Myoglobin in Brown Adipose Tissue: A Multifaceted Player in Thermogenesis
Abstract
:1. Introduction
2. Structure, Location, and Classical Functions of MB
2.1. Structure
2.2. Location
2.3. Classic Gas-Binding Functions
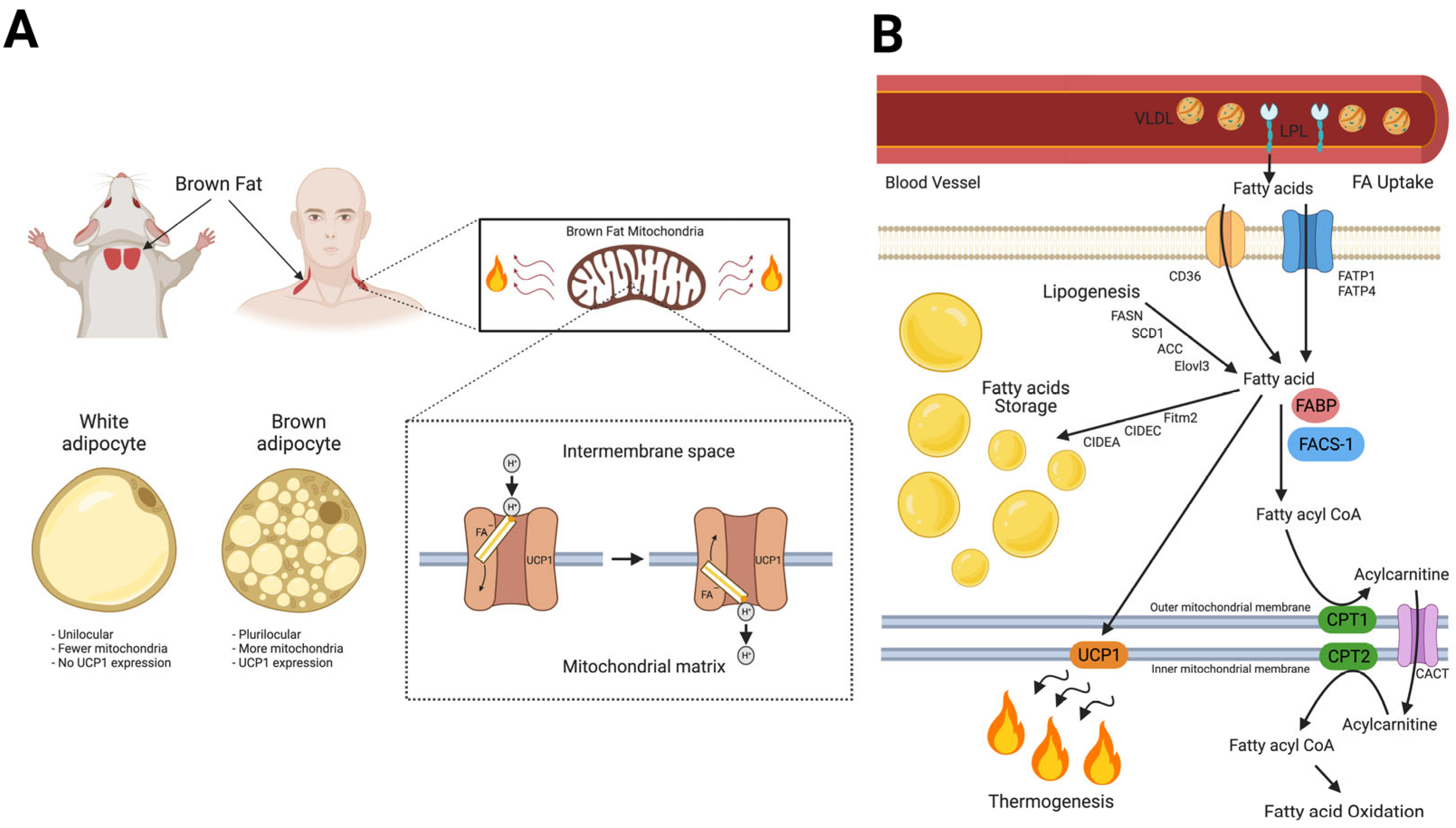
3. Fatty Acid Homeostasis in BAT Thermogenesis and Novel Roles of MB in Lipid Metabolism
4. Myoglobin in BAT; State-of-the-Art Research
4.1. Expression
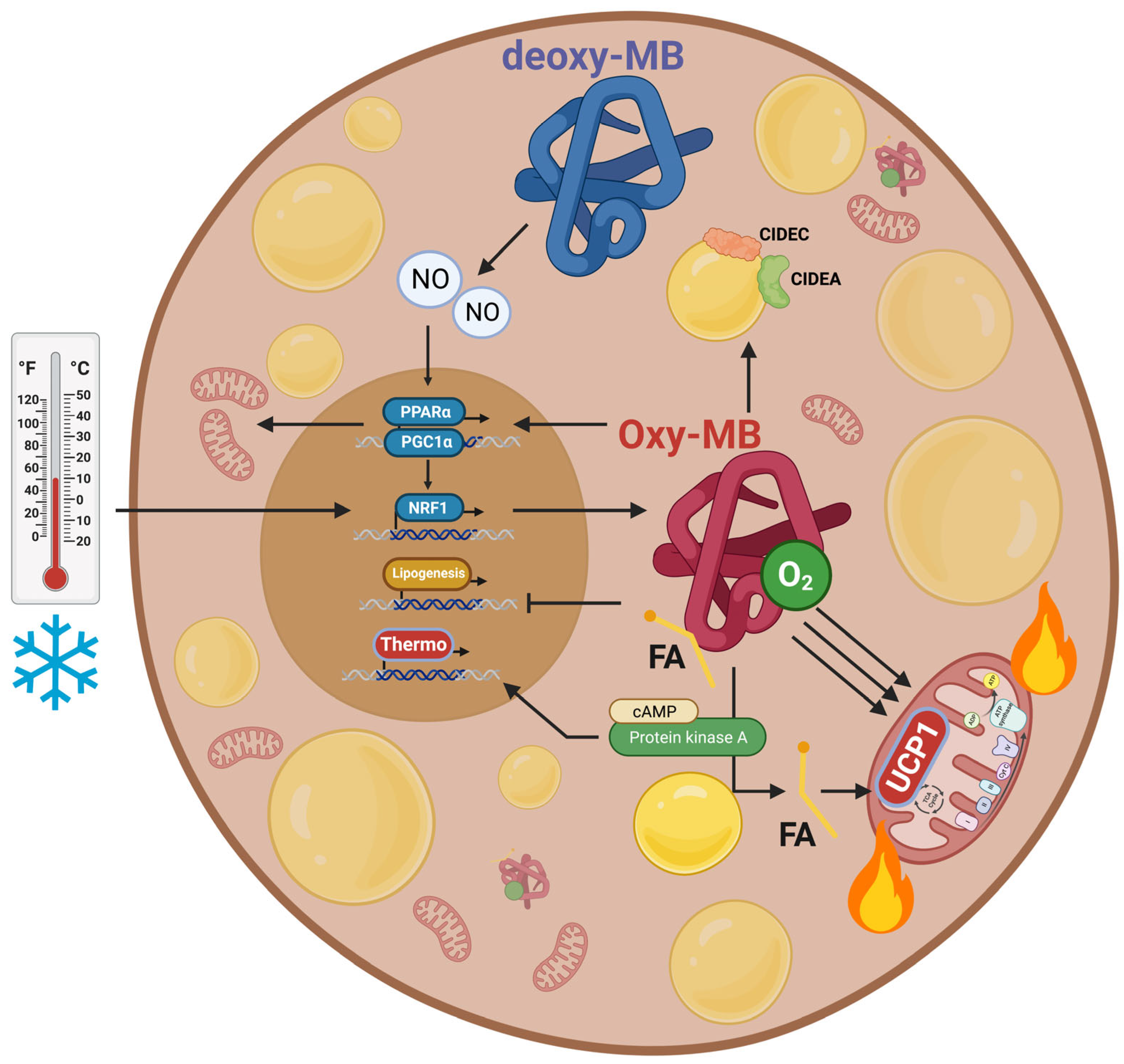
4.2. Regulation of Mitochondrial Metabolism and UCP1 Expression
4.3. Regulation of Lipid Metabolism
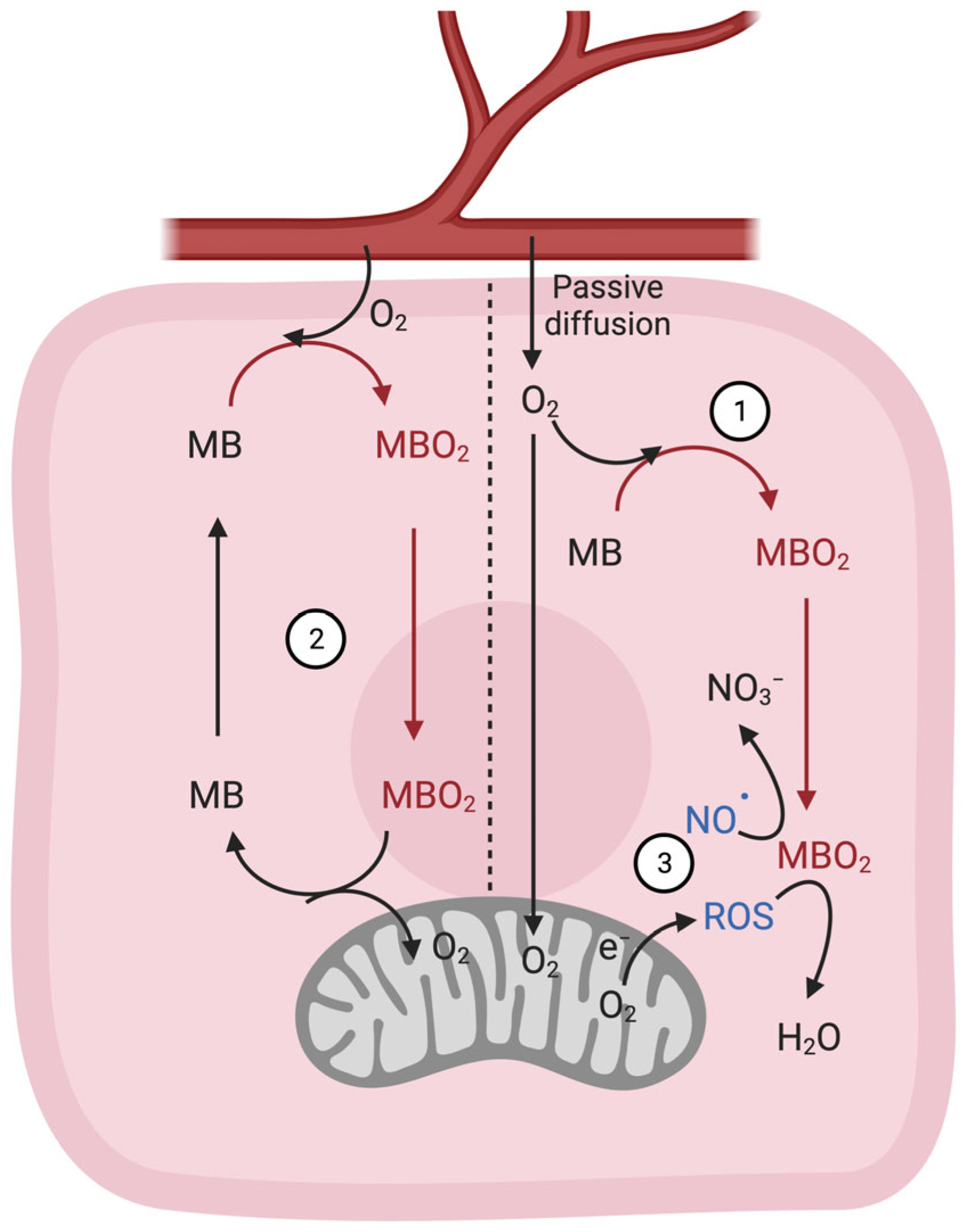
4.4. Regulation of NO• Metabolism
4.5. Regulation of Energy Expenditure and Clinical Implications
5. Bioinformatics Visions
5.1. Protein-Protein Interaction (PPI) Databases
5.2. Transcriptomics Insights
6. Sexual Dimorphism and MB Outcomes
7. Potential Therapeutic Implications of MB in BAT-Mediated Energy Metabolism
7.1. Targeting MB-FA Binding Pharmacologically
7.2. Promoting BAT Activation and WAT Browning via Regulating MB Expression
7.3. Gene Therapy
7.4. Exosomes
8. Limitations of the Current Studies
9. Summary and Conclusions
10. Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lankester, E.R. Ueber das Vorkommen von Haemoglobin in den Muskeln der Mollusken und die Verbreitung desselben in den lebendigen Organismen. Arch. Gesamte Physiol. Menschen Tiere 1871, 4, 315–320. [Google Scholar] [CrossRef]
- Günther, H. Über den Muskelfarbstoff. Virchows Arch. Pathol. Anat. Physiol. Klin. Med. 1921, 230, 146–178. [Google Scholar] [CrossRef]
- Kendrew, J.C.; Dickerson, R.E.; Strandberg, B.E.; Hart, R.G.; Davies, D.R.; Phillips, D.C.; Shore, V.C. Structure of myoglobin: A three-dimensional Fourier synthesis at 2 A. resolution. Nature 1960, 185, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Kendrew, J.C.; Bodo, G.; Dintzis, H.M.; Parrish, R.G.; Wyckoff, H.; Phillips, D.C. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 1958, 181, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Endeward, V.; Gros, G.; Jürgens, K.D. Significance of myoglobin as an oxygen store and oxygen transporter in the intermittently perfused human heart: A model study. Cardiovasc. Res. 2010, 87, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Gödecke, A.; Flögel, U.; Zanger, K.; Ding, Z.; Hirchenhain, J.; Decking, U.K.; Schrader, J. Disruption of myoglobin in mice induces multiple compensatory mechanisms. Proc. Natl. Acad. Sci. USA 1999, 96, 10495–10500. [Google Scholar] [CrossRef] [PubMed]
- Grange, R.W.; Meeson, A.; Chin, E.; Lau, K.S.; Stull, J.T.; Shelton, J.M.; Williams, R.S.; Garry, D.J. Functional and molecular adaptations in skeletal muscle of myoglobin-mutant mice. Am. J. Physiol.-Cell Physiol. 2001, 281, C1487–C1494. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Matsushita, M.; Yoneshiro, T.; Okamatsu-Ogura, Y. Brown Adipose Tissue, Diet-Induced Thermogenesis, and Thermogenic Food Ingredients: From Mice to Men. Front. Endocrinol. 2020, 11, 222. [Google Scholar] [CrossRef]
- Din, M.U.; Saari, T.; Raiko, J.; Kudomi, N.; Maurer, S.F.; Lahesmaa, M.; Fromme, T.; Amri, E.Z.; Klingenspor, M.; Solin, O.; et al. Postprandial Oxidative Metabolism of Human Brown Fat Indicates Thermogenesis. Cell Metab. 2018, 28, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Forner, F.; Kumar, C.; Luber, C.A.; Fromme, T.; Klingenspor, M.; Mann, M. Proteome Differences between Brown and White Fat Mitochondria Reveal Specialized Metabolic Functions. Cell Metab. 2009, 10, 324–335. [Google Scholar] [CrossRef]
- Timmons, J.A.; Wennmalm, K.; Larsson, O.; Walden, T.B.; Lassmann, T.; Petrovic, N.; Hamilton, D.L.; Gimeno, R.E.; Wahlestedt, C.; Baar, K.; et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. Proc. Natl. Acad. Sci. USA 2007, 104, 4401–4406. [Google Scholar] [CrossRef] [PubMed]
- Aboouf, M.A.; Armbruster, J.; Thiersch, M.; Gassmann, M.; Goedecke, A.; Gnaiger, E.; Kristiansen, G.; Bicker, A.; Hankeln, T.; Zhu, H.; et al. Myoglobin, expressed in brown adipose tissue of mice, regulates the content and activity of mitochondria and lipid droplets. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 2021, 1866, 159026. [Google Scholar] [CrossRef]
- Armbruster, J.; Aboouf, M.A.; Gassmann, M.; Egert, A.; Schorle, H.; Hornung, V.; Schmidt, T.; Schmid-Burgk, J.L.; Kristiansen, G.; Bicker, A.; et al. Myoglobin regulates fatty acid trafficking and lipid metabolism in mammary epithelial cells. PLoS ONE 2022, 17, e0275725. [Google Scholar] [CrossRef]
- Chintapalli, S.V.; Jayanthi, S.; Mallipeddi, P.L.; Gundampati, R.; Kumar, T.K.S.; van Rossum, D.B.; Anishkin, A.; Adams, S.H. Novel Molecular Interactions of Acylcarnitines and Fatty Acids with Myoglobin. J. Biol. Chem. 2016, 291, 251133–251143. [Google Scholar] [CrossRef] [PubMed]
- Hendgen-Cotta, U.B.; Esfeld, S.; Coman, C.; Ahrends, R.; Klein-Hitpass, L.; Flögel, U.; Rassaf, T.; Totzeck, M. A novel physiological role for cardiac myoglobin in lipid metabolism. Sci. Rep. 2017, 7, 43219. [Google Scholar] [CrossRef] [PubMed]
- Schlater, A.E.; De Miranda, M.A.; Frye, M.A.; Trumble, S.J.; Kanatous, S.B. Changing the paradigm for myoglobin: A novel link between lipids and myoglobin. J. Appl. Physiol. 2014, 117, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Schlater, A.E.; De Miranda, M.A.; Corley, A.M.; Kanatous, S.B. Lipid stimulates myoglobin expression in skeletal muscle cells. FASEB J. 2012, 26, 1078.16. [Google Scholar] [CrossRef]
- Blackburn, M.L.; Wankhade, U.D.; Ono-Moore, K.D.; Chintapalli, S.V.; Fox, R.; Rutkowsky, J.M.; Willis, B.J.; Tolentino, T.; Lloyd, K.C.K.; Adams, S.H. On the potential role of globins in brown adipose tissue: A novel conceptual model and studies in myoglobin knockout mice. Am. J. Physiol.-Endocrinol. Metab. 2021, 321, E47–E62. [Google Scholar] [CrossRef]
- Christen, L.; Broghammer, H.; Rapohn, I.; Mohlis, K.; Strehlau, C.; Ribas-Latre, A.; Gebhardt, C.; Roth, L.; Krause, K.; Landgraf, K.; et al. Myoglobin-mediated lipid shuttling increases adrenergic activation of brown and white adipocyte metabolism and is as a marker of thermogenic adipocytes in humans. Clin. Transl. Med. 2022, 12, e1108. [Google Scholar] [CrossRef]
- Gotz, F.M.; Hertel, M.; Groschelstewart, U. Fatty-Acid-Binding of Myoglobin Depends on Its Oxygenation. Biol. Chem. Hoppe-Seyler 1994, 375, 387–392. [Google Scholar] [CrossRef]
- Jue, T.; Simond, G.; Wright, T.J.; Shih, L.F.; Chung, Y.R.; Sriram, R.; Kreutzer, U.; Davis, R.W. Effect of fatty acid interaction on myoglobin oxygen affinity and triglyceride metabolism. J. Physiol. Biochem. 2016, 73, 359–370. [Google Scholar] [CrossRef]
- Gorr, T.A.; Wichmann, D.; Pilarsky, C.; Theurillat, J.P.; Fabrizius, A.; Laufs, T.; Bauer, T.; Koslowski, M.; Horn, S.; Burmester, T.; et al. Old proteins–new locations: Myoglobin, haemoglobin, neuroglobin and cytoglobin in solid tumours and cancer cells. Acta Physiol. 2011, 202, 563–581. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.L.; Cox, M.M.; Lehninger, A.L. Lehninger Principles of Biochemistry, 7th ed.; W.H. Freeman and Company: New York, NY, USA; Basingstoke, UK, 2017; I45p. [Google Scholar]
- Springer, B.A.; Egeberg, K.D.; Sligar, S.G.; Rohlfs, R.J.; Mathews, A.J.; Olson, J.S. Discrimination between oxygen and carbon monoxide and inhibition of autooxidation by myoglobin. Site-directed mutagenesis of the distal histidine. J. Biol. Chem. 1989, 264, 3057–3060. [Google Scholar] [CrossRef]
- Fraser, J.; de Mello, L.V.; Ward, D.; Rees, H.H.; Williams, D.R.; Fang, Y.; Brass, A.; Gracey, A.Y.; Cossins, A.R. Hypoxia-inducible myoglobin expression in nonmuscle tissues. Proc. Natl. Acad. Sci. USA 2006, 103, 2977–2981. [Google Scholar] [CrossRef] [PubMed]
- Burmester, T.; Weich, B.; Reinhardt, S.; Hankeln, T. A vertebrate globin expressed in the brain. Nature 2000, 407, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Cossins, A.R.; Williams, D.R.; Foulkes, N.S.; Berenbrink, M.; Kipar, A. Diverse cell-specific expression of myoglobin isoforms in brain, kidney, gill and liver of the hypoxia-tolerant carp and zebrafish. J. Exp. Biol. 2009, 212 Pt 5, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Bicker, A.; Dietrich, D.; Gleixner, E.; Kristiansen, G.; Gorr, T.A.; Hankeln, T. Extensive transcriptional complexity during hypoxia-regulated expression of the myoglobin gene in cancer. Hum. Mol. Genet. 2014, 23, 479–490. [Google Scholar] [CrossRef] [PubMed]
- Aboouf, M.A.; Armbruster, J.; Guscetti, F.; Thiersch, M.; Gassmann, M.; Gorr, T.A. The role of myoglobin in breast cancer. Cancer Res. 2021, 81, PS19-17. [Google Scholar] [CrossRef]
- Aboouf, M.A.; Armbruster, J.; Thiersch, M.; Guscetti, F.; Kristiansen, G.; Schraml, P.; Bicker, A.; Petry, R.; Hankeln, T.; Gassmann, M.; et al. Pro-Apoptotic and Anti-Invasive Properties Underscore the Tumor-Suppressing Impact of Myoglobin on a Subset of Human Breast Cancer Cells. Int. J. Mol. Sci. 2022, 23, 11483. [Google Scholar] [CrossRef]
- Behnes, C.L.; Bedke, J.; Schneider, S.; Kuffer, S.; Strauss, A.; Bremmer, F.; Strobel, P.; Radzun, H.J. Myoglobin expression in renal cell carcinoma is regulated by hypoxia. Exp. Mol. Pathol. 2013, 95, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Bicker, A.; Nauth, T.; Gerst, D.; Aboouf, M.A.; Fandrey, J.; Kristiansen, G.; Gorr, T.A.; Hankeln, T. The role of myoglobin in epithelial cancers: Insights from transcriptomics. Int. J. Mol. Med. 2020, 45, 385–400. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.J. Immunohistochemistry of soft tissue tumors. Myoglobin as a tumor marker for rhabdomyosarcoma. Cancer 1982, 50, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Emoto, M.; Iwasaki, H.; Kikuchi, M.; Shirakawa, K. Characteristics of cloned cells of mixed mullerian tumor of the human uterus. Carcinoma cells showing myogenic differentiation in vitro. Cancer 1993, 71, 3065–3075. [Google Scholar] [CrossRef]
- Eusebi, V.; Bondi, A.; Rosai, J. Immunohistochemical localization of myoglobin in nonmuscular cells. Am. J. Surg. Pathol. 1984, 8, 51–55. [Google Scholar] [CrossRef]
- Flonta, S.E.; Arena, S.; Pisacane, A.; Michieli, P.; Bardelli, A. Expression and functional regulation of myoglobin in epithelial cancers. Am. J. Pathol. 2009, 175, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, G.; Rose, M.; Geisler, C.; Fritzsche, F.R.; Gerhardt, J.; Luke, C.; Ladhoff, A.M.; Knuchel, R.; Dietel, M.; Moch, H.; et al. Endogenous myoglobin in human breast cancer is a hallmark of luminal cancer phenotype. Br. J. Cancer 2010, 102, 1736–1745. [Google Scholar] [CrossRef]
- Lamovec, J.; Zidar, A.; Bracko, M.; Golouh, R. Primary bone sarcoma with rhabdomyosarcomatous component. Pathol. Res. Pract. 1994, 190, 51–60. [Google Scholar] [CrossRef]
- Meller, S.; Bicker, A.; Montani, M.; Ikenberg, K.; Rostamzadeh, B.; Sailer, V.; Wild, P.; Dietrich, D.; Uhl, B.; Sulser, T.; et al. Myoglobin expression in prostate cancer is correlated to androgen receptor expression and markers of tumor hypoxia. Virchows Arch. 2014, 465, 419–427. [Google Scholar] [CrossRef]
- Meller, S.; Van Ellen, A.; Gevensleben, H.; Bicker, A.; Hankeln, T.; Gorr, T.A.; Sailer, V.; Droge, F.; Schrock, F.; Bootz, F.; et al. Ectopic Myoglobin Expression Is Associated with a Favourable Outcome in Head and Neck Squamous Cell Carcinoma Patients. Anticancer. Res. 2016, 36, 6235–6241. [Google Scholar] [CrossRef] [PubMed]
- Oleksiewicz, U.; Daskoulidou, N.; Liloglou, T.; Tasopoulou, K.; Bryan, J.; Gosney, J.R.; Field, J.K.; Xinarianos, G. Neuroglobin and myoglobin in non-small cell lung cancer: Expression, regulation and prognosis. Lung Cancer 2011, 74, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Kristiansen, G.; Hu, J.M.; Wichmann, D.; Stiehl, D.P.; Rose, M.; Gerhardt, J.; Bohnert, A.; ten Haaf, A.; Moch, H.; Raleigh, J.; et al. Endogenous Myoglobin in Breast Cancer Is Hypoxia-inducible by Alternative Transcription and Functions to Impair Mitochondrial Activity. A role in tumor suppression? J. Biol. Chem. 2011, 286, 43417–43428. [Google Scholar] [CrossRef]
- Aboouf, M.A.; Armbruster, J.; Guscetti, F.; Thiersch, M.; Boss, A.; Gödecke, A.; Winning, S.; Padberg, C.; Fandrey, J.; Kristiansen, G.; et al. Endogenous myoglobin expression in mouse models of mammary carcinoma reduces hypoxia and metastasis in PyMT mice. Sci. Rep. 2023, 13, 7530. [Google Scholar] [CrossRef] [PubMed]
- Garry, D.J.; Ordway, G.A.; Lorenz, L.N.; Radford, N.B.; Chin, E.R.; Grange, R.W.; Bassel-Duby, R.; Williams, R.S. Mice without myoglobin. Nature 1998, 395, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Merx, M.W.; Gödecke, A.; Flögel, U.; Schrader, J. Oxygen supply and nitric oxide scavenging by myoglobin contribute to exercise endurance and cardiac function. FASEB J. 2005, 19, 1015–1017. [Google Scholar] [CrossRef] [PubMed]
- Flogel, U.; Godecke, A.; Klotz, L.O.; Schrader, J. Role of myoglobin in the antioxidant defense of the heart. FASEB J. 2004, 18, 1156–1158. [Google Scholar] [CrossRef]
- Flogel, U.; Merx, M.W.; Godecke, A.; Decking, U.K.; Schrader, J. Myoglobin: A scavenger of bioactive NO. Proc. Natl. Acad. Sci. USA 2001, 98, 735–740. [Google Scholar] [CrossRef]
- Hendgen-Cotta, U.B.; Merx, M.W.; Shiva, S.; Schmitz, J.; Becher, S.; Klare, J.P.; Steinhoff, H.J.; Goedecke, A.; Schrader, J.; Gladwin, M.T.; et al. Nitrite reductase activity of myoglobin regulates respiration and cellular viability in myocardial ischemia-reperfusion injury. Proc. Natl. Acad. Sci. USA 2008, 105, 10256–10261. [Google Scholar] [CrossRef]
- Cosby, K.; Partovi, K.S.; Crawford, J.H.; Patel, R.P.; Reiter, C.D.; Martyr, S.; Yang, B.K.; Waclawiw, M.A.; Zalos, G.; Xu, X.; et al. Nitrite reduction to nitric oxide by deoxyhemoglobin vasodilates the human circulation. Nat. Med. 2003, 9, 1498–1505. [Google Scholar] [CrossRef]
- Gladwin, M.T.; Kim-Shapiro, D.B. The functional nitrite reductase activity of the heme-globins. Blood 2008, 112, 2636–2647. [Google Scholar] [CrossRef]
- Huang, Z.; Shiva, S.; Kim-Shapiro, D.B.; Patel, R.P.; Ringwood, L.A.; Irby, C.E.; Huang, K.T.; Ho, C.; Hogg, N.; Schechter, A.N.; et al. Enzymatic function of hemoglobin as a nitrite reductase that produces NO under allosteric control. J. Clin. Investig. 2005, 115, 2099–2107. [Google Scholar] [CrossRef]
- Brunori, M. Nitric oxide, cytochrome-c oxidase and myoglobin. Trends Biochem. Sci. 2001, 26, 21–23. [Google Scholar] [CrossRef]
- Brunori, M. Myoglobin strikes back. Protein Sci. 2010, 19, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Shiva, S.; Brookes, P.S.; Patel, R.P.; Anderson, P.G.; Darley-Usmar, V.M. Nitric oxide partitioning into mitochondrial membranes and the control of respiration at cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2001, 98, 7212–7217. [Google Scholar] [CrossRef] [PubMed]
- Beaudry, J.L.; Kaur, K.D.; Varin, E.M.; Baggio, L.L.; Cao, X.; Mulvihill, E.E.; Stem, J.H.; Campbell, J.E.; Scherer, P.E.; Drucker, D.J. The brown adipose tissue glucagon receptor is functional but not essential for control of energy homeostasis in mice. Mol. Metab. 2019, 22, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Beaudry, J.L.; Kaur, K.D.; Varin, E.M.; Baggio, L.L.; Cao, X.M.; Mulvihill, E.E.; Bates, H.E.; Campbell, J.E.; Drucker, D.J. Physiological roles of the GIP receptor in murine brown adipose tissue. Mol. Metab. 2019, 28, 14–25. [Google Scholar] [CrossRef]
- Kobilka, B.K. G protein coupled receptor structure and activation. Biochim. Biophys. Acta-Biomembr. 2007, 1768, 794–807. [Google Scholar] [CrossRef]
- Wettschureck, N.; Offermanns, S. Mammalian G proteins and their cell type specific functions. Physiol. Rev. 2005, 85, 1159–1204. [Google Scholar] [CrossRef]
- Chondronikola, M.; Volpi, E.; Borsheim, E.; Porter, C.; Saraf, M.K.; Annamalai, P.; Yfanti, C.; Chao, T.; Wong, D.; Shinoda, K.; et al. Brown Adipose Tissue Activation Is Linked to Distinct Systemic Effects on Lipid Metabolism in Humans. Cell Metab. 2016, 23, 1200–1206. [Google Scholar] [CrossRef]
- Bartelt, A.; Bruns, O.T.; Reimer, R.; Hohenberg, H.; Ittrich, H.; Peldschus, K.; Kaul, M.G.; Tromsdorf, U.I.; Weller, H.; Waurisch, C.; et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 2011, 17, 200–205. [Google Scholar] [CrossRef]
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef]
- Mills, E.L.; Pierce, K.A.; Jedrychowski, M.P.; Garrity, R.; Winther, S.; Vidoni, S.; Yoneshiro, T.; Spinelli, J.B.; Lu, G.Z.; Kazak, L.; et al. Accumulation of succinate controls activation of adipose tissue thermogenesis. Nature 2018, 560, 102–106. [Google Scholar] [CrossRef]
- Golozoubova, V.; Hohtola, E.; Matthias, A.; Jacobsson, A.; Cannon, B.; Nedergaard, J. Only UCP1 can mediate adaptive nonshivering thermogenesis in the cold. FASEB J. 2001, 15, 2048–2050. [Google Scholar] [CrossRef]
- Feldmann, H.M.; Golozoubova, V.; Cannon, B.; Nedergaard, J. UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab. 2009, 9, 203–209. [Google Scholar] [CrossRef]
- Divakaruni, A.S.; Humphrey, D.M.; Brand, M.D. Fatty acids change the conformation of uncoupling protein 1 (UCP1). J. Biol. Chem. 2012, 287, 36845–36853. [Google Scholar] [CrossRef]
- Fedorenko, A.; Lishko, P.V.; Kirichok, Y. Mechanism of fatty-acid-dependent UCP1 uncoupling in brown fat mitochondria. Cell 2012, 151, 400–413. [Google Scholar] [CrossRef]
- Bertholet, A.M.; Kirichok, Y. The Mechanism FA-Dependent H(+) Transport by UCP1. Handb. Exp. Pharmacol. 2019, 251, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Davis, T.R.; Johnston, D.R.; Bell, F.C.; Cremer, B.J. Regulation of shivering and non-shivering heat production during acclimation of rats. Am. J. Physiol. 1960, 198, 471–475. [Google Scholar] [CrossRef] [PubMed]
- Depocas, F.; Hart, J.S.; Heroux, O. Cold acclimation and the electromyogram of unanesthetized rats. J. Appl. Physiol. 1956, 9, 404–408. [Google Scholar] [CrossRef] [PubMed]
- Frontini, A.; Cinti, S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metab. 2010, 11, 253–256. [Google Scholar] [CrossRef]
- Cypess, A.M.; Lehman, S.; Williams, G.; Tal, I.; Rodman, D.; Goldfine, A.B.; Kuo, F.C.; Palmer, E.L.; Tseng, Y.H.; Doria, A.; et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009, 360, 1509–1517. [Google Scholar] [CrossRef]
- Virtanen, K.A.; Lidell, M.E.; Orava, J.; Heglind, M.; Westergren, R.; Niemi, T.; Taittonen, M.; Laine, J.; Savisto, N.J.; Enerback, S.; et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 2009, 360, 1518–1525. [Google Scholar] [CrossRef]
- Zingaretti, M.C.; Crosta, F.; Vitali, A.; Guerrieri, M.; Frontini, A.; Cannon, B.; Nedergaard, J.; Cinti, S. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 2009, 23, 3113–3120. [Google Scholar] [CrossRef]
- Cinti, S. Between brown and white: Novel aspects of adipocyte differentiation. Ann. Med. 2011, 43, 104–115. [Google Scholar] [CrossRef]
- Kozak, L.P.; Anunciado-Koza, R. UCP1: Its involvement and utility in obesity. Int. J. Obes. 2008, 32 (Suppl. S7), S32–S38. [Google Scholar] [CrossRef] [PubMed]
- Nicholls, D.G.; Rial, E. A history of the first uncoupling protein, UCP1. J. Bioenerg. Biomembr. 1999, 31, 399–406. [Google Scholar] [CrossRef]
- Calderon-Dominguez, M.; Mir, J.F.; Fucho, R.; Weber, M.; Serra, D.; Herrero, L. Fatty acid metabolism and the basis of brown adipose tissue function. Adipocyte 2016, 5, 98–118. [Google Scholar] [CrossRef] [PubMed]
- Chintapalli, S.V.; Bhardwaj, G.; Patel, R.; Shah, N.; Patterson, R.L.; van Rossum, D.B.; Anishkin, A.; Adams, S.H. Molecular dynamic simulations reveal the structural determinants of Fatty Acid binding to oxy-myoglobin. PLoS ONE 2015, 10, e0128496. [Google Scholar] [CrossRef] [PubMed]
- Shih, L.; Chung, Y.; Sriram, R.; Jue, T. Palmitate interaction with physiological states of myoglobin. Biochim. Biophys. Acta 2014, 1840, 656–666. [Google Scholar] [CrossRef]
- Sriram, R.; Kreutzer, U.; Shih, L.; Jue, T. Interaction of fatty acid with myoglobin. FEBS Lett. 2008, 582, 3643–3649. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Bernlohr, D.A. Metabolic functions of FABPs-mechanisms and therapeutic implications. Nat. Rev. Endocrinol. 2015, 11, 592–605. [Google Scholar] [CrossRef]
- Koundouros, N.; Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 2020, 122, 4–22. [Google Scholar] [CrossRef]
- Available online: http://biogps.org/ (accessed on 20 July 2023).
- Fournier, B.; Murray, B.; Gutzwiller, S.; Marcaletti, S.; Marcellin, D.; Bergling, S.; Brachat, S.; Persohn, E.; Pierrel, E.; Bombard, F.; et al. Blockade of the activin receptor IIb activates functional brown adipogenesis and thermogenesis by inducing mitochondrial oxidative metabolism. Mol. Cell Biol. 2012, 32, 2871–2879. [Google Scholar] [CrossRef]
- Watanabe, M.; Yamamoto, T.; Kakuhata, R.; Okada, N.; Kajimoto, K.; Yamazaki, N.; Kataoka, M.; Baba, Y.; Tamaki, T.; Shinohara, Y. Synchronized changes in transcript levels of genes activating cold exposure-induced thermogenesis in brown adipose tissue of experimental animals. Biochim. Biophys. Acta 2008, 1777, 104–112. [Google Scholar] [CrossRef]
- Klein, J.; Fasshauer, M.; Klein, H.H.; Benito, M.; Kahn, C.R. Novel adipocyte lines from brown fat: A model system for the study of differentiation, energy metabolism, and insulin action. Bioessays 2002, 24, 382–388. [Google Scholar] [CrossRef]
- Sveidahl Johansen, O.; Ma, T.; Hansen, J.B.; Markussen, L.K.; Schreiber, R.; Reverte-Salisa, L.; Dong, H.; Christensen, D.P.; Sun, W.; Gnad, T.; et al. Lipolysis drives expression of the constitutively active receptor GPR3 to induce adipose thermogenesis. Cell 2021, 184, 3502–3518.e33. [Google Scholar] [CrossRef]
- Gnad, T.; Scheibler, S.; von Kugelgen, I.; Scheele, C.; Kilic, A.; Glode, A.; Hoffmann, L.S.; Reverte-Salisa, L.; Horn, P.; Mutlu, S.; et al. Adenosine activates brown adipose tissue and recruits beige adipocytes via A2A receptors. Nature 2014, 516, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Wu, H.; Tarr, P.T.; Zhang, C.Y.; Wu, Z.; Boss, O.; Michael, L.F.; Puigserver, P.; Isotani, E.; Olson, E.N.; et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature 2002, 418, 797–801. [Google Scholar] [CrossRef]
- Bartelt, A.; Widenmaier, S.B.; Schlein, C.; Johann, K.; Goncalves, R.L.S.; Eguchi, K.; Fischer, A.W.; Parlakgul, G.; Snyder, N.A.; Nguyen, T.B.; et al. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nat. Med. 2018, 24, 292–303. [Google Scholar] [CrossRef] [PubMed]
- Wittenberg, B.A. Both hypoxia and work are required to enhance expression of myoglobin in skeletal muscle. Focus on “Hypoxia reprograms calcium signaling and regulates myoglobin expression”. Am. J. Physiol.-Cell Physiol. 2009, 296, C390–C392. [Google Scholar] [CrossRef] [PubMed]
- Koma, R.; Shibaguchi, T.; Perez Lopez, C.; Oka, T.; Jue, T.; Takakura, H.; Masuda, K. Localization of myoglobin in mitochondria: Implication in regulation of mitochondrial respiration in rat skeletal muscle. Physiol. Rep. 2021, 9, e14769. [Google Scholar] [CrossRef]
- Yamada, T.; Takakura, H.; Jue, T.; Hashimoto, T.; Ishizawa, R.; Furuichi, Y.; Kato, Y.; Iwanaka, N.; Masuda, K. Myoglobin and the regulation of mitochondrial respiratory chain complex IV. J. Physiol. 2016, 594, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.D.; Puigserver, P.; Andersson, U.; Zhang, C.Y.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Wicksteed, B.; Dickson, L.M. PKA Differentially Regulates Adipose Depots to Control Energy Expenditure. Endocrinology 2017, 158, 464–466. [Google Scholar] [CrossRef] [PubMed]
- Balkow, A.; Jagow, J.; Haas, B.; Siegel, F.; Kilic, A.; Pfeifer, A. A novel crosstalk between Alk7 and cGMP signaling differentially regulates brown adipocyte function. Mol. Metab. 2015, 4, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Mitschke, M.M.; Hoffmann, L.S.; Gnad, T.; Scholz, D.; Kruithoff, K.; Mayer, P.; Haas, B.; Sassmann, A.; Pfeifer, A.; Kilic, A. Increased cGMP promotes healthy expansion and browning of white adipose tissue. FASEB J. 2013, 27, 1621–1630. [Google Scholar] [CrossRef]
- Nisoli, E.; Clementi, E.; Tonello, C.; Sciorati, C.; Briscini, L.; Carruba, M.O. Effects of nitric oxide on proliferation and differentiation of rat brown adipocytes in primary cultures. Br. J. Pharmacol. 1998, 125, 888–894. [Google Scholar] [CrossRef]
- Ceddia, R.P.; Collins, S. A compendium of G-protein-coupled receptors and cyclic nucleotide regulation of adipose tissue metabolism and energy expenditure. Clin. Sci. 2020, 134, 473–512. [Google Scholar] [CrossRef]
- Movafagh, S.; Crook, S.; Vo, K. Regulation of Hypoxia-Inducible Factor-1a by Reactive Oxygen Species: New Developments in an Old Debate. J. Cell Biochem. 2015, 116, 696–703. [Google Scholar] [CrossRef]
- Carraway, M.S.; Suliman, H.B.; Jones, W.S.; Chen, C.W.; Babiker, A.; Piantadosi, C.A. Erythropoietin Activates Mitochondrial Biogenesis and Couples Red Cell Mass to Mitochondrial Mass in the Heart. Circ. Res. 2010, 106, 1722-U1111. [Google Scholar] [CrossRef]
- Aboouf, M.A.; Guscetti, F.; von Buren, N.; Armbruster, J.; Ademi, H.; Ruetten, M.; Melendez-Rodriguez, F.; Rulicke, T.; Seymer, A.; Jacobs, R.A.; et al. Erythropoietin receptor regulates tumor mitochondrial biogenesis through iNOS and pAKT. Front. Oncol. 2022, 12, 976961. [Google Scholar] [CrossRef]
- Nisoli, E.; Falcone, S.; Tonello, C.; Cozzi, V.; Palomba, L.; Fiorani, M.; Pisconti, A.; Brunelli, S.; Cardile, A.; Francolini, M.; et al. Mitochondrial biogenesis by NO yields functionally active mitochondria in mammals. Proc. Natl. Acad. Sci. USA 2004, 101, 16507–16512. [Google Scholar] [CrossRef]
- Szelenyi, Z. Effect of cold exposure on oxygen tension in brown adipose tissue in the non-cold adapted adult rat. Acta Physiol. Acad. Sci. Hung. 1968, 33, 311–316. [Google Scholar] [PubMed]
- Xue, Y.; Petrovic, N.; Cao, R.; Larsson, O.; Lim, S.; Chen, S.; Feldmann, H.M.; Liang, Z.; Zhu, Z.; Nedergaard, J.; et al. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metab. 2009, 9, 99–109. [Google Scholar] [CrossRef]
- Saha, S.K.; Kuroshima, A. Nitric oxide and thermogenic function of brown adipose tissue in rats. Jpn. J. Physiol. 2000, 50, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi-Utsumi, K.; Gao, B.H.; Ohinata, H.; Hashimoto, M.; Yamamoto, N.; Kuroshima, A. Enhanced gene expression of endothelial nitric oxide synthase in brown adipose tissue during cold exposure. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2002, 282, R623–R626. [Google Scholar] [CrossRef]
- Stone, J.R.; Marletta, M.A. Spectral and kinetic studies on the activation of soluble guanylate cyclase by nitric oxide. Biochemistry 1996, 35, 1093–1099. [Google Scholar] [CrossRef] [PubMed]
- Sebag, S.C.; Zhang, Z.Y.; Qian, Q.W.; Li, M.; Zhu, Z.Y.; Harata, M.; Li, W.X.; Zingman, L.V.; Liu, L.M.; Lira, V.A.; et al. ADH5-mediated NO bioactivity maintains metabolic homeostasis in brown adipose tissue. Cell Rep. 2021, 37, 110003. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Orozco, C.; Boyer, J.; Leglise, M.; Goodale, J.; Batalov, S.; Hodge, C.L.; Haase, J.; Janes, J.; Huss, J.W.; et al. BioGPS: An extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009, 10, R130. [Google Scholar] [CrossRef]
- Nagashima, T.; Ohinata, H.; Kuroshima, A. Involvement of Nitric-Oxide in Noradrenaline-Induced Increase in Blood-Flow through Brown Adipose-Tissue. Life Sci. 1994, 54, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Ono-Moore, K.D.; Olfert, I.M.; Rutkowsky, J.M.; Chintapalli, S.V.; Willis, B.J.; Blackburn, M.L.; Williams, D.K.; O’Reilly, J.; Tolentino, T.; Lloyd, K.C.K.; et al. Metabolic physiology and skeletal muscle phenotypes in male and female myoglobin knockout mice. Am. J. Physiol. Endocrinol. Metab. 2021, 321, E63–E79. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Fatunmbi, O.; Abzalimov, R.R.; Savinov, S.N.; Gershenson, A.; Kaltashov, I.A. Interactions of Haptoglobin with Monomeric Globin Species: Insights from Molecular Modeling and Native Electrospray Ionization Mass Spectrometry. Biochemistry 2016, 55, 1918–1928. [Google Scholar] [CrossRef]
- Kazak, L.; Chouchani, E.T.; Jedrychowski, M.P.; Erickson, B.K.; Shinoda, K.; Cohen, P.; Vetrivelan, R.; Lu, G.Z.; Laznik-Bogoslavski, D.; Hasenfuss, S.C.; et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 2015, 163, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Rahbani, J.F.; Roesler, A.; Hussain, M.F.; Samborska, B.; Dykstra, C.B.; Tsai, L.; Jedrychowski, M.P.; Vergnes, L.; Reue, K.; Spiegelman, B.M.; et al. Creatine kinase B controls futile creatine cycling in thermogenic fat. Nature 2021, 590, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Oughtred, R.; Rust, J.; Chang, C.; Breitkreutz, B.J.; Stark, C.; Willems, A.; Boucher, L.; Leung, G.; Kolas, N.; Zhang, F.; et al. The BioGRID database: A comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021, 30, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Norreen-Thorsen, M.; Struck, E.C.; Oling, S.; Zwahlen, M.; Von Feilitzen, K.; Odeberg, J.; Lindskog, C.; Ponten, F.; Uhlen, M.; Dusart, P.J.; et al. A human adipose tissue cell-type transcriptome atlas. Cell Rep. 2022, 40, 111046. [Google Scholar] [CrossRef]
- Perdikari, A.; Leparc, G.G.; Balaz, M.; Pires, N.D.; Lidell, M.E.; Sun, W.F.; Fernandez-Albert, F.; Muller, S.; Akchiche, N.; Dong, H.; et al. BATLAS: Deconvoluting Brown Adipose Tissue. Cell Rep. 2018, 25, 784–797.e4. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nagane, M.; Suzuki, M.; Yamauchi, A.; Kato, K.; Kawashima, N.; Nemoto, Y.; Maruo, T.; Kawakami, Y.; Yamashita, T. Tumor hypoxia regulates ganglioside GM3 synthase, which contributes to oxidative stress resistance in malignant melanoma. Biochim. Biophys. Acta-Gen. Subj. 2020, 1864, 129723. [Google Scholar] [CrossRef]
- Mikami, H.; Saito, Y.; Okamoto, N.; Kakihana, A.; Kuga, T.; Nakayama, Y. Requirement of Hsp105 in CoCl(2)-induced HIF-1alpha accumulation and transcriptional activation. Exp. Cell Res. 2017, 352, 225–233. [Google Scholar] [CrossRef]
- Xiong, G.F.; Stewart, R.L.; Chen, J.; Gao, T.Y.; Scott, T.L.; Samayoa, L.M.; O’Connor, K.; Lane, A.N.; Xu, R. Collagen prolyl 4-hydroxylase 1 is essential for HIF-1 alpha stabilization and TNBC chemoresistance. Nat. Commun. 2018, 9, 4456. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Van Norstrand, D.W.; Medeiros-Domingo, A.; Valdivia, C.; Tan, B.H.; Ye, B.; Kroboth, S.; Vatta, M.; Tester, D.J.; January, C.T.; et al. Alpha1-syntrophin mutations identified in sudden infant death syndrome cause an increase in late cardiac sodium current. Circ. Arrhythm. Electrophysiol. 2009, 2, 667–676. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Valdivia, C.; Medeiros-Domingo, A.; Tester, D.J.; Vatta, M.; Farrugia, G.; Ackerman, M.J.; Makielski, J.C. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc. Natl. Acad. Sci. USA 2008, 105, 9355–9360. [Google Scholar] [CrossRef]
- Whitlock, N.A.; Agarwal, N.; Ma, J.X.; Crosson, C.E. Hsp27 upregulation by HIF-1 signaling offers protection against retinal ischemia in rats. Invest. Ophth Vis. Sci. 2005, 46, 1092–1098. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Lv, Z.; Weng, X.Y.; Ye, S.Y.; Shen, K.Z.; Li, M.S.; Qin, Y.S.; Hu, C.; Zhang, C.X.; Wu, J.; et al. Hsp27 Acts as a Master Molecular Chaperone and Plays an Essential Role in Hepatocellular Carcinoma Progression. Digestion 2015, 92, 192–202. [Google Scholar] [CrossRef]
- Taipale, M.; Tucker, G.; Peng, J.; Krykbaeva, I.; Lin, Z.Y.; Larsen, B.; Choi, H.; Berger, B.; Gingras, A.C.; Lindquist, S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 2014, 158, 434–448. [Google Scholar] [CrossRef]
- Lu, K.T.; Huang, T.C.; Wang, J.Y.; You, Y.S.; Chou, J.L.; Chan, M.W.; Wo, P.Y.; Amstislavskaya, T.G.; Tikhonova, M.A.; Yang, Y.L. NKCC1 mediates traumatic brain injury-induced hippocampal neurogenesis through CREB phosphorylation and HIF-1alpha expression. Pflugers Arch. 2015, 467, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, S.F.; O’Donnell, M.E.; Anderson, S.E.; Cala, P.M. Physiology and pathophysiology of Na+/H+ exchange and Na+ -K+ -2Cl- cotransport in the heart, brain, and blood. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2006, 291, R1–R25. [Google Scholar] [CrossRef]
- Cho, Y.; Hazen, B.C.; Gandra, P.G.; Ward, S.R.; Schenk, S.; Russell, A.P.; Kralli, A. Perm1 enhances mitochondrial biogenesis, oxidative capacity, and fatigue resistance in adult skeletal muscle. FASEB J. 2016, 30, 674–687. [Google Scholar] [CrossRef]
- Cho, Y.; Hazen, B.C.; Russell, A.P.; Kralli, A. Peroxisome Proliferator-activated Receptor gamma Coactivator 1 (PGC-1)- and Estrogen-related Receptor (ERR)-induced Regulator in Muscle 1 (PERM1) Is a Tissue-specific Regulator of Oxidative Capacity in Skeletal Muscle Cells. J. Biol. Chem. 2013, 288, 25207–25218. [Google Scholar] [CrossRef]
- Oka, S.; Sabry, A.D.; Horiuchi, A.K.; Cawley, K.M.; O’Very, S.A.; Zaitsev, M.A.; Shankar, T.S.; Byun, J.; Mukai, R.; Xu, X.Y.; et al. Perm1 regulates cardiac energetics as a downstream target of the histone methyltransferase Smyd1. PLoS ONE 2020, 15, e0234913. [Google Scholar] [CrossRef]
- Haitina, T.; Lindblom, J.; Renstrom, T.; Fredriksson, R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics 2006, 88, 779–790. [Google Scholar] [CrossRef]
- Li, Y.; Wang, D.; Ping, X.; Zhang, Y.; Zhang, T.; Wang, L.; Jin, L.; Zhao, W.; Guo, M.; Shen, F.; et al. Local hyperthermia therapy induces browning of white fat and treats obesity. Cell 2022, 185, 949–966.e19. [Google Scholar] [CrossRef]
- Wang, T.; Feugang, J.M.; Crenshaw, M.A.; Regmi, N.; Blanton, J.R.; Liao, S.F. A Systems Biology Approach Using Transcriptomic Data Reveals Genes and Pathways in Porcine Skeletal Muscle Affected by Dietary Lysine. Int. J. Mol. Sci. 2017, 18, 885. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.C.; Zhang, M.R.; Tian, A.W.; Chen, L.L.; Sun, Z.W.; Wang, L.Y.; Chen, P. Exploring the genetic correlation between obesity-related traits and regional brain volumes: Evidence from UK Biobank cohort. Neuroimage-Clin. 2022, 33, 102870. [Google Scholar] [CrossRef] [PubMed]
- Klemm, R.W.; Norton, J.P.; Cole, R.A.; Li, C.S.; Park, S.H.; Crane, M.M.; Li, L.Y.; Jin, D.; Boye-Doe, A.; Liu, T.Y.; et al. A Conserved Role for Atlastin GTPases in Regulating Lipid Droplet Size. Cell Rep. 2013, 3, 1465–1475. [Google Scholar] [CrossRef]
- Le Guillou, S.; Laubier, J.; Pechoux, C.; Aujean, E.; Castille, J.; Leroux, C.; Le Provost, F. Defects of the endoplasmic reticulum and changes to lipid droplet size in mammary epithelial cells due to miR-30b-5p overexpression are correlated to a reduction in Atlastin 2 expression. Biochem. Biophys. Res. Commun. 2019, 512, 283–288. [Google Scholar] [CrossRef]
- Patwari, P.; Emilsson, V.; Schadt, E.E.; Chutkow, W.A.; Lee, S.; Marsili, A.; Zhang, Y.Z.; Dobrin, R.; Cohen, D.E.; Larsen, P.R.; et al. The Arrestin Domain-Containing 3 Protein Regulates Body Mass and Energy Expenditure. Cell Metab. 2011, 14, 671–683. [Google Scholar] [CrossRef]
- Raulerson, C.K.; Ko, A.; Kidd, J.C.; Currin, K.W.; Brotman, S.M.; Cannon, M.E.; Wu, Y.; Spracklen, C.N.; Jackson, A.U.; Stringham, H.M.; et al. Adipose Tissue Gene Expression Associations Reveal Hundreds of Candidate Genes for Cardiometabolic Traits. Am. J. Hum. Genet. 2019, 105, 773–787. [Google Scholar] [CrossRef]
- Morita, S.; Nakabayashi, K.; Kawai, T.; Hayashi, K.; Horii, T.; Kimura, M.; Kamei, Y.; Ogawa, Y.; Hata, K.; Hatada, I. Gene expression profiling of white adipose tissue reveals paternal transmission of proneness to obesity. Sci. Rep. 2016, 6, 21693. [Google Scholar] [CrossRef]
- Zhang, Z.D.; Li, H.X.; Gan, H.; Tang, Z.; Guo, Y.Y.; Yao, S.Q.; Liuyu, T.; Zhong, B.; Lin, D. RNF115 Inhibits the Post-ER Trafficking of TLRs and TLRs-Mediated Immune Responses by Catalyzing K11-Linked Ubiquitination of RAB1A and RAB13. Adv. Sci. 2022, 9, e2105391. [Google Scholar] [CrossRef]
- Wu, X.T.; Wang, Y.H.; Cai, X.Y.; Dong, Y.; Cui, Q.; Zhou, Y.N.; Yang, X.W.; Lu, W.F.; Zhang, M. RNF115 promotes lung adenocarcinoma through Wnt/beta-catenin pathway activation by mediating APC ubiquitination. Cancer Metab. 2021, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Xiong, X.; Nam, D.; Yechoor, V.; Ma, K. SRF-MRTF signaling suppresses brown adipocyte development by modulating TGF-beta/BMP pathway. Mol. Cell Endocrinol. 2020, 515, 110920. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, A.A.; Zohn, I.E. Hectd1 regulates intracellular localization and secretion of Hsp90 to control cellular behavior of the cranial mesenchyme. J. Cell Biol. 2012, 196, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Pascual, M.; Vicente, M.; Monferrer, L.; Artero, R. The Muscleblind family of proteins: An emerging class of regulators of developmentally programmed alternative splicing. Differentiation 2006, 74, 65–80. [Google Scholar] [CrossRef]
- Collura, K.M.; Niu, J.; Sanders, S.S.; Montersino, A.; Holland, S.M.; Thomas, G.M. The palmitoyl acyltransferases ZDHHC5 and ZDHHC8 are uniquely present in DRG axons and control retrograde signaling via the Gp130/JAK/STAT3 pathway. J. Biol. Chem. 2020, 295, 15427–15437. [Google Scholar] [CrossRef] [PubMed]
- Golomb, L.; Bublik, D.R.; Wilder, S.; Nevo, R.; Kiss, V.; Grabusic, K.; Volarevic, S.; Oren, M. Importin 7 and exportin 1 link c-Myc and p53 to regulation of ribosomal biogenesis. Mol. Cell 2012, 45, 222–232. [Google Scholar] [CrossRef]
- Tachibana, M.; Kiyokawa, E.; Hara, S.; Iemura, S.; Natsume, T.; Manabe, T.; Matsuda, M. Ankyrin repeat domain 28 (ANKRD28), a novel binding partner of DOCK180, promotes cell migration by regulating focal adhesion formation. Exp. Cell Res. 2009, 315, 863–876. [Google Scholar] [CrossRef]
- Zhang, T.; Gao, H.; Sahana, G.; Zan, Y.; Fan, H.; Liu, J.; Shi, L.; Wang, H.; Du, L.; Wang, L.; et al. Genome-wide association studies revealed candidate genes for tail fat deposition and body size in the Hulun Buir sheep. J. Anim. Breed. Genet. 2019, 136, 362–370. [Google Scholar] [CrossRef]
- Matsunaga, H.; Ito, K.; Akiyama, M.; Takahashi, A.; Koyama, S.; Nomura, S.; Ieki, H.; Ozaki, K.; Onouchi, Y.; Sakaue, S.; et al. Transethnic Meta-Analysis of Genome-Wide Association Studies Identifies Three New Loci and Characterizes Population-Specific Differences for Coronary Artery Disease. Circ. Genom. Precis. Med. 2020, 13, e002670. [Google Scholar] [CrossRef]
- Hudson, B.H.; Frederick, J.P.; Drake, L.Y.; Megosh, L.C.; Irving, R.P.; York, J.D. Role for cytoplasmic nucleotide hydrolysis in hepatic function and protein synthesis. Proc. Natl. Acad. Sci. USA 2013, 110, 5040–5045. [Google Scholar] [CrossRef] [PubMed]
- Ladraa, S.; Zerbib, L.; Bayard, C.; Fraissenon, A.; Venot, Q.; Morin, G.; Garneau, A.P.; Isnard, P.; Chapelle, C.; Hoguin, C.; et al. PIK3CA gain-of-function mutation in adipose tissue induces metabolic reprogramming with Warburg-like effect and severe endocrine disruption. Sci. Adv. 2022, 8, eade7823. [Google Scholar] [CrossRef] [PubMed]
- Bennett, C.N.; Ross, S.E.; Longo, K.A.; Bajnok, L.; Hemati, N.; Johnson, K.W.; Harrison, S.D.; MacDougald, O.A. Regulation of Wnt signaling during adipogenesis. J. Biol. Chem. 2002, 277, 30998–31004. [Google Scholar] [CrossRef] [PubMed]
- Coyne, E.S.; Bedard, N.; Gong, Y.J.; Faraj, M.; Tchernof, A.; Wing, S.S. The deubiquitinating enzyme USP19 modulates adipogenesis and potentiates high-fat-diet-induced obesity and glucose intolerance in mice. Diabetologia 2019, 62, 136–146. [Google Scholar] [CrossRef] [PubMed]
- Bugler-Lamb, A.R.; Hasib, A.; Weng, X.; Hennayake, C.K.; Lin, C.; McCrimmon, R.J.; Stimson, R.H.; Ashford, M.L.J.; Wasserman, D.H.; Kang, L. Adipocyte integrin-linked kinase plays a key role in the development of diet-induced adipose insulin resistance in male mice. Mol. Metab. 2021, 49, 101197. [Google Scholar] [CrossRef] [PubMed]
- Sztalryd, C.; Brasaemle, D.L. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 1221–1232. [Google Scholar] [CrossRef] [PubMed]
- Fino, K.K.; Yang, L.L.; Silveyra, P.; Hu, S.M.; Umstead, T.M.; DiAngelo, S.; Halstead, E.S.; Cooper, T.K.; Abraham, T.; Takahashi, Y.; et al. SH3GLB2/endophilin B2 regulates lung homeostasis and recovery from severe influenza A virus infection. Sci. Rep. 2017, 7, 7262. [Google Scholar] [CrossRef] [PubMed]
- Manke, M.C.; Geue, S.; Coman, C.; Peng, B.; Kollotzek, F.; Munzer, P.; Walker, B.; Huber, S.M.; Rath, D.; Sickmann, A.; et al. ANXA7 Regulates Platelet Lipid Metabolism and Ca2+ Release in Arterial Thrombosis. Circ. Res. 2021, 129, 494–507. [Google Scholar] [CrossRef] [PubMed]
- Li, H.Y.; Huang, S.Y.; Wang, S.Q.; Wang, L.; Qi, L.; Zhang, Y.; Zhang, S.L.; Zhao, B.X.; Miao, J.Y. Relationship between annexin A7 and integrin beta 4 in autophagy. Int. J. Biochem. Cell Biol. 2013, 45, 2605–2611. [Google Scholar] [CrossRef] [PubMed]
- Mears, D.; Zimliki, C.L.; Atwater, I.; Rojas, E.; Glassman, M.; Leighton, X.; Pollard, H.B.; Srivastava, M. The Anx7(+/-) Knockout Mutation Alters Electrical and Secretory Responses to Ca2+-Mobilizing Agents in Pancreatic beta-cells. Cell Physiol. Biochem. 2012, 29, 697–704. [Google Scholar] [CrossRef]
- Fu, T.; Guan, Y.; Xu, J.; Wang, Y. APP, APLP2 and LRP1 interact with PCSK9 but are not required for PCSK9-mediated degradation of the LDLR in vivo. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 883–889. [Google Scholar] [CrossRef] [PubMed]
- Mauro, M.S.; Celma, G.; Zimyanin, V.; Magaj, M.M.; Gibson, K.H.; Redemann, S.; Bahmanyar, S. Ndc1 drives nuclear pore complex assembly independent of membrane biogenesis to promote nuclear formation and growth. eLife 2022, 11, e75513. [Google Scholar] [CrossRef] [PubMed]
- Diane, A.; Nikolic, N.; Rudecki, A.P.; King, S.M.; Bowie, D.J.; Gray, S.L. PACAP is essential for the adaptive thermogenic response of brown adipose tissue to cold exposure. J. Endocrinol. 2014, 222, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Burwinkel, B.; Scott, J.W.; Buhrer, C.; van Landeghem, F.K.H.; Cox, G.F.; Wilson, C.J.; Hardie, D.G.; Kilimann, M.W. Fatal congenital heart glycogenosis caused by a recurrent activating R531Q mutation in the gamma 2-subunit of AMP-activated protein kinase (PRKAG2), not by phosphorylase kinase deficiency. Am. J. Human. Genet. 2005, 76, 1034–1049. [Google Scholar] [CrossRef]
- Wu, L.Y.; Zhang, L.N.; Li, B.H.; Jiang, H.W.; Duan, Y.N.; Xie, Z.F.; Shuai, L.; Li, J.; Li, J.Y. AMP-Activated Protein Kinase (AMPK) Regulates Energy Metabolism through Modulating Thermogenesis in Adipose Tissue. Front. Physiol. 2018, 9, 122. [Google Scholar] [CrossRef]
- Singh, S.; Brocker, C.; Koppaka, V.; Chen, Y.; Jackson, B.C.; Matsumoto, A.; Thompson, D.C.; Vasiliou, V. Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radic. Biol. Med. 2013, 56, 89–101. [Google Scholar] [CrossRef]
- Dharan, R.; Huang, Y.; Cheppali, S.K.; Goren, S.; Shendrik, P.; Wang, W.; Qiao, J.; Kozlov, M.M.; Yu, L.; Sorkin, R. Tetraspanin 4 stabilizes membrane swellings and facilitates their maturation into migrasomes. Nat. Commun. 2023, 14, 1037. [Google Scholar] [CrossRef]
- Strzyz, P. Migrasomes promote angiogenesis. Nat. Rev. Mol. Cell Biol. 2023, 24, 84. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, W.; Bi, M.; Liu, W.; Zhou, L.; Liu, H.; Yan, F.; Guan, L.; Zhang, J.; Xu, J. Migrasomes: From Biogenesis, Release, Uptake, Rupture to Homeostasis and Diseases. Oxid. Med. Cell Longev. 2022, 2022, 4525778. [Google Scholar] [CrossRef]
- Grewal, T.; Enrich, C.; Rentero, C.; Buechler, C. Annexins in Adipose Tissue: Novel Players in Obesity. Int. J. Mol. Sci. 2019, 20, 3449. [Google Scholar] [CrossRef]
- Huang, K.P.; Ronveaux, C.C.; Knotts, T.A.; Rutkowsky, J.R.; Ramsey, J.J.; Raybould, H.E. Sex differences in response to short-term high fat diet in mice. Physiol. Behav. 2020, 221, 112894. [Google Scholar] [CrossRef]
- Norheim, F.; Hasin-Brumshtein, Y.; Vergnes, L.; Chella Krishnan, K.; Pan, C.; Seldin, M.M.; Hui, S.T.; Mehrabian, M.; Zhou, Z.; Gupta, S.; et al. Gene-by-Sex Interactions in Mitochondrial Functions and Cardio-Metabolic Traits. Cell Metab. 2019, 29, 932–949 e934. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.M.; Quevedo-Coli, S.; Roca, P.; Palou, A. Sex-dependent dietary obesity, induction of UCPs, and leptin expression in rat adipose tissues. Obes. Res. 2001, 9, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Mauvais-Jarvis, F. Sex differences in metabolic homeostasis, diabetes, and obesity. Biol. Sex. Differ. 2015, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Cypess, A.M.; Kahn, C.R. Brown fat as a therapy for obesity and diabetes. Curr. Opin. Endocrinol. Diabetes Obes. 2010, 17, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Tsagkaraki, E.; Nicoloro, S.M.; DeSouza, T.; Solivan-Rivera, J.; Desai, A.; Lifshitz, L.M.; Shen, Y.F.; Kelly, M.; Guilherme, A.; Henriques, F.; et al. CRISPR-enhanced human adipocyte browning as cell therapy for metabolic disease. Nat. Commun. 2021, 12, 6931. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Natsume, Y.; Yamanaka, H.; Fukuda, M.; Yao, R. A protocol for efficient CRISPR-Cas9-mediated knock-in in colorectal cancer patient-derived organoids. STAR Protoc. 2021, 2, 100780. [Google Scholar] [CrossRef]
- Seleit, A.; Aulehla, A.; Paix, A. Endogenous protein tagging in medaka using a simplified CRISPR/Cas9 knock-in approach. eLife 2021, 10, e75050. [Google Scholar] [CrossRef]
- Chin, E.R.; Olson, E.N.; Richardson, J.A.; Yano, Q.; Humphries, C.; Shelton, J.M.; Wu, H.; Zhu, W.G.; Bassel-Duby, R.; Williams, R.S. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Gene Dev. 1998, 12, 2499–2509. [Google Scholar] [CrossRef]
- Chen, B.K.; Huang, C.C.; Chang, W.C.; Chen, Y.J.; Kikkawa, U.; Nakahama, K.; Morita, I.; Chang, W.C. PP2B-mediated dephosphorylation of c-Jun C terminus regulates phorbol ester-induced c-Jun/Sp1 interaction in A431 cells. Mol. Biol. Cell 2007, 18, 1118–1127. [Google Scholar] [CrossRef]
- Schaeffer, P.J.; Wende, A.R.; Magee, C.J.; Neilson, J.R.; Leone, T.C.; Chen, F.; Kelly, D.P. Calcineurin and calcium/calmodulin-dependent protein kinase activate distinct metabolic gene regulatory programs in cardiac muscle. J. Biol. Chem. 2004, 279, 39593–39603. [Google Scholar] [CrossRef]
- Olson, E.N.; Williams, R.S. Calcineurin signaling and muscle remodeling. Cell 2000, 101, 689–692. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Lu, J.R.; Antos, C.L.; Markham, B.; Richardson, J.; Robbins, J.; Grant, S.R.; Olson, E.N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 1998, 93, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Tai, P.W.L.; Gao, G.P. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://maayanlab.cloud/Harmonizome/gene_set/extracellular+vesicular+exosome/COMPARTMENTS+Curated+Protein+Localization+Evidence+Scores (accessed on 4 August 2023).
- Seale, P.; Bjork, B.; Yang, W.L.; Kajimura, S.; Chin, S.; Kuang, S.H.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef] [PubMed]
- Li, F.N.; Li, Y.H.; Duan, Y.H.; Hu, C.A.A.; Tang, Y.L.; Yin, Y.L. Myokines and adipokines: Involvement in the crosstalk between skeletal muscle and adipose tissue. Cytokine Growth Factor Rev. 2017, 33, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, A.; Becerril, S.; Ezquerro, S.; Mendez-Gimenez, L.; Fruhbeck, G. Crosstalk between adipokines and myokines in fat browning. Acta Physiol. 2017, 219, 362–381. [Google Scholar] [CrossRef] [PubMed]
- Trayhurn, P.; Drevon, C.A.; Eckel, J. Secreted proteins from adipose tissue and skeletal muscle-adipokines, myokines and adipose/muscle cross-talk. Arch. Physiol. Biochem. 2011, 117, 47–56. [Google Scholar] [CrossRef]
- Stanford, K.I.; Middelbeek, R.J.W.; Townsend, K.L.; An, D.; Nygaard, E.B.; Hitchcox, K.M.; Markan, K.R.; Nakano, K.; Hirshman, M.F.; Tseng, Y.H.; et al. Brown adipose tissue regulates glucose homeostasis and insulin sensitivity. J. Clin. Investig. 2013, 123, 215–223. [Google Scholar] [CrossRef]


| NCBI ID | Mapped Ids: Gene Symbol; Gene Name/Panther Family/Panther Protein Class | Panther GO-SLIM Molecular Functions | Panther GO-SLIM Biological Functions | Role Concerning the Metabolic Phenotype | Ref. | |
|---|---|---|---|---|---|---|
| 20454 | St3gal5; ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (Encoding Ganglioside GM synthase GM3S)//transferase | Catalytic activity | Metabolic and cellular process | Hypoxia-induced regulation of sialic acid-containing glycerophospholipids | HIF-1α target gene | [121] |
| 15505 | Hsph1:Hsp105; Heat shock 105 kDa protein 1//chaperone | Binding | - | Transcriptional activation of HIF-1α | - | [122] |
| 18451 | P4ha1; Proline 4-hydroxylase, alpha 1//protein modifying enzyme | Catalytic activity | Metabolic and cellular process | Increases HIF-1α protein stabilization | - | [123] |
| 20648 | Snta1; Syntrophin 1//scaffold/adaptor protein | Unclassified | Unclassified | Forms a complex between nNOS and the nNOS inhibitor plasma membrane Ca-ATPase 4 b (PMCA4b) | S-nitrosylation of at least one sodium channel and changes in sodium current | [124,125] |
| 15507 | Hspb1: Hsp25 or 27; Heat shock protein 1/Heat shock protein beta-1/chaperone | - | Response to stimulus, cellular process | Target gene for HIF1α | Interdependence expression in cancer cells | [126,127] |
| 67072 | CDC37L1; Cell division cycle 37-like 1//Chaperone | Binding | Biological regulation, cellular process | Interacts with HSP90 | Enhance the binding of client proteins to HSP90 | [128] |
| 20496 | Slc12a2: NKCC1; Ischemia- and cell stress-responsive protein solute carrier family 12, member 2//- | - | - | Activation of HIF-1α | Regulation of sodium influx | [129,130] |
| 74183 | Perm1; PGC-1 and ERR-induced regulator in muscle protein 1: Peroxisome proliferator-activated receptor coactivator 1- and estrogen-related receptor-induced regulator in muscle//- | Unclassified | Response to stimulus, metabolic process, biological regulation, cellular process | Supporting mitochondrial oxidative machinery in a PGC1-α-dependent manner (BAT) Triggering oxidative metabolism, mitochondrial biogenesis and muscle performance (muscles) | Upregulated in muscle with exercise | [131,132,133] |
| 384071 | Slc25a34; Solute carrier family 25, member 34//- | Unclassified | Unclassified | Transports molecules over the mitochondrial membrane | - | [134] |
| 66917 | CHORDC1; Cysteine and histidine-rich domain (CHORD) containing 1//scaffold/adaptor protein | Binding | Cellular process | Hsp-interacting protein, target to HSF1, differentially expressed in pigs’ skeletal muscle fed three different diets | Regulates beige adipocyte activation in mice and humans | [135,136] |
| 237860 | Ssh2; Slingshot protein phosphatase 2//Protein phosphatase | Binding, catalytic activity | Biological regulation, cellular process | Obesity occurs if mutated | Alterations in regional brain volumes | [137] |
| 56298 | Atl2; Atlastin GTPase 2//heterotrimeric G-protein | Binding, catalytic activity | Cellular process | Regulating lipid droplet size | Low expression is associated with enlarged lipid droplets and changed milk FA composition | [138,139] |
| 105171 | Arrdc3; Arrestin Domain-Containing 3 Protein//ubiquitin-protein ligase | Unclassified | - | Highly expressed in adipose and muscle tissues and correlated to obesity | Regulates body mass index and energy expenditure | [140] |
| 13690 | Eif4g2; Eukaryotic translation initiation factor 4, gamma 2 (DAP5)//translation initiation factor | Binding, translation regulator activity | Unclassified | Associated with fasting free FA and insulin sensitivity | - | [141] |
| 329260 | Dennd1b; DENN/MADD domain containing 1B//- | Binding | Localization, cellular process | One of the maternity-expressed imprinted gene candidates | Positively associated with obesity | [142] |
| 67845 | Rnf115; Ring finger protein 115/E3 ubiquitin-protein ligase RNF115/ubiquitin-protein ligase | - | Metabolic and cellular process | E3 ubiquitin-protein ligase | Involved in negative regulation of EGFR signaling pathway in cancer cells | [143,144] |
| 239719 | Mrtfb; Myocardin related transcription factor B//DNA-binding transcription factor | Unclassified | Developmental process, cellular process | SRF-MRTF signaling suppresses brown adipocyte development | Modulating TGF-β/BMP pathway | [145] |
| 207304 | Hectd1; HECT domain E3 ubiquitin protein ligase 1//ubiquitin-protein ligase | Catalytic activity | Metabolic and cellular process | E3 ubiquitin protein ligase for ABCA1-mediated cholesterol export from macrophages | Regulates the secretion of Hsp90 | [146] |
| 105559 | Mbnl2; Muscle blind like splicing factor 2//RNA splicing factor | Unclassified | Unclassified | Participates in integrin-α3 subcellular localization | Required for muscle differentiation | [147] |
| 228136 | Zdhhc5; Zinc finger, DHHC domain containing 5/Palmitoyltransferase ZDHHC5/- | Catalytic activity | Metabolic and cellular process | Palmitoyl acyltransferase catalyzes palmitoylation | Acts via Gp130/JAK/STAT3 pathway | [148] |
| 233726 | Ipo7; Importin 7//transporter | Unclassified | Cellular process | Plays a role in ferroptosis, promotes translocation of substrates via nuclear pore complex, triggers p53-dependent growth arrest, ribosomal biogenesis stress and nucleolar morphology changes | Negatively regulated by perlipin2 | [149] |
| 105522 | Ankrd28; Ankyrin repeat domain 28/Serine/threonine-protein phosphatase 6 regulatory ankyrin repeat subunit A/scaffold/adaptor protein | Unclassified | Unclassified | Enables protein binding and promotes cell migration | Regulating focal adhesion formation | [150] |
| 93762 | Smarca5; SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5//DNA helicase | ATP-dependent activity, binding, catalytic activity | Metabolic process, Biological regulation, cellular process | Mutation associated with ovine fat deposition and body size | - | [151] |
| 19684 | Rdx; Radixin//actin or actin-binding cytoskeletal protein | Binding | Localization, developmental process, Biological regulation, cellular process | Cytoskeletal protein plays a role in connecting actin to the plasma membrane | Plays a role in atherosclerosis | [152] |
| 242291 | Impad1:Bpnt2; 3′(2′), 5′-bis-phosphate nucleotidase 2 | - | - | An enzyme that hydrolyzes pAp to 5′-AMP and Pi | - | [153] |
| 18706 | Pik3ca; Phosphatidyl inositol-4,5-bisphosphate 3-kinase catalytic subunit alpha/Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform/Kinase | Catalytic activity | Signaling, response to stimulus, metabolic process, locomotion, localization, biological regulation, cellular process | PIK3CA gain-of-function mutation associated with adipose tissue overgrowth | Induces metabolic reprogramming with Warburg-like effect and severe endocrine disruption | [154] |
| 17847 | Usp34; Ubiquitin specific peptidase 34//cysteine protease | Catalytic activity | Metabolic and cellular process | In the Wnt pathway | Role in adipogenesis regulation | [155] |
| 71472 | Usp19; Ubiquitin specific peptidase 1/Ubiquitin carboxyl-terminal hydrolase 19/cysteine protease | Unclassified | Unclassified | A deubiquitinating enzyme which potentiates high-fat-diet-induced obesity | Modulates adipogenesis and glucose intolerance in mice | [156] |
| NCBI ID | Mapped IDs: Gene Symbol; Gene Name/Panther Family/Panther Protein Class | Panther GO-SLIM Molecular Functions | Panther GO-SLIM Biological Functions | Role Concerning Metabolic Phenotype | Ref. | |
|---|---|---|---|---|---|---|
| 16202 | Ilk; Integrin linked protein kinase/Integrin linked protein kinase/non-receptor serine/threonine protein kinase | Catalytic | Biological adhesion and regulation, cellular and developmental process, response to stimulus, signaling | Role in development of diet-induced adipose insulin resistance in male mice | - | [157] |
| 103968 | Plin1; Perilipin 1/PERILIPIN 1/- | No Panther-assigned category | Unclassified | Abundant lipid droplet coat protein, restricting lipolysis under basal or fed conditions | Role in lipid metabolism regulation | [158] |
| 227700 | Sh3glb2; SH3-domain GRB2-like endophilin B2/DREBRIN-LIKE PROTEIN-RELATED/scaffold/adaptor protein | No Panther-assigned category | Cellular process | Regulates lung homeostasis | Upregulated in PGC1b-FAT-KO mice | [159] |
| 11750 | Anxa7; Annexin A7/ANNEXIN A7/calcium-binding protein | Binding | Unclassified | Enables activities of calcium-dependent phospholipid binding, calcium-dependent protein binding, integrin binding activity | Role in insulin sensitivity and glucose uptake | [160,161,162] |
| 11804 | Aplp2; Amyloid beta (A4) precursor-like protein 2/Amyloid-like protein 2/Protease inhibitor | No Panther-assigned category | Cellular and developmental process, multicellular organismal process | Interacts with proprotein convertase subtilisin/kexin type 9 (PCSK9) | Post-transcriptionally regulates hepatic low-density lipoprotein receptors | [163] |
| 72787 | Ndc1; NDC1transmembrane nucleoporin/NUCLEOPORIN NDC1/- | Molecular adaptor, binding | Cellular process | In nuclear pore complex assembly and localization | - | [164] |
| 16890 | HSL; Hormone sensitive lipase/HORMONE SENSITIVE LIPASE/lipase | Catalytic | Cellular and metabolic process | Thermogenesis gene | Regulate lipolysis under cold | [165] |
| 108099 | Prkag2; 5′-AMP-activated, protein kinase subunit gamma 2/5′-AMP-ACTIVATED PROTEIN KINASE SUBUNIT GAMMA-2/kinase modulator | No Panther-assigned category | Unclassified | Required for manufacture of gamma-2 subunit of AMP-Activated Protein Kinase (AMPK), modulating thermogenesis in adipose t. | Regulates energy metabolism | [166,167] |
| 26358 | Aldh1a7; Aldehyde dehydrogenase cytosolic 1, subfamily A7/ALDEHYDE DEHYDROGENASE, CYTOSOLIC 1/dehydrogenase | Catalytic | Unclassified | Protects against high metabolic aldehydes from lipid oxidation | - | [168] |
| 64540 | Tspan4; Tetraspanin-4/TERTRASPANIN-4/scaffold/adaptor protein | No Panther-assigned category | Unclassified | Role in migrasome formation, promote angiogenesis | Role in oxidative stress by discarding damaged mitochondria | [169,170,171] |
| 12306 | Anxa2; Annexin A2/Annexin A2/calcium-binding protein | Binding | Unclassified | Role in GLUT4 translocation, insulin response, glucose uptake, CD36-mediated FA uptake, inflammation, macrophage infiltration | HSL activation | [172] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aboouf, M.A.; Gorr, T.A.; Hamdy, N.M.; Gassmann, M.; Thiersch, M. Myoglobin in Brown Adipose Tissue: A Multifaceted Player in Thermogenesis. Cells 2023, 12, 2240. https://doi.org/10.3390/cells12182240
Aboouf MA, Gorr TA, Hamdy NM, Gassmann M, Thiersch M. Myoglobin in Brown Adipose Tissue: A Multifaceted Player in Thermogenesis. Cells. 2023; 12(18):2240. https://doi.org/10.3390/cells12182240
Chicago/Turabian StyleAboouf, Mostafa A., Thomas A. Gorr, Nadia M. Hamdy, Max Gassmann, and Markus Thiersch. 2023. "Myoglobin in Brown Adipose Tissue: A Multifaceted Player in Thermogenesis" Cells 12, no. 18: 2240. https://doi.org/10.3390/cells12182240
APA StyleAboouf, M. A., Gorr, T. A., Hamdy, N. M., Gassmann, M., & Thiersch, M. (2023). Myoglobin in Brown Adipose Tissue: A Multifaceted Player in Thermogenesis. Cells, 12(18), 2240. https://doi.org/10.3390/cells12182240








