Topology and Function of the S. cerevisiae Autophagy Protein Atg15
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Plasmids
| Plasmid | Genotype | Reference |
|---|---|---|
| PMET-GFP-Atg15 | pUG34 CEN PMET17-GFP-ATG15-TCYC1 | This study |
| PATG15-GFP-Atg15 | pRS313 CEN PAtg15-GFP-ATG15-TATG15 | This study |
| GFP | pUG34 CEN PMET17-GFP-TCYC1 | [22] |
| 2xCitrine-Atg15 | pRS313 CEN PATG15-2xyECitrine-ATG15-TAtg15 | This study |
| Atg15-3xHA | pRS316 CEN PATG15-ATG15-3xHA | [6] |
| Atg15no glyc-3xHA | pRS316 CEN PATG15-ATG15N173/202/208A-3xHA | This study |
| Atg151glyc-3xHA | pRS316 CEN PATG15-ATG15N173/208A-3xHA | This study |
| mCherry-Atg8 | pRS316 CEN PATG8-mCherry-ATG8-TAtg8 | [25] |
| PAtg15-Atg15∆TMD-3xHA | pRS426 2µ PATG15-ATG15∆TMD-3xHA-TCYC1 | This study |
| PMET17-Atg15∆TMD-3xHA | ||
| PGAL1-Atg15∆TMD-3xHA | pYES2 2µ PGAL1-ATG15∆TMD-3xHA-TCYC1 | This study |
| ALP-Atg15∆TMD-3xHA | pYES2 2µ PGAL1-ALP(1-58)-ATG15∆TMD-3xHA-TCYC1 | This study |
| CPY-Atg15∆TMD-3xHA | pYES2 2µ PGAL1-CPY(1-58)-ATG15∆TMD-3xHA-TCYC1 | This study |
| Atg15-HA | ||
| Sec63-RFP | pJK59 PSec63-Sec63-RFP | This study |
| GFP-Atg18 | pRS316 CEN PATG8-GFP-ATG8-TATG8 | [26] |
| Pgk1-GFP | pRS316 CEN PPGK1-PGK1-GFP-TADH1 | [27] |
| Atg15(1-520)-Suc2His4 | YEp352 2µ ATG151−520-SUC2HIS4C | This study |
2.3. Western Blot Analysis
2.4. Fluorescence Microscopy
2.5. Deglycosylation Assay
2.6. Membrane Topology
2.7. Statistical Analyses
3. Results
3.1. Atg15 Is Not Localized at the Phagophore or at Autophagic Bodies
3.2. Generation and Characterization of Atg15-Chimeras
3.3. Experimental Dissection of the Membrane Topology of Atg15
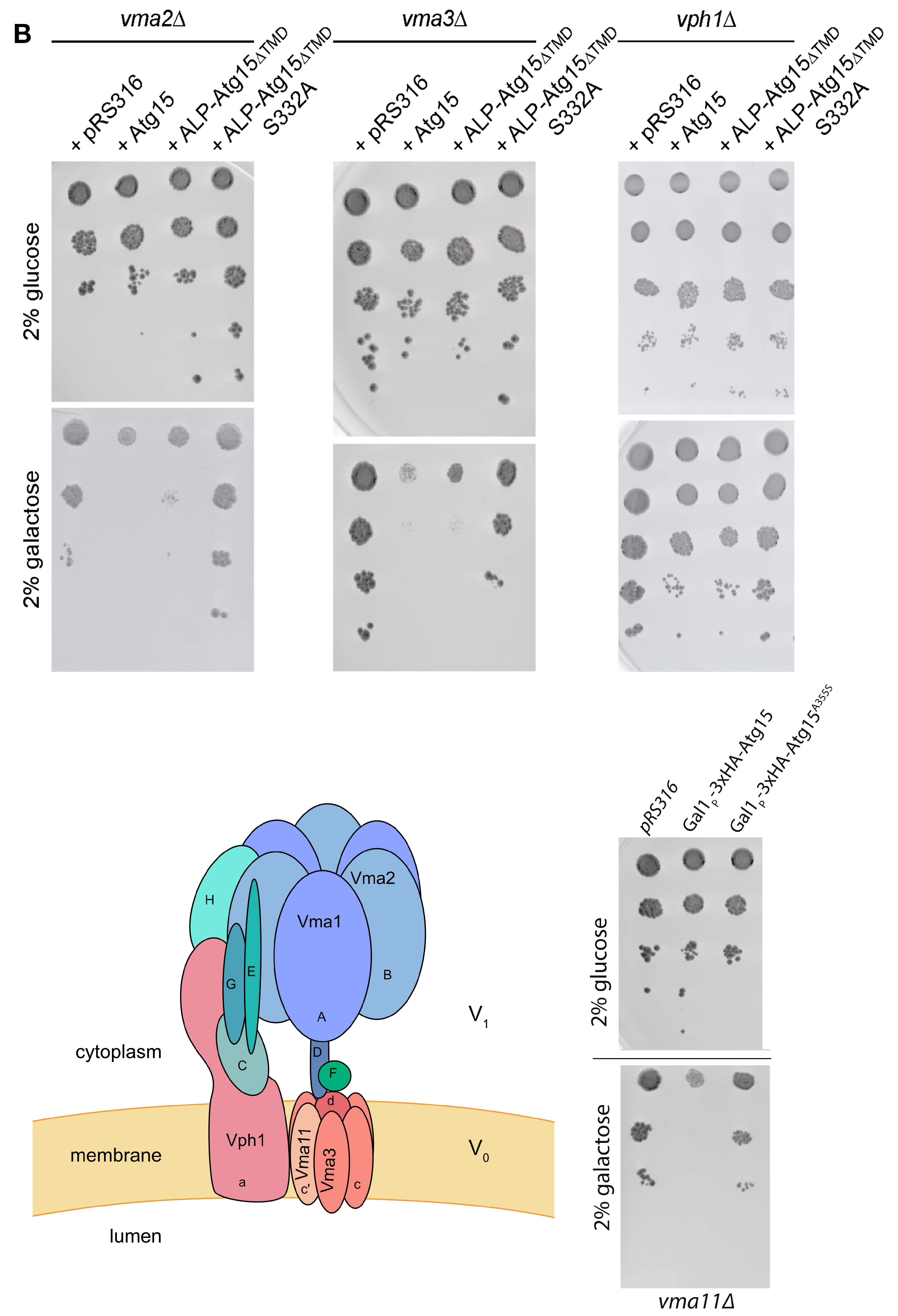
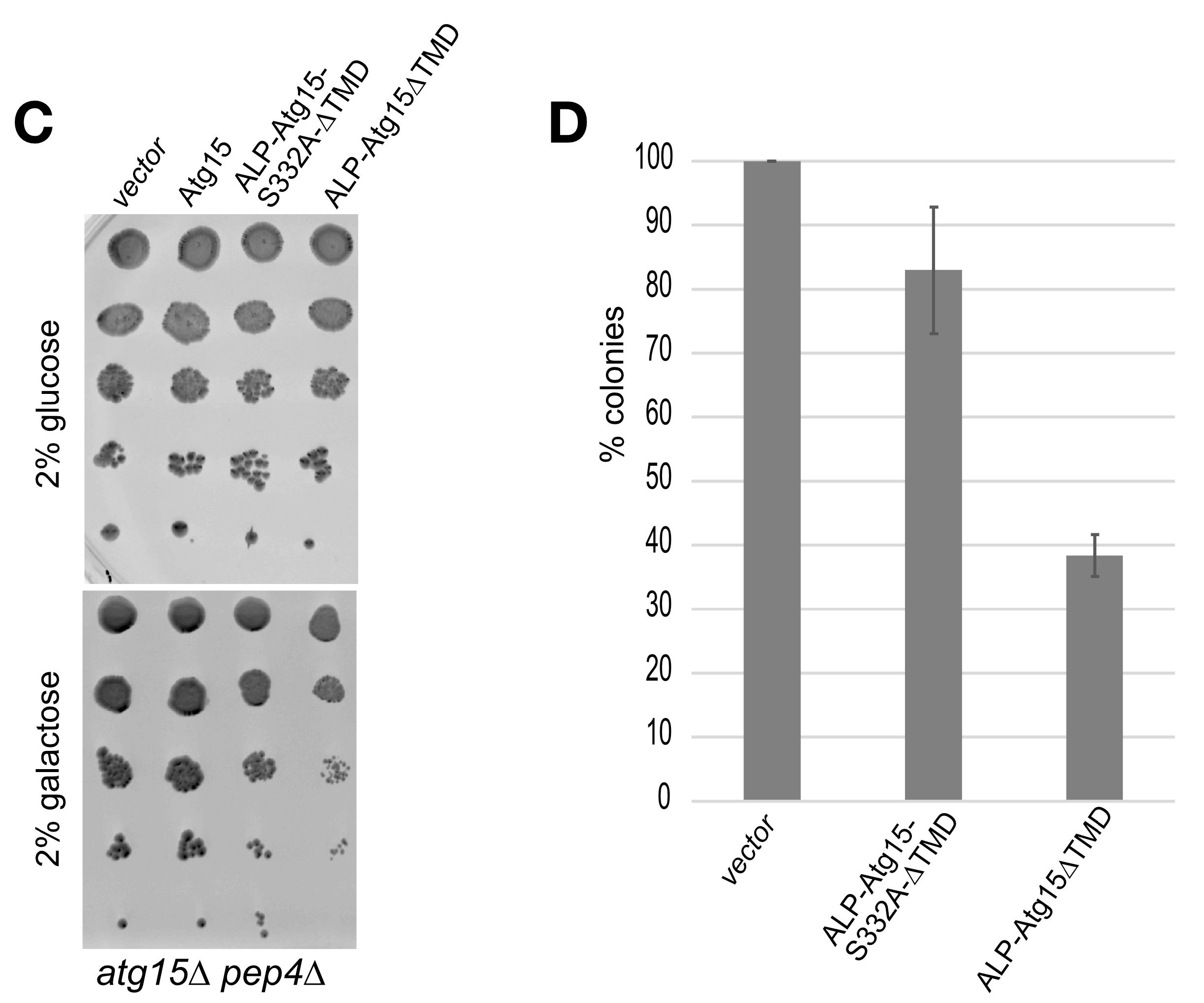
3.4. Overexpression of Membrane-Embedded Atg15 Variants Affects Growth, Dependent on the Active Site Serine 332
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wen, X.; Yang, Y.; Klionsky, D.J. Moments in autophagy and disease: Past and present. Mol. Aspects Med. 2021, 82, 100966. [Google Scholar] [CrossRef]
- Hu, Y.; Reggiori, F. Molecular regulation of autophagosome formation. Biochem. Soc. Trans. 2022, 50, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Zhang, S.; Mizushima, N. Autophagy genes in biology and disease. Nat. Rev. Genet. 2023, 24, 382–400. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, L.; Thumm, M. Autophagic and non-autophagic functions of the Saccharomyces cerevisiae PROPPINs Atg18, Atg21 and Hsv2. Biol. Chem. 2023. [Google Scholar] [CrossRef]
- Epple, U.D.; Eskelinen, E.L.; Thumm, M. Intravacuolar membrane lysis in Saccharomyces cerevisiae. Does vacuolar targeting of Cvt17/Aut5p affect its function? J. Biol. Chem. 2003, 278, 7810–7821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epple, U.D.; Suriapranata, I.; Eskelinen, E.L.; Thumm, M. Aut5/Cvt17p, a putative lipase essential for disintegration of autophagic bodies inside the vacuole. J. Bacteriol. 2001, 183, 5942–5955. [Google Scholar] [CrossRef] [Green Version]
- Teter, S.A.; Eggerton, K.P.; Scott, S.V.; Kim, J.; Fischer, A.M.; Klionsky, D.J. Degradation of lipid vesicles in the yeast vacuole requires function of Cvt17, a putative lipase. J. Biol. Chem. 2001, 276, 2083–2087. [Google Scholar] [CrossRef] [Green Version]
- Hirata, E.; Shirai, K.; Kawaoka, T.; Sato, K.; Kodama, F.; Suzuki, K. Atg15 in Saccharomyces cerevisiae consists of two functionally distinct domains. Mol. Biol. Cell 2021, 32, 645–663. [Google Scholar] [CrossRef]
- Krick, R.; Muehe, Y.; Prick, T.; Bremer, S.; Schlotterhose, P.; Eskelinen, E.L.; Millen, J.; Goldfarb, D.S.; Thumm, M. Piecemeal microautophagy of the nucleus requires the core macroautophagy genes. Mol. Biol. Cell 2008, 19, 4492–4505. [Google Scholar] [CrossRef] [Green Version]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef] [Green Version]
- Otto, F.B.; Thumm, M. Mechanistic dissection of macro- and micronucleophagy. Autophagy 2021, 17, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Otto, F.B.; Thumm, M. Nucleophagy-Implications for Microautophagy and Health. Int. J. Mol. Sci. 2020, 21, 4506. [Google Scholar] [CrossRef]
- van Zutphen, T.; Todde, V.; de Boer, R.; Kreim, M.; Hofbauer, H.F.; Wolinski, H.; Veenhuis, M.; van der Klei, I.J.; Kohlwein, S.D. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 25, 290–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maeda, Y.; Oku, M.; Sakai, Y. A defect of the vacuolar putative lipase Atg15 accelerates degradation of lipid droplets through lipolysis. Autophagy 2015, 11, 1247–1258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, L.N.; Bormann, J.; Le, G.T.; Starkel, C.; Olsson, S.; Nosanchuk, J.D.; Giese, H.; Schafer, W. Autophagy-related lipase FgATG15 of Fusarium graminearum is important for lipid turnover and plant infection. Fungal Genet. Biol. 2011, 48, 217–224. [Google Scholar] [CrossRef]
- Ramya, V.; Rajasekharan, R. ATG15 encodes a phospholipase and is transcriptionally regulated by YAP1 in Saccharomyces cerevisiae. FEBS Lett. 2016, 590, 3155–3167. [Google Scholar] [CrossRef]
- Yamakuchi, Y.; Kurokawa, Y.; Konishi, R.; Fukuda, K.; Masatani, T.; Fujita, A. Selective increment of phosphatidylserine on the autophagic body membrane in the yeast vacuole. FEBS Lett. 2021, 595, 2197–2207. [Google Scholar] [CrossRef]
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992, 119, 301–311. [Google Scholar] [CrossRef]
- Nakamura, N.; Matsuura, A.; Wada, Y.; Ohsumi, Y. Acidification of vacuoles is required for autophagic degradation in the yeast, Saccharomyces cerevisiae. J. Biochem. 1997, 121, 338–344. [Google Scholar] [CrossRef]
- Thumm, M.; Egner, R.; Koch, B.; Schlumpberger, M.; Straub, M.; Veenhuis, M.; Wolf, D.H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994, 349, 275–280. [Google Scholar] [CrossRef] [Green Version]
- Janke, C.; Magiera, M.M.; Rathfelder, N.; Taxis, C.; Reber, S.; Maekawa, H.; Moreno-Borchart, A.; Doenges, G.; Schwob, E.; Schiebel, E.; et al. A versatile toolbox for PCR-based tagging of yeast genes: New fluorescent proteins, more markers and promoter substitution cassettes. Yeast 2004, 21, 947–962. [Google Scholar] [CrossRef] [PubMed]
- Gueldener, U.; Heinisch, J.; Koehler, G.J.; Voss, D.; Hegemann, J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002, 30, e23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheff, M.A.; Thorn, K.S. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 2004, 21, 661–670. [Google Scholar] [CrossRef] [PubMed]
- Strahl-Bolsinger, S.; Scheinost, A. Transmembrane topology of pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 1999, 274, 9068–9075. [Google Scholar] [CrossRef] [Green Version]
- Munzel, L.; Neumann, P.; Otto, F.B.; Krick, R.; Metje-Sprink, J.; Kroppen, B.; Karedla, N.; Enderlein, J.; Meinecke, M.; Ficner, R.; et al. Atg21 organizes Atg8 lipidation at the contact of the vacuole with the phagophore. Autophagy 2021, 17, 1458–1478. [Google Scholar] [CrossRef]
- Suzuki, K.; Kirisako, T.; Kamada, Y.; Mizushima, N.; Noda, T.; Ohsumi, Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001, 20, 5971–5981. [Google Scholar] [CrossRef]
- Welter, E.; Thumm, M.; Krick, R. Quantification of nonselective bulk autophagy in S. cerevisiae using Pgk1-GFP. Autophagy 2010, 6, 794–797. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Meiling-Wesse, K.; Barth, H.; Thumm, M. Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett. 2002, 526, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Akioka, M.; Kondo-Kakuta, C.; Yamamoto, H.; Ohsumi, Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J. Cell Sci. 2013, 126, 2534–2544. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.B.; Ho, C.T.; Winkler, J.; Khokhrina, M.; Neuner, A.; Mohamed, M.Y.; Guilbride, D.L.; Richter, K.; Lisby, M.; Schiebel, E.; et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. EMBO J. 2015, 34, 778–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, H.E.; Samant, R.S. Alternative systems for misfolded protein clearance: Life beyond the proteasome. FEBS J. 2021, 288, 4464–4487. [Google Scholar] [CrossRef] [PubMed]
- Deak, P.M.; Wolf, D.H. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 2001, 276, 10663–10669. [Google Scholar] [CrossRef] [Green Version]
- Sengstag, C.; Stirling, C.; Schekman, R.; Rine, J. Genetic and biochemical evaluation of eucaryotic membrane protein topology: Multiple transmembrane domains of Saccharomyces cerevisiae 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol. Cell Biol. 1990, 10, 672–680. [Google Scholar] [CrossRef] [PubMed]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Huang, J.; Geng, J.; Nair, U.; Klionsky, D.J. Atg22 recycles amino acids to link the degradative and recycling functions of autophagy. Mol. Biol. Cell 2006, 17, 5094–5104. [Google Scholar] [CrossRef] [Green Version]
- Balderhaar, H.J.; Ungermann, C. CORVET and HOPS tethering complexes—Coordinators of endosome and lysosome fusion. J. Cell Sci. 2013, 126, 1307–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkel, O.; Oskolkova, O.V.; Raab, F.; El-Toukhy, R.; Paltauf, F. Regulation of activity in vitro and in vivo of three phospholipases B from Saccharomyces cerevisiae. Biochem. J. 2005, 387, 489–496. [Google Scholar] [CrossRef] [Green Version]







| Strain | Genotype | Reference |
|---|---|---|
| WCG atg15∆ | atg15∆::KAN | [6] |
| WCG atg15∆ atg1∆ | atg15∆::KAN atg1∆::KAN | [6] |
| WCG atg15∆ vps4∆ | atg15∆::KAN vps4∆::KAN | This study |
| WCG atg15∆ atg1∆ vps4∆ | atg15∆::KAN atg1∆::natNT2 vps4∆::KAN | This study |
| WCG atg15∆ pep4∆ | atg15∆::KAN pep4∆::KAN | This study |
| WCG atg15∆ pep4∆ vps4∆ | atg15∆::KAN pep4∆::hphNT1 vps4∆::KAN | This study |
| WCG atg15∆atg1∆ pep4∆ vps4∆ | atg15∆::KAN atg1∆::natNT2 pep4∆::hphNT1 vps4∆::KAN | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquardt, L.; Montino, M.; Mühe, Y.; Schlotterhose, P.; Thumm, M. Topology and Function of the S. cerevisiae Autophagy Protein Atg15. Cells 2023, 12, 2056. https://doi.org/10.3390/cells12162056
Marquardt L, Montino M, Mühe Y, Schlotterhose P, Thumm M. Topology and Function of the S. cerevisiae Autophagy Protein Atg15. Cells. 2023; 12(16):2056. https://doi.org/10.3390/cells12162056
Chicago/Turabian StyleMarquardt, Lisa, Marco Montino, Yvonne Mühe, Petra Schlotterhose, and Michael Thumm. 2023. "Topology and Function of the S. cerevisiae Autophagy Protein Atg15" Cells 12, no. 16: 2056. https://doi.org/10.3390/cells12162056
APA StyleMarquardt, L., Montino, M., Mühe, Y., Schlotterhose, P., & Thumm, M. (2023). Topology and Function of the S. cerevisiae Autophagy Protein Atg15. Cells, 12(16), 2056. https://doi.org/10.3390/cells12162056





