Variable Cre Recombination Efficiency in Placentas of Cyp19-Cre ROSAmT/mG Transgenic Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Collection
2.3. Evaluation of Cyp19-Cre Activity
2.4. Gene Excision Analysis and Determination of Recombination
2.5. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.6. Statistical Analysis
3. Results and Discussion
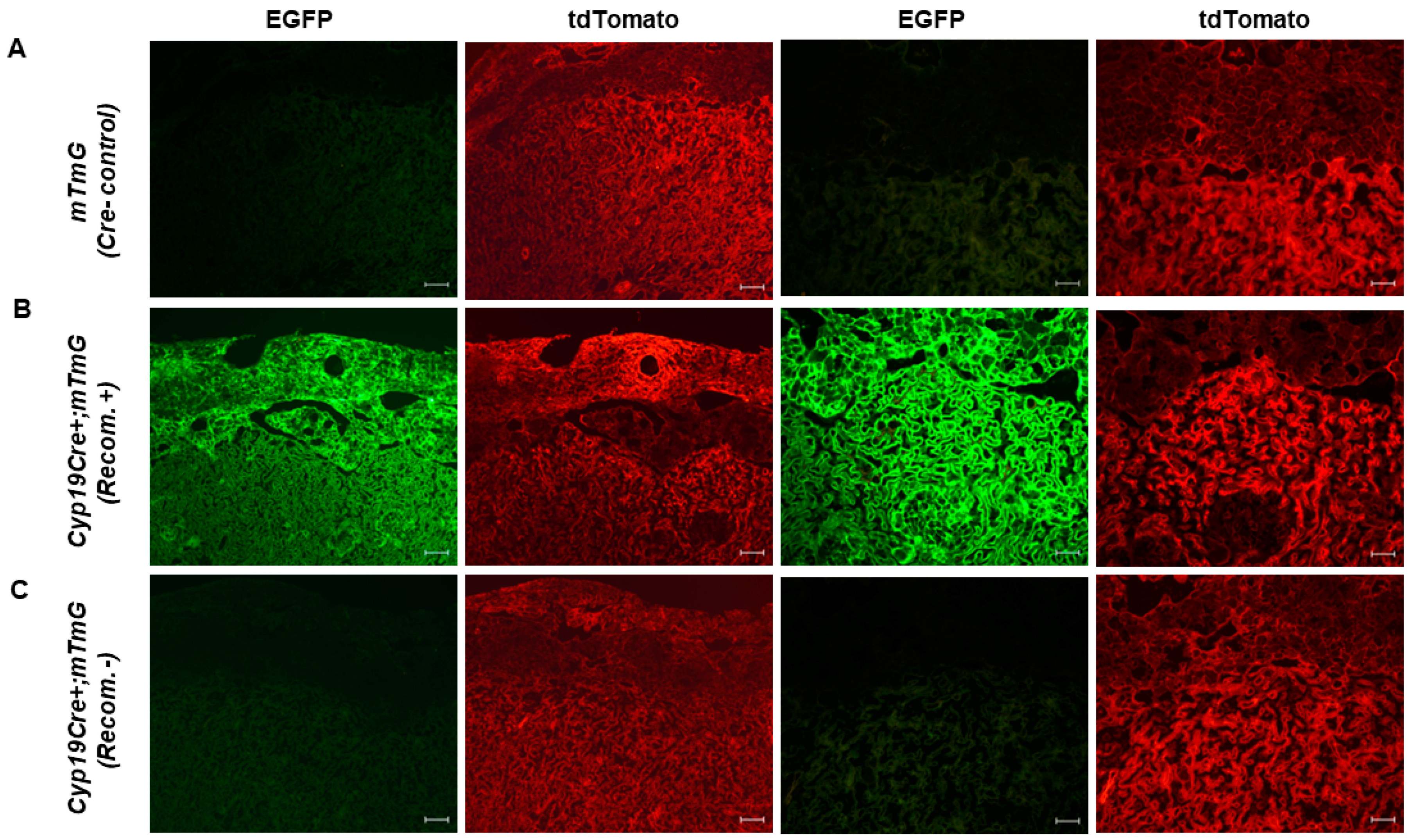
3.1. Variable Cre Recombinase Activity in Cyp19-Cre;ROSAmT/mG Placentas
3.2. Lack of Cre-Mediated Recombination at loxP Sites in Cyp19-Cre;ROSAmT/mG Placentas
3.3. High Level of Recombination Efficiency with the Maternally Inherited Cre Allele in Cyp19-Cre;ROSAmT/mG Placentas
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wenzel, P.L.; Leone, G. Expression of Cre recombinase in early diploid trophoblast cells of the mouse placenta. Genesis 2007, 45, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, P.L.; Wu, L.; de Bruin, A.; Chong, J.L.; Chen, W.Y.; Dureska, G.; Sites, E.; Pan, T.; Sharma, A.; Huang, K.; et al. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 2007, 21, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Ouseph, M.M.; Li, J.; Chen, H.Z.; Pecot, T.; Wenzel, P.; Thompson, J.C.; Comstock, G.; Chokshi, V.; Byrne, M.; Forde, B.; et al. Atypical E2F repressors and activators coordinate placental development. Dev. Cell 2012, 22, 849–862. [Google Scholar] [CrossRef]
- Howerton, C.L.; Morgan, C.P.; Fischer, D.B.; Bale, T.L. O-GlcNAc transferase (OGT) as a placental biomarker of maternal stress and reprogramming of CNS gene transcription in development. Proc. Natl. Acad. Sci. USA 2013, 110, 5169–5174. [Google Scholar] [CrossRef] [PubMed]
- Howerton, C.L.; Bale, T.L. Targeted placental deletion of OGT recapitulates the prenatal stress phenotype including hypothalamic mitochondrial dysfunction. Proc. Natl. Acad. Sci. USA 2014, 111, 9639–9644. [Google Scholar] [CrossRef] [PubMed]
- Bronson, S.L.; Chan, J.C.; Bale, T.L. Sex-Specific Neurodevelopmental Programming by Placental Insulin Receptors on Stress Reactivity and Sensorimotor Gating. Biol. Psychiatry 2017, 82, 127–138. [Google Scholar] [CrossRef]
- Nugent, B.M.; O’Donnell, C.M.; Epperson, C.N.; Bale, T.L. Placental H3K27me3 establishes female resilience to prenatal insults. Nat. Commun. 2018, 9, 2555. [Google Scholar] [CrossRef]
- Shawber, C.J.; Brown-Grant, D.A.; Wu, T.; Kitajewski, J.K.; Douglas, N.C. Dominant-negative inhibition of canonical Notch signaling in trophoblast cells does not disrupt placenta formation. Biol. Open 2019, 8, bio037721. [Google Scholar] [CrossRef]
- Bao, H.; Liu, D.; Xu, Y.; Sun, Y.; Mu, C.; Yu, Y.; Wang, C.; Han, Q.; Liu, S.; Cai, H.; et al. Hyperactivated Wnt-beta-catenin signaling in the absence of sFRP1 and sFRP5 disrupts trophoblast differentiation through repression of Ascl2. BMC Biol. 2020, 18, 151. [Google Scholar] [CrossRef]
- Lu, J.; Wu, W.; Xin, Q.; Zhou, C.; Wang, J.; Ni, Z.; Liu, D.; Xu, Y.; Yu, Y.; Yang, N.; et al. Spatiotemporal coordination of trophoblast and allantoic Rbpj signaling directs normal placental morphogenesis. Cell Death Dis. 2019, 10, 438. [Google Scholar] [CrossRef]
- Akhaphong, B.; Baumann, D.C.; Beetch, M.; Lockridge, A.D.; Jo, S.; Wong, A.; Zemanovic, T.; Mohan, R.; Fondevilla, D.L.; Sia, M.; et al. Placental mTOR complex 1 regulates fetal programming of obesity and insulin resistance in mice. JCI Insight 2021, 6, e149271. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Sacharidou, A.; Nguyen, A.; Li, C.; Chambliss, K.L.; Salmon, J.E.; Shen, Y.M.; Lo, J.; Leone, G.W.; Herz, J.; et al. Protein Phosphatase 2A Activation Via ApoER2 in Trophoblasts Drives Preeclampsia in a Mouse Model of the Antiphospholipid Syndrome. Circ. Res. 2021, 129, 735–750. [Google Scholar] [CrossRef] [PubMed]
- Sung, D.C.; Chen, X.; Chen, M.; Yang, J.; Schultz, S.; Babu, A.; Xu, Y.; Gao, S.; Keller, T.C.S.T.; Mericko-Ishizuka, P.; et al. VE-cadherin enables trophoblast endovascular invasion and spiral artery remodeling during placental development. eLife 2022, 11, e77241. [Google Scholar] [CrossRef] [PubMed]
- Moreau, J.L.; Artap, S.T.; Shi, H.; Chapman, G.; Leone, G.; Sparrow, D.B.; Dunwoodie, S.L. Cited2 is required in trophoblasts for correct placental capillary patterning. Dev. Biol. 2014, 392, 62–79. [Google Scholar] [CrossRef]
- Lopez-Tello, J.; Perez-Garcia, V.; Khaira, J.; Kusinski, L.C.; Cooper, W.N.; Andreani, A.; Grant, I.; Fernandez de Liger, E.; Lam, B.Y.; Hemberger, M.; et al. Fetal and trophoblast PI3K p110alpha have distinct roles in regulating resource supply to the growing fetus in mice. eLife 2019, 8, e45282. [Google Scholar] [CrossRef]
- Sandovici, I.; Georgopoulou, A.; Perez-Garcia, V.; Hufnagel, A.; Lopez-Tello, J.; Lam, B.Y.H.; Schiefer, S.N.; Gaudreau, C.; Santos, F.; Hoelle, K.; et al. The imprinted Igf2-Igf2r axis is critical for matching placental microvasculature expansion to fetal growth. Dev. Cell 2022, 57, 63–79.e68. [Google Scholar] [CrossRef]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef]
- Crish, J.; Conti, M.A.; Sakai, T.; Adelstein, R.S.; Egelhoff, T.T. Keratin 5-Cre-driven excision of nonmuscle myosin IIA in early embryo trophectoderm leads to placenta defects and embryonic lethality. Dev. Biol. 2013, 382, 136–148. [Google Scholar] [CrossRef]
- Kong, S.; Liang, G.; Tu, Z.; Chen, D.; Wang, H.; Lu, J. Generation of Elf5-Cre knockin mouse strain for trophoblast-specific gene manipulation. Genesis 2018, 56, e23101. [Google Scholar] [CrossRef]
- Wattez, J.S.; Qiao, L.; Lee, S.; Natale, D.R.C.; Shao, J. The platelet-derived growth factor receptor alpha promoter-directed expression of cre recombinase in mouse placenta. Dev. Dyn. 2019, 248, 363–374. [Google Scholar] [CrossRef]
- Wieczorek, A.; Perani, C.V.; Nixon, M.; Constancia, M.; Sandovici, I.; Zazara, D.E.; Leone, G.; Zhang, M.Z.; Arck, P.C.; Solano, M.E. Sex-specific regulation of stress-induced fetal glucocorticoid surge by the mouse placenta. Am. J. Physiol. Endocrinol. Metab. 2019, 317, E109–E120. [Google Scholar] [CrossRef] [PubMed]
- Ain, R.; Canham, L.N.; Soares, M.J. Gestation stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: Novel endocrine phenotype and regulation. Dev. Biol. 2003, 260, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Rai, A.; Kambham, N.; Sung, J.F.; Singh, N.; Petitt, M.; Dhal, S.; Agrawal, R.; Sutton, R.E.; Druzin, M.L.; et al. Endometrial VEGF induces placental sFLT1 and leads to pregnancy complications. J. Clin. Investig. 2014, 124, 4941–4952. [Google Scholar] [CrossRef] [PubMed]
- Vacher, C.M.; Lacaille, H.; O’Reilly, J.J.; Salzbank, J.; Bakalar, D.; Sebaoui, S.; Liere, P.; Clarkson-Paredes, C.; Sasaki, T.; Sathyanesan, A.; et al. Placental endocrine function shapes cerebellar development and social behavior. Nat. Neurosci. 2021, 24, 1392–1401. [Google Scholar] [CrossRef]
- Lawson, H.A.; Cheverud, J.M.; Wolf, J.B. Genomic imprinting and parent-of-origin effects on complex traits. Nat. Rev. Genet 2013, 14, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Mott, R.; Yuan, W.; Kaisaki, P.; Gan, X.; Cleak, J.; Edwards, A.; Baud, A.; Flint, J. The architecture of parent-of-origin effects in mice. Cell 2014, 156, 332–342. [Google Scholar] [CrossRef]
- Macias-Velasco, J.F.; St Pierre, C.L.; Wayhart, J.P.; Yin, L.; Spears, L.; Miranda, M.A.; Carson, C.; Funai, K.; Cheverud, J.M.; Semenkovich, C.F.; et al. Parent-of-origin effects propagate through networks to shape metabolic traits. eLife 2022, 11, e72989. [Google Scholar] [CrossRef]
- Redrup, L.; Branco, M.R.; Perdeaux, E.R.; Krueger, C.; Lewis, A.; Santos, F.; Nagano, T.; Cobb, B.S.; Fraser, P.; Reik, W. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 2009, 136, 525–530. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anamthathmakula, P.; Shallie, P.D.; Nayak, N.; Dhal, S.; Vivian, J.L.; Mor, G.; Soares, M.J.; Nayak, N.R. Variable Cre Recombination Efficiency in Placentas of Cyp19-Cre ROSAmT/mG Transgenic Mice. Cells 2023, 12, 2096. https://doi.org/10.3390/cells12162096
Anamthathmakula P, Shallie PD, Nayak N, Dhal S, Vivian JL, Mor G, Soares MJ, Nayak NR. Variable Cre Recombination Efficiency in Placentas of Cyp19-Cre ROSAmT/mG Transgenic Mice. Cells. 2023; 12(16):2096. https://doi.org/10.3390/cells12162096
Chicago/Turabian StyleAnamthathmakula, Prashanth, Philemon D. Shallie, Neha Nayak, Sabita Dhal, Jay L. Vivian, Gil Mor, Michael J. Soares, and Nihar R. Nayak. 2023. "Variable Cre Recombination Efficiency in Placentas of Cyp19-Cre ROSAmT/mG Transgenic Mice" Cells 12, no. 16: 2096. https://doi.org/10.3390/cells12162096





