | logFC | FDR | logFC | FDR | logFC | FDR | |||
| Glycogen metabolism | Phosphorylase b kinase | Phkb | −0.41 | ** | −0.38 | * | ||||
| Glycogen phosphorylase | Pygm | 0.44 | ** | |||||||
| Glycogen debranching enzyme | Agl | −0.48 | ** | −0.35 | * | |||||
| Glycogenin | Gyg | 0.28 | ** | |||||||
| UTP-glucose-1-phosphate uridylyltransferase | Ugp2 | −0.28 | * | |||||||
| Glycogen synthase | Gys1 | 0.23 | * | |||||||
| Glycolysis | Glucose transporter Glut1 | Slc2a1 | ||||||||
| Glucose transporter Glut4 | Slc2a4 | 0.23 | * | 0.36 | **** | 0.27 | * | |||
| Hexokinase | Hk2 | −0.50 | *** | 0.75 | *** | 0.41 | * | |||
| Phosphoglucoisomerase | Gpi1 | 0.38 | *** | 0.58 | **** | 0.26 | 0.057 | |||
| Phosphofructokinase | Pfkm | 0.30 | * | |||||||
| Aldolase | Aldoa | 0.36 | ** | 0.51 | **** | |||||
| Phosphotriose isomerase | Tpi1 | 0.45 | **** | 0.53 | **** | |||||
| Glyceraldehyde 3-P dehydrogenase | Gapdh | 0.66 | **** | 0.84 | **** | 0.39 | * | |||
| Phosphoglycerate kinase | Pgk1 | 0.29 | * | 0.27 | * | |||||
| Phosphoglycerate mutase | Pgam1 | 0.73 | * | |||||||
| Pgam2 | 0.52 | **** | 0.61 | **** | ||||||
| Enolase | Eno3 | 0.62 | **** | 0.55 | **** | |||||
| Pyruvate kinase | Pkm | 0.30 | * | 0.39 | *** | |||||
| Lactate dehydrogenase | Ldha | 0.41 | *** | 0.40 | *** | |||||
| Ldhd | 0.26 | * | 0.25 | * | ||||||
| Pyruvate Transporter | Mpc1 | 0.43 | **** | 0.59 | **** | |||||
| Mpc2 | 0.47 | **** | 0.29 | ** | ||||||
| Lactate transporter MCT1 | Slc16a1 | −0.43 | * | |||||||
| Lactate transporter MCT2 | Slc16a3 | 0.40 | * | 0.40 | * | |||||
| Fatty acid (FA) oxidation | FA transporter CD36 | Cd36 | ||||||||
| FA transportprotein 1 | Slc27a1 | 0.58 | ** | 1.03 | **** | |||||
| FA transportprotein 4 | Slc27a4 | 0.39 | *** | 0.32 | * | |||||
| FA binding protein 3 | Fabp3 | 0.54 | * | 1.14 | **** | |||||
| Carnitine-O-palmitoyltransferase 2 | Cpt2 | 0.45 | ** | 0.63 | **** | |||||
| Acyl-CoA dehydrogenase | Acads | 0.62 | **** | 0.77 | **** | |||||
| Acadm | 0.52 | ** | 0.53 | ** | ||||||
| Acadl | 0.83 | **** | ||||||||
| Acadvl | 0.57 | *** | 0.84 | **** | ||||||
| Enoyl-CoA hydratase | Hadha | 0.32 | * | 0.68 | **** | |||||
| Hydroxy acyl-CoA dehydrogenase | Hsd17b10 | 0.58 | **** | 0.75 | **** | |||||
| Ketoacyl-CoA thiolase | Acaa2 | 0.77 | **** | 1.05 | **** | |||||
| TCA cycle | Citrate synthase | Cs | 0.43 | *** | 0.50 | **** | ||||
| Aconitate hydratase | Aco2 | 0.38 | ** | 0.64 | **** | |||||
| Isocitrate dehydrogenase | Idh2 | 0.86 | ** | 1.35 | **** | |||||
| Isocitrate dehydrogenase | Idh3b | 0.42 | **** | 0.55 | **** | |||||
| Isocitrate dehydrogenase | Idh3g | 0.45 | **** | 0.59 | **** | |||||
| a-ketoglutarate dehydrogenase | Ogdh | 0.43 | **** | 0.24 | 0.062 | |||||
| Succinyl-CoA synthetase | Suclg1 | 0.45 | **** | 0.50 | **** | |||||
| Suclg2 | 0.31 | ** | 0.28 | ** | ||||||
| Sucla2 | 0.27 | ** | ||||||||
| Succinate dehydrogenase | Sdhc | 0.63 | **** | 0.53 | **** | |||||
| Sdhd | 0.45 | **** | 0.46 | **** | ||||||
| Sdhb | 0.47 | ** | 0.72 | **** | ||||||
| Sdha | 0.38 | *** | ||||||||
| Fumarase | Fh1 | 0.45 | **** | 0.52 | **** | |||||
| Malate dehydrogenase | Mdh2 | 0.53 | **** | 0.83 | **** | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brenmoehl, J.; Brosig, E.; Trakooljul, N.; Walz, C.; Ohde, D.; Noce, A.; Walz, M.; Langhammer, M.; Petkov, S.; Röntgen, M.; et al. Metabolic Pathway Modeling in Muscle of Male Marathon Mice (DUhTP) and Controls (DUC)—A Possible Role of Lactate Dehydrogenase in Metabolic Flexibility. Cells 2023, 12, 1925. https://doi.org/10.3390/cells12151925
Brenmoehl J, Brosig E, Trakooljul N, Walz C, Ohde D, Noce A, Walz M, Langhammer M, Petkov S, Röntgen M, et al. Metabolic Pathway Modeling in Muscle of Male Marathon Mice (DUhTP) and Controls (DUC)—A Possible Role of Lactate Dehydrogenase in Metabolic Flexibility. Cells. 2023; 12(15):1925. https://doi.org/10.3390/cells12151925
Chicago/Turabian StyleBrenmoehl, Julia, Elli Brosig, Nares Trakooljul, Christina Walz, Daniela Ohde, Antonia Noce, Michael Walz, Martina Langhammer, Stefan Petkov, Monika Röntgen, and et al. 2023. "Metabolic Pathway Modeling in Muscle of Male Marathon Mice (DUhTP) and Controls (DUC)—A Possible Role of Lactate Dehydrogenase in Metabolic Flexibility" Cells 12, no. 15: 1925. https://doi.org/10.3390/cells12151925






