Overexpression of FRA1 (FOSL1) Leads to Global Transcriptional Perturbations, Reduced Cellular Adhesion and Altered Cell Cycle Progression
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Culture
2.2. Design and Cloning of CRISPRa gRNA Sequences into Lentiviral Transfer Plasmid
2.3. Preparation of CRISPRa Lentiviral Particles via Calcium Phosphate Transfection
2.4. CGL1 Lentiviral Infection for Producing CRISPRa Mediated FRA1 Overexpression
2.5. RNA Extraction
2.6. cDNA Synthesis
2.7. RT-qPCR
2.8. Protein Extraction and Western Blot Analysis
2.9. Cell Growth Assay
2.10. RNA-seq Whole Transcriptome Analysis
2.11. Cell Adhesion Assay
2.12. Cell Cycle Flow-Cytometry Analysis
2.13. Quantification and Statistical Analysis
3. Results
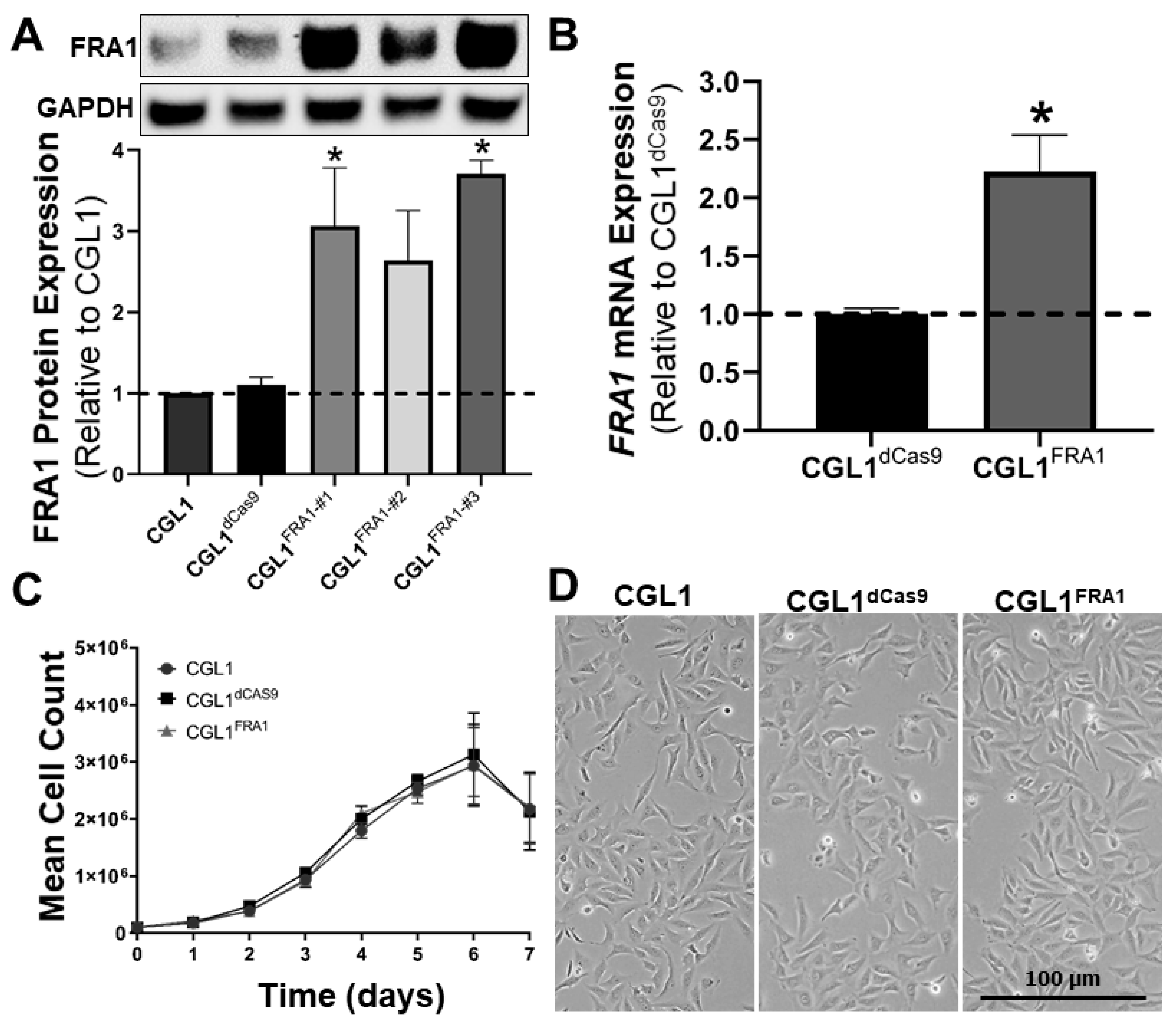
3.1. Establishing FRA1-Overexpressing CGL1 Cells
3.2. Whole Transcriptome RNA-seq Analysis of CGL1FRA1 Cells Relative to CGL1dCas9 Cells
3.3. Gene Ontology (GO) Enrichment Analysis of DEGs in CGL1FRA1 Cells
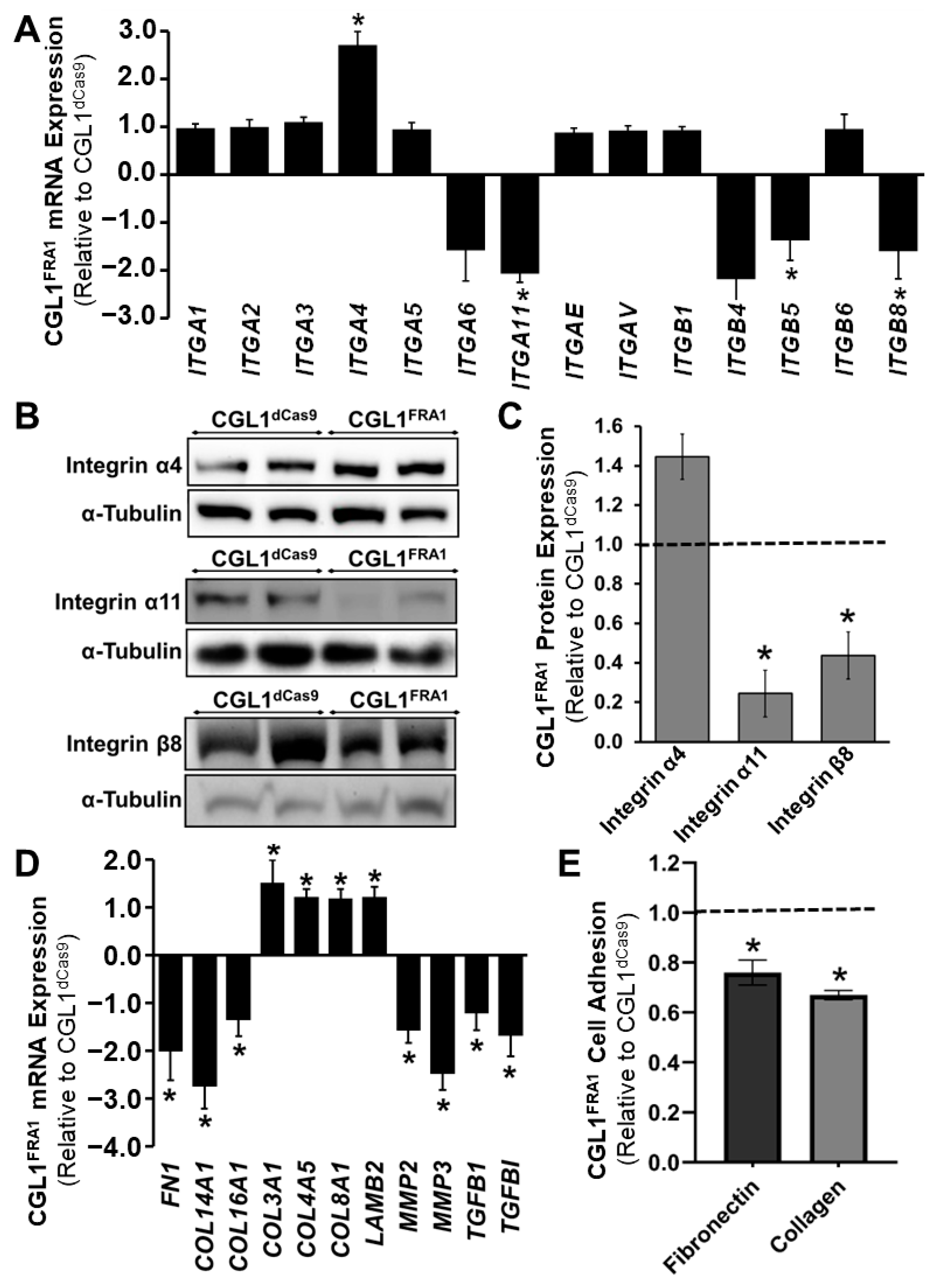
3.4. FRA1 Upregulation Alters the Expression of Core AP-1 Complex Members
3.5. FRA1 Upregulation Alters the Adhesion Profile of CGL1 Cells
3.6. Cell Cycle Analysis of CGL1FRA1 and CGL1dCas9 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sobolev, V.V.; Khashukoeva, A.Z.; Evina, O.E.; Geppe, N.A.; Chebysheva, S.N.; Korsunskaya, I.M.; Tchepourina, E.; Mezentsev, A. Role of the Transcription Factor FOSL1 in Organ Development and Tumorigenesis. Int. J. Mol. Sci. 2022, 23, 1521. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Talotta, F.; Cimmino, A.; Verde, P. The Fra-1/AP-1 Oncoprotein: From the “Undruggable” Transcription Factor to Therapeutic Targeting. Cancers 2022, 14, 1480. [Google Scholar] [CrossRef] [PubMed]
- Galvagni, F.; Orlandini, M.; Oliviero, S. Role of the AP-1 transcription factor FOSL1 in endothelial cells adhesion and migration. Cell Adhes. Migr. 2013, 7, 408–411. [Google Scholar] [CrossRef]
- Matsuo, K.; Owens, J.M.; Tonko, M.; Elliott, C.; Chambers, T.J.; Wagner, E.F. Fosl1 is a transcriptional target of c-Fos during osteoclast differentiation. Nat. Genet. 2000, 24, 184–187. [Google Scholar] [CrossRef]
- Wang, C.; Li, Z.; Shao, F.; Yang, X.; Feng, X.; Shi, S.; Gao, Y.; He, J. High expression of Collagen Triple Helix Repeat Containing 1 (CTHRC1) facilitates progression of oesophageal squamous cell carcinoma through MAPK/MEK/ERK/FRA-1 activation. J. Exp. Clin. Cancer Res. CR 2017, 36, 84. [Google Scholar] [CrossRef] [PubMed]
- Bejjani, F.; Evanno, E.; Zibara, K.; Piechaczyk, M.; Jariel-Encontre, I. The AP-1 transcriptional complex: Local switch or remote command? Biochim. Et Biophys. Acta. Rev. Cancer 2019, 1872, 11–23. [Google Scholar] [CrossRef]
- John, M.; Leppik, R.; Busch, S.J.; Granger-Schnarr, M.; Schnarr, M. DNA binding of Jun and Fos bZip domains: Homodimers and heterodimers induce a DNA conformational change in solution. Nucleic Acids Res. 1996, 24, 4487–4494. [Google Scholar] [CrossRef]
- Lee, W.; Mitchell, P.; Tjian, R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell 1987, 49, 741–752. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.G.; Nesterova, M.; Agrawal, S.; Cho-Chung, Y.S. Dual blockade of cyclic AMP response element- (CRE) and AP-1-directed transcription by CRE-transcription factor decoy oligonucleotide. gene-specific inhibition of tumor growth. J. Biol. Chem. 1999, 274, 1573–1580. [Google Scholar] [CrossRef]
- Karin, M.; Liu, Z.; Zandi, E. AP-1 function and regulation. Curr. Opin. Cell Biol. 1997, 9, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Wisdom, R. AP-1: One switch for many signals. Exp. Cell Res. 1999, 253, 180–185. [Google Scholar] [CrossRef]
- Zeng, F.; He, J.; Jin, X.; Liao, Q.; Chen, Z.; Peng, H.; Zhou, Y. FRA-1: A key factor regulating signal transduction of tumor cells and a potential target molecule for tumor therapy. Biomed. Pharmacother. Biomed. Pharmacother. 2022, 150, 113037. [Google Scholar] [CrossRef]
- Glover, J.N.; Harrison, S.C. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 1995, 373, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Georgopoulos, K.; Greenberg, M.E.; Leder, P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell 1988, 55, 917–924. [Google Scholar] [CrossRef] [PubMed]
- Fan, F.; Podar, K. The Role of AP-1 Transcription Factors in Plasma Cell Biology and Multiple Myeloma Pathophysiology. Cancers 2021, 13, 2326. [Google Scholar] [CrossRef] [PubMed]
- Nishina, T.; Deguchi, Y.; Miura, R.; Yamazaki, S.; Shinkai, Y.; Kojima, Y.; Okumura, K.; Kumagai, Y.; Nakano, H. Critical Contribution of Nuclear Factor Erythroid 2-related Factor 2 (NRF2) to Electrophile-induced Interleukin-11 Production. The J. Biol. Chem. 2017, 292, 205–216. [Google Scholar] [CrossRef]
- Takada, Y.; Gresh, L.; Bozec, A.; Ikeda, E.; Kamiya, K.; Watanabe, M.; Kobayashi, K.; Asano, K.; Toyama, Y.; Wagner, E.F.; et al. Interstitial lung disease induced by gefitinib and toll-like receptor ligands is mediated by Fra-1. Oncogene 2011, 30, 3821–3832. [Google Scholar] [CrossRef]
- Niidome, K.; Taniguchi, R.; Yamazaki, T.; Tsuji, M.; Itoh, K.; Ishihara, Y. FosL1 Is a Novel Target of Levetiracetam for Suppressing the Microglial Inflammatory Reaction. Int. J. Mol. Sci. 2021, 22, 10962. [Google Scholar] [CrossRef]
- Wu, H.Y.; Zhou, Y.M.; Liao, Z.Q.; Zhong, J.W.; Liu, Y.B.; Zhao, H.; Liang, C.Q.; Huang, R.J.; Park, K.S.; Feng, S.S.; et al. Fosl1 is vital to heart regeneration upon apex resection in adult Xenopus tropicalis. NPJ Regen. Med. 2021, 6, 36. [Google Scholar] [CrossRef]
- Kakumoto, K.; Sasai, K.; Sukezane, T.; Oneyama, C.; Ishimaru, S.; Shibutani, K.; Mizushima, H.; Mekada, E.; Hanafusa, H.; Akagi, T. FRA1 is a determinant for the difference in RAS-induced transformation between human and rat fibroblasts. Proc. Natl. Acad. Sci. USA 2006, 103, 5490–5495. [Google Scholar] [CrossRef]
- Evellin, S.; Galvagni, F.; Zippo, A.; Neri, F.; Orlandini, M.; Incarnato, D.; Dettori, D.; Neubauer, S.; Kessler, H.; Wagner, E.F.; et al. FOSL1 controls the assembly of endothelial cells into capillary tubes by direct repression of alphav and beta3 integrin transcription. Mol. Cell. Biol. 2013, 33, 1198–1209. [Google Scholar] [CrossRef]
- Bakiri, L.; Macho-Maschler, S.; Custic, I.; Niemiec, J.; Guio-Carrion, A.; Hasenfuss, S.C.; Eger, A.; Muller, M.; Beug, H.; Wagner, E.F. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFbeta expression. Cell Death Differ. 2015, 22, 336–350. [Google Scholar] [CrossRef]
- Shao, S.; Liu, Y.; Hong, W.; Mo, Y.; Shu, F.; Jiang, L.; Tan, N. Influence of FOSL1 Inhibition on Vascular Calcification and ROS Generation through Ferroptosis via P53-SLC7A11 Axis. Biomedicines 2023, 11, 635. [Google Scholar] [CrossRef] [PubMed]
- Luther, J.; Driessler, F.; Megges, M.; Hess, A.; Herbort, B.; Mandic, V.; Zaiss, M.M.; Reichardt, A.; Zech, C.; Tuckermann, J.P.; et al. Elevated Fra-1 expression causes severe lipodystrophy. J. Cell Sci. 2011, 124, 1465–1476. [Google Scholar] [CrossRef] [PubMed]
- Gillies, T.E.; Pargett, M.; Minguet, M.; Davies, A.E.; Albeck, J.G. Linear Integration of ERK Activity Predominates over Persistence Detection in Fra-1 Regulation. Cell Syst. 2017, 5, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Talotta, F.; Casalino, L.; Verde, P. The nuclear oncoprotein Fra-1: A transcription factor knocking on therapeutic applications’ door. Oncogene 2020, 39, 4491–4506. [Google Scholar] [CrossRef] [PubMed]
- Young, M.R.; Nair, R.; Bucheimer, N.; Tulsian, P.; Brown, N.; Chapp, C.; Hsu, T.C.; Colburn, N.H. Transactivation of Fra-1 and consequent activation of AP-1 occur extracellular signal-regulated kinase dependently. Mol. Cell. Biol. 2002, 22, 587–598. [Google Scholar] [CrossRef]
- Cursons, J.; Leuchowius, K.J.; Waltham, M.; Tomaskovic-Crook, E.; Foroutan, M.; Bracken, C.P.; Redfern, A.; Crampin, E.J.; Street, I.; Davis, M.J.; et al. Stimulus-dependent differences in signalling regulate epithelial-mesenchymal plasticity and change the effects of drugs in breast cancer cell lines. Cell Commun. Signal. CCS 2015, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Catani, M.V.; Rossi, A.; Costanzo, A.; Sabatini, S.; Levrero, M.; Melino, G.; Avigliano, L. Induction of gene expression via activator protein-1 in the ascorbate protection against UV-induced damage. Biochem. J. 2001, 356, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Limoge, M.; Safina, A.; Beattie, A.; Kapus, L.; Truskinovsky, A.M.; Bakin, A.V. Tumor-fibroblast interactions stimulate tumor vascularization by enhancing cytokine-driven production of MMP9 by tumor cells. Oncotarget 2017, 8, 35592–35608. [Google Scholar] [CrossRef]
- Eferl, R.; Hoebertz, A.; Schilling, A.F.; Rath, M.; Karreth, F.; Kenner, L.; Amling, M.; Wagner, E.F. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004, 23, 2789–2799. [Google Scholar] [CrossRef]
- Dai, C.; Rennhack, J.P.; Arnoff, T.E.; Thaker, M.; Younger, S.T.; Doench, J.G.; Huang, A.Y.; Yang, A.; Aguirre, A.J.; Wang, B.; et al. SMAD4 represses FOSL1 expression and pancreatic cancer metastatic colonization. Cell Rep. 2021, 36, 109443. [Google Scholar] [CrossRef]
- Terasawa, K.; Okazaki, K.; Nishida, E. Regulation of c-Fos and Fra-1 by the MEK5-ERK5 pathway. Genes Cells Devoted Mol. Cell. Mech. 2003, 8, 263–273. [Google Scholar] [CrossRef]
- Basbous, J.; Chalbos, D.; Hipskind, R.; Jariel-Encontre, I.; Piechaczyk, M. Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Mol. Cell. Biol. 2007, 27, 3936–3950. [Google Scholar] [CrossRef]
- Talotta, F.; Mega, T.; Bossis, G.; Casalino, L.; Basbous, J.; Jariel-Encontre, I.; Piechaczyk, M.; Verde, P. Heterodimerization with Fra-1 cooperates with the ERK pathway to stabilize c-Jun in response to the RAS oncoprotein. Oncogene 2010, 29, 4732–4740. [Google Scholar] [CrossRef] [PubMed]
- Belguise, K.; Kersual, N.; Galtier, F.; Chalbos, D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 2005, 24, 1434–1444. [Google Scholar] [CrossRef]
- Bakiri, L.; Takada, Y.; Radolf, M.; Eferl, R.; Yaniv, M.; Wagner, E.F.; Matsuo, K. Role of heterodimerization of c-Fos and Fra1 proteins in osteoclast differentiation. Bone 2007, 40, 867–875. [Google Scholar] [CrossRef]
- Shirsat, N.V.; Shaikh, S.A. Overexpression of the immediate early gene fra-1 inhibits proliferation, induces apoptosis, and reduces tumourigenicity of c6 glioma cells. Exp. Cell Res. 2003, 291, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Rattanasinchai, C.; Llewellyn, B.J.; Conrad, S.E.; Gallo, K.A. MLK3 regulates FRA-1 and MMPs to drive invasion and transendothelial migration in triple-negative breast cancer cells. Oncogenesis 2017, 6, e345. [Google Scholar] [CrossRef] [PubMed]
- Jochum, W.; David, J.P.; Elliott, C.; Wutz, A.; Plenk, H., Jr.; Matsuo, K.; Wagner, E.F. Increased bone formation and osteosclerosis in mice overexpressing the transcription factor Fra-1. Nat. Med. 2000, 6, 980–984. [Google Scholar] [CrossRef]
- Jiang, X.; Xie, H.; Dou, Y.; Yuan, J.; Zeng, D.; Xiao, S. Expression and function of FRA1 protein in tumors. Mol. Biol. Rep. 2020, 47, 737–752. [Google Scholar] [CrossRef] [PubMed]
- Sayan, A.E.; Stanford, R.; Vickery, R.; Grigorenko, E.; Diesch, J.; Kulbicki, K.; Edwards, R.; Pal, R.; Greaves, P.; Jariel-Encontre, I.; et al. Fra-1 controls motility of bladder cancer cells via transcriptional upregulation of the receptor tyrosine kinase AXL. Oncogene 2012, 31, 1493–1503. [Google Scholar] [CrossRef]
- Gao, X.Q.; Ge, Y.S.; Shu, Q.H.; Ma, H.X. Expression of Fra-1 in human hepatocellular carcinoma and its prognostic significance. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317709635. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Oh, J.H.; Kim, N.H.; Choi, K.M.; Kim, S.J.; Baik, S.H.; Choi, D.S.; Lee, E.S. Fra-1 expression in malignant and benign thyroid tumor. Korean J. Intern. Med. 2001, 16, 93–97. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhu, G.; Gao, L.; Chen, P.; Long, Y.; Liao, S.; Yi, H.; Yi, W.; Pei, Z.; Wu, M.; et al. Fra-1 is upregulated in gastric cancer tissues and affects the PI3K/Akt and p53 signaling pathway in gastric cancer. Int. J. Oncol. 2015, 47, 1725–1734. [Google Scholar] [CrossRef] [PubMed]
- Kimura, R.; Ishikawa, C.; Rokkaku, T.; Janknecht, R.; Mori, N. Phosphorylated c-Jun and Fra-1 induce matrix metalloproteinase-1 and thereby regulate invasion activity of 143B osteosarcoma cells. Biochim. Et Biophys. Acta 2011, 1813, 1543–1553. [Google Scholar] [CrossRef]
- Iskit, S.; Schlicker, A.; Wessels, L.; Peeper, D.S. Fra-1 is a key driver of colon cancer metastasis and a Fra-1 classifier predicts disease-free survival. Oncotarget 2015, 6, 43146–43161. [Google Scholar] [CrossRef]
- Dikshit, A.; Jin, Y.J.; Degan, S.; Hwang, J.; Foster, M.W.; Li, C.Y.; Zhang, J.Y. UBE2N Promotes Melanoma Growth via MEK/FRA1/SOX10 Signaling. Cancer Res. 2018, 78, 6462–6472. [Google Scholar] [CrossRef]
- Hanson, R.L.; Brown, R.B.; Steele, M.M.; Grandgenett, P.M.; Grunkemeyer, J.A.; Hollingsworth, M.A. Identification of FRA-1 as a novel player in pancreatic cancer in cooperation with a MUC1: ERK signaling axis. Oncotarget 2016, 7, 39996–40011. [Google Scholar] [CrossRef]
- Ma, K.; Chang, D.; Gong, M.; Ding, F.; Luo, A.; Tian, F.; Liu, Z.; Wang, T. Expression and significance of FRA-1 in non-small-cell lung cancer. Cancer Investig. 2009, 27, 353–359. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, L.; He, J.; He, Z.; Yue, C.; Jin, X.; Gao, M.; Xiao, S.; Zhou, Y. Fra-1 Inhibits Cell Growth and the Warburg Effect in Cervical Cancer Cells via STAT1 Regulation of the p53 Signaling Pathway. Front. Cell Dev. Biol. 2020, 8, 579629. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Zhou, Y.; Yi, W.; Luo, G.; Jiang, B.; Tian, Q.; Li, Y.; Xue, M. Fra-1 is downregulated in cervical cancer tissues and promotes cervical cancer cell apoptosis by p53 signaling pathway in vitro. Int. J. Oncol. 2015, 46, 1677–1684. [Google Scholar] [CrossRef] [PubMed]
- Pirkkanen, J.S.; Boreham, D.R.; Mendonca, M.S. The CGL1 (HeLa x Normal Skin Fibroblast) Human Hybrid Cell Line: A History of Ionizing Radiation Induced Effects on Neoplastic Transformation and Novel Future Directions in SNOLAB. Radiat. Res. 2017, 188, 512–524. [Google Scholar] [CrossRef] [PubMed]
- Pirkkanen, J.; Tharmalingam, S.; Thome, C.; Sinex, H.C.; Benjamin, L.V.; Losch, A.C.; Borgmann, A.J.; Dhaemers, R.M.; Gordon, G.; Boreham, D.R.; et al. Genomic Loss and Epigenetic Silencing of the FOSL1 Tumor Suppressor Gene in Radiation-induced Neoplastic Transformation of Human CGL1 Cells Alters the Tumorigenic Phenotype In Vitro and In Vivo. Radiat. Res. 2023, 200, 48–64. [Google Scholar] [CrossRef]
- Pirkkanen, J.; Tharmalingam, S.; Morais, I.H.; Lam-Sidun, D.; Thome, C.; Zarnke, A.M.; Benjamin, L.V.; Losch, A.C.; Borgmann, A.J.; Sinex, H.C.; et al. Transcriptomic profiling of gamma ray induced mutants from the CGL1 human hybrid cell system reveals novel insights into the mechanisms of radiation-induced carcinogenesis. Free. Radic. Biol. Med. 2019, 145, 300–311. [Google Scholar] [CrossRef]
- Hannemann, N.; Cao, S.; Eriksson, D.; Schnelzer, A.; Jordan, J.; Eberhardt, M.; Schleicher, U.; Rech, J.; Ramming, A.; Uebe, S.; et al. Transcription factor Fra-1 targets arginase-1 to enhance macrophage-mediated inflammation in arthritis. J. Clin. Investig. 2019, 129, 2669–2684. [Google Scholar] [CrossRef]
- Morishita, H.; Saito, F.; Kayama, H.; Atarashi, K.; Kuwata, H.; Yamamoto, M.; Takeda, K. Fra-1 negatively regulates lipopolysaccharide-mediated inflammatory responses. Int. Immunol. 2009, 21, 457–465. [Google Scholar] [CrossRef]
- Cao, S.; Schnelzer, A.; Hannemann, N.; Schett, G.; Soulat, D.; Bozec, A. The Transcription Factor FRA-1/AP-1 Controls Lipocalin-2 Expression and Inflammation in Sepsis Model. Front. Immunol. 2021, 12, 701675. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, M.S.; Antoniono, R.J.; Latham, K.M.; Stanbridge, E.J.; Redpath, J.L. Characterization of intestinal alkaline phosphatase expression and the tumorigenic potential of gamma-irradiated HeLa x fibroblast cell hybrids. Cancer Res. 1991, 51, 4455–4462. [Google Scholar]
- Redpath, J.L.; Sun, C.; Colman, M.; Stanbridge, E.J. Radiobiological studies of human hybrid cells (skin fibroblasts X HeLa) and their tumourigenic segregants. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1985, 48, 479–483. [Google Scholar] [CrossRef]
- Redpath, J.L.; Sun, C.; Colman, M.; Stanbridge, E.J. Neoplastic transformation of human hybrid cells by gamma radiation: A quantitative assay. Radiat. Res. 1987, 110, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, M.S.; Farrington, D.L.; Mayhugh, B.M.; Qin, Y.; Temples, T.; Comerford, K.; Chakrabarti, R.; Zainabadi, K.; Redpath, J.L.; Stanbridge, E.J.; et al. Homozygous deletions within the 11q13 cervical cancer tumor-suppressor locus in radiation-induced, neoplastically transformed human hybrid cells. Genes Chromosomes Cancer 2004, 39, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Mendonca, M.S.; Howard, K.; Desmond, L.A.; Derrow, C.W. Previous loss of chromosome 11 containing a suppressor locus increases radiosensitivity, neoplastic transformation frequency and delayed death in HeLa x fibroblast human hybrid cells. Mutagenesis 1999, 14, 483–490. [Google Scholar] [CrossRef]
- Maeder, M.L.; Linder, S.J.; Cascio, V.M.; Fu, Y.; Ho, Q.H.; Joung, J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 2013, 10, 977–979. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Casas-Mollano, J.A.; Zinselmeier, M.H.; Erickson, S.E.; Smanski, M.J. CRISPR-Cas Activators for Engineering Gene Expression in Higher Eukaryotes. CRISPR J. 2020, 3, 350–364. [Google Scholar] [CrossRef]
- Khurana, S.; Grandbois, J.; Tharmalingam, S.; Murray, A.; Graff, K.; Nguyen, P.; Tai, T.C. Fetal programming of adrenal PNMT and hypertension by glucocorticoids in WKY rats is dose and sex-dependent. PLoS ONE 2019, 14, e0221719. [Google Scholar] [CrossRef] [PubMed]
- Lamothe, J.; Khurana, S.; Tharmalingam, S.; Williamson, C.; Byrne, C.J.; Khaper, N.; Mercier, S.; Tai, T.C. The Role of DNMT and HDACs in the Fetal Programming of Hypertension by Glucocorticoids. Oxid. Med. Cell Longev. 2020, 2020, 5751768. [Google Scholar] [CrossRef]
- Vigneux, G.; Pirkkanen, J.; Laframboise, T.; Prescott, H.; Tharmalingam, S.; Thome, C. Radiation-Induced Alterations in Proliferation, Migration, and Adhesion in Lens Epithelial Cells and Implications for Cataract Development. Bioengineering 2022, 9, 29. [Google Scholar] [CrossRef]
- Davidson, C.Q.; Tharmalingam, S.; Niccoli, S.; Nemec-Bakk, A.; Khurana, S.; Murray, A.; Tai, T.C.; Boreham, D.R.; Khaper, N.; Lees, S.J. Dose threshold for radiation induced fetal programming in a mouse model at 4 months of age: Hepatic expression of genes and proteins involved in glucose metabolism and glucose uptake in brown adipose tissue. PLoS ONE 2020, 15, e0231650. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.; Tharmalingam, S.; Khurana, S.; Lalonde, C.; Nguyen, P.; Tai, T.C. Effect of Prenatal Glucocorticoid Exposure on Circadian Rhythm Gene Expression in the Brains of Adult Rat Offspring. Cells 2022, 11, 1613. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Puukila, S.; Tharmalingam, S.; Al-Khayyat, W.; Peterson, J.; Hooker, A.M.; Muise, S.; Boreham, D.R.; Dixon, D.L. Transcriptomic Response in the Spleen after Whole-Body Low-Dose X-Ray Irradiation. Radiat. Res. 2021, 196, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Tharmalingam, S.; Daulat, A.M.; Antflick, J.E.; Ahmed, S.M.; Nemeth, E.F.; Angers, S.; Conigrave, A.D.; Hampson, D.R. Calcium-sensing receptor modulates cell adhesion and migration via integrins. J. Biol. Chem. 2011, 286, 40922–40933. [Google Scholar] [CrossRef]
- Tharmalingam, S.; Wu, C.; Hampson, D.R. The calcium-sensing receptor and integrins modulate cerebellar granule cell precursor differentiation and migration. Dev. Neurobiol. 2016, 76, 375–389. [Google Scholar] [CrossRef]
- Peterson, J.; McTiernan, C.D.; Thome, C.; Khaper, N.; Lees, S.J.; Boreham, D.R.; Tai, T.C.; Tharmalingam, S. Identification of Radiation-Induced miRNA Biomarkers Using the CGL1 Cell Model System. Bioengineering 2022, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Tharmalingam, S.; Khurana, S.; Murray, A.; Lamothe, J.; Tai, T.C. Whole transcriptome analysis of adrenal glands from prenatal glucocorticoid programmed hypertensive rodents. Sci. Rep. 2020, 10, 18755. [Google Scholar] [CrossRef]
- Lai, L.; Shin, G.Y.; Qiu, H. The Role of Cell Cycle Regulators in Cell Survival-Dual Functions of Cyclin-Dependent Kinase 20 and p21(Cip1/Waf1). Int. J. Mol. Sci. 2020, 21, 8504. [Google Scholar] [CrossRef]
- Ju, Y.T.; Chang, A.C.; She, B.R.; Tsaur, M.L.; Hwang, H.M.; Chao, C.C.; Cohen, S.N.; Lin-Chao, S. gas7: A gene expressed preferentially in growth-arrested fibroblasts and terminally differentiated Purkinje neurons affects neurite formation. Proc. Natl. Acad. Sci. USA 1998, 95, 11423–11428. [Google Scholar] [CrossRef]
- Yang, L.; Karin, M. Roles of tumor suppressors in regulating tumor-associated inflammation. Cell Death Differ. 2014, 21, 1677–1686. [Google Scholar] [CrossRef]
- Cohen, D.R.; Ferreira, P.C.; Gentz, R.; Franza, B.R., Jr.; Curran, T. The product of a fos-related gene, fra-1, binds cooperatively to the AP-1 site with Jun: Transcription factor AP-1 is comprised of multiple protein complexes. Genes Dev. 1989, 3, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Pecce, V.; Verrienti, A.; Fiscon, G.; Sponziello, M.; Conte, F.; Abballe, L.; Durante, C.; Farina, L.; Filetti, S.; Paci, P. The role of FOSL1 in stem-like cell reprogramming processes. Sci. Rep. 2021, 11, 14677. [Google Scholar] [CrossRef] [PubMed]
- Irie, A.; Kamata, T.; Takada, Y. Multiple loop structures critical for ligand binding of the integrin alpha4 subunit in the upper face of the beta-propeller mode 1. Proc. Natl. Acad. Sci. USA 1997, 94, 7198–7203. [Google Scholar] [CrossRef] [PubMed]
- Rose, D.M.; Han, J.; Ginsberg, M.H. Alpha4 integrins and the immune response. Immunol. Rev. 2002, 186, 118–124. [Google Scholar] [CrossRef]
- Zhang, K.; Myllymaki, S.M.; Gao, P.; Devarajan, R.; Kytola, V.; Nykter, M.; Wei, G.H.; Manninen, A. Oncogenic K-Ras upregulates ITGA6 expression via FOSL1 to induce anoikis resistance and synergizes with alphaV-Class integrins to promote EMT. Oncogene 2017, 36, 5681–5694. [Google Scholar] [CrossRef]
- Ge, J.; Apicella, M.; Mills, J.A.; Garcon, L.; French, D.L.; Weiss, M.J.; Bessler, M.; Mason, P.J. Dysregulation of the Transforming Growth Factor beta Pathway in Induced Pluripotent Stem Cells Generated from Patients with Diamond Blackfan Anemia. PLoS ONE 2015, 10, e0134878. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.K.; Sheppard, D.; Chapman, H.A. TGF-beta1 Signaling and Tissue Fibrosis. Cold Spring Harb. Perspect. Biol. 2018, 10, a022293. [Google Scholar] [CrossRef]
- Esparza, J.; Vilardell, C.; Calvo, J.; Juan, M.; Vives, J.; Urbano-Marquez, A.; Yague, J.; Cid, M.C. Fibronectin upregulates gelatinase B (MMP-9) and induces coordinated expression of gelatinase A (MMP-2) and its activator MT1-MMP (MMP-14) by human T lymphocyte cell lines. A process repressed through RAS/MAP kinase signaling pathways. Blood 1999, 94, 2754–2766. [Google Scholar] [CrossRef]
- Birkedal-Hansen, H.; Moore, W.G.; Bodden, M.K.; Windsor, L.J.; Birkedal-Hansen, B.; DeCarlo, A.; Engler, J.A. Matrix metalloproteinases: A review. Crit. Rev. Oral Biol. Med. Off. Publ. Am. Assoc. Oral Biol. 1993, 4, 197–250. [Google Scholar] [CrossRef]
- Lee, J.M.; Dedhar, S.; Kalluri, R.; Thompson, E.W. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 2006, 172, 973–981. [Google Scholar] [CrossRef]
- Li, Y.; Peart, M.J.; Prives, C. Stxbp4 regulates DeltaNp63 stability by suppression of RACK1-dependent degradation. Mol. Cell. Biol. 2009, 29, 3953–3963. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Qing, O.; Ji, Z.; Chengqu, Z.; Ying, C.; Hao, C.; Minhui, X.; Lunshan, X. Downregulation of miR-16 via URGCP pathway contributes to glioma growth. Sci. Rep. 2017, 7, 13470. [Google Scholar] [CrossRef] [PubMed]
- Eldridge, A.G.; Loktev, A.V.; Hansen, D.V.; Verschuren, E.W.; Reimann, J.D.; Jackson, P.K. The evi5 oncogene regulates cyclin accumulation by stabilizing the anaphase-promoting complex inhibitor emi1. Cell 2006, 124, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Ghatak, D.; Das, P.; Roychoudhury, S. Transcriptional control of mitosis: Deregulation and cancer. Front. Endocrinol. 2015, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Casalino, L.; Bakiri, L.; Talotta, F.; Weitzman, J.B.; Fusco, A.; Yaniv, M.; Verde, P. Fra-1 promotes growth and survival in RAS-transformed thyroid cells by controlling cyclin A transcription. EMBO J. 2007, 26, 1878–1890. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, P.; Nair, A.; Patidar, A.; Dandapat, J.; Sarkar, A.; Saha, B. A primer on cytokines. Cytokine 2021, 145, 155458. [Google Scholar] [CrossRef]
- Bignold, R.; Johnson, J.R. Effects of cytokine signaling inhibition on inflammation-driven tissue remodeling. Curr. Res. Pharmacol. Drug Discov. 2021, 2, 100023. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Murray, P.J. Cytokine signaling modules in inflammatory responses. Immunity 2008, 28, 477–487. [Google Scholar] [CrossRef]
- Hanada, T.; Yoshimura, A. Regulation of cytokine signaling and inflammation. Cytokine Growth Factor Rev. 2002, 13, 413–421. [Google Scholar] [CrossRef]
- Barrat, F.J.; Crow, M.K.; Ivashkiv, L.B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 2019, 20, 1574–1583. [Google Scholar] [CrossRef]
- Singh, H.; Khan, A.A.; Dinner, A.R. Gene regulatory networks in the immune system. Trends Immunol. 2014, 35, 211–218. [Google Scholar] [CrossRef]
- Li, Q.R.; Ni, W.P.; Lei, N.J.; Yang, J.Y.; Xuan, X.Y.; Liu, P.P.; Gong, G.M.; Yan, F.; Feng, Y.S.; Zhao, R.; et al. The overexpression of Fra1 disorders the inflammatory cytokine secretion by mTEC of myasthenia gravis thymus. Scand. J. Immunol. 2018, 88, e12676. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, R.; Lv, Y.; Fu, R. The regulatory role of Fos related antigen-1 in inflammatory bowel disease. Mol. Med. Rep. 2018, 17, 1979–1985. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, X.; Zhang, C.; Hou, B.; Zhao, H. FOSL1 knockdown ameliorates DSS-induced inflammation and barrier damage in ulcerative colitis via MMP13 downregulation. Exp. Ther. Med. 2022, 24, 551. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tang, M.; Chen, Z.; Nie, L.; Li, S.; Xiong, Y.; Szymonowicz, K.A.; Park, J.M.; Zhang, H.; Feng, X.; et al. Genetic vulnerabilities upon inhibition of DNA damage response. Nucleic Acids Res. 2021, 49, 8214–8231. [Google Scholar] [CrossRef]
- Lou, J.; Chen, H.; Han, J.; He, H.; Huen, M.S.Y.; Feng, X.H.; Liu, T.; Huang, J. AUNIP/C1orf135 directs DNA double-strand breaks towards the homologous recombination repair pathway. Nat. Commun. 2017, 8, 985. [Google Scholar] [CrossRef]
- Moura, D.S.; Pena-Chilet, M.; Cordero Varela, J.A.; Alvarez-Alegret, R.; Agra-Pujol, C.; Izquierdo, F.; Ramos, R.; Ortega-Medina, L.; Martin-Davila, F.; Castilla-Ramirez, C.; et al. A DNA damage repair gene-associated signature predicts responses of patients with advanced soft-tissue sarcoma to treatment with trabectedin. Mol. Oncol. 2021, 15, 3691–3705. [Google Scholar] [CrossRef]
- Stoyanova, T.; Roy, N.; Kopanja, D.; Raychaudhuri, P.; Bagchi, S. DDB2 (damaged-DNA binding protein 2) in nucleotide excision repair and DNA damage response. Cell Cycle 2009, 8, 4067–4071. [Google Scholar] [CrossRef]
- Wang, W.; Mani, A.M.; Wu, Z.H. DNA damage-induced nuclear factor-kappa B activation and its roles in cancer progression. J. Cancer Metastasis Treat. 2017, 3, 45–59. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, L.P.; Himmels, S.F.; Trenner, A.; Walker, C.; von Aesch, C.; Eggenschwiler, A.; Murina, O.; Enchev, R.I.; Peter, M.; Freire, R.; et al. Cullin3-KLHL15 ubiquitin ligase mediates CtIP protein turnover to fine-tune DNA-end resection. Nat. Commun. 2016, 7, 12628. [Google Scholar] [CrossRef]
- Tharmalingam, S.; Sreetharan, S.; Brooks, A.L.; Boreham, D.R. Re-evaluation of the linear no-threshold (LNT) model using new paradigms and modern molecular studies. Chem. Biol. Interact. 2019, 301, 54–67. [Google Scholar] [CrossRef]
- Matsuoka, K.; Bakiri, L.; Wolff, L.I.; Linder, M.; Mikels-Vigdal, A.; Patino-Garcia, A.; Lecanda, F.; Hartmann, C.; Sibilia, M.; Wagner, E.F. Wnt signaling and Loxl2 promote aggressive osteosarcoma. Cell Res. 2020, 30, 885–901. [Google Scholar] [CrossRef]
- Yook, J.I.; Li, X.Y.; Ota, I.; Hu, C.; Kim, H.S.; Kim, N.H.; Cha, S.Y.; Ryu, J.K.; Choi, Y.J.; Kim, J.; et al. A Wnt-Axin2-GSK3beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006, 8, 1398–1406. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; Han, L.; Yi, Y. Reassessing the Potential of Myb-targeted Anti-cancer Therapy. J. Cancer 2018, 9, 1259–1266. [Google Scholar] [CrossRef]
- Yook, J.I.; Li, X.Y.; Ota, I.; Fearon, E.R.; Weiss, S.J. Wnt-dependent regulation of the E-cadherin repressor snail. J. Biol. Chem. 2005, 280, 11740–11748. [Google Scholar] [CrossRef] [PubMed]
- Soza-Ried, C.; Hess, I.; Netuschil, N.; Schorpp, M.; Boehm, T. Essential role of c-myb in definitive hematopoiesis is evolutionarily conserved. Proc. Natl. Acad. Sci. USA 2010, 107, 17304–17308. [Google Scholar] [CrossRef] [PubMed]
- Ciciro, Y.; Sala, A. MYB oncoproteins: Emerging players and potential therapeutic targets in human cancer. Oncogenesis 2021, 10, 19. [Google Scholar] [CrossRef]
- Hu, R.J.; Lee, M.P.; Connors, T.D.; Johnson, L.A.; Burn, T.C.; Su, K.; Landes, G.M.; Feinberg, A.P. A 2.5-Mb transcript map of a tumor-suppressing subchromosomal transferable fragment from 11p15.5, and isolation and sequence analysis of three novel genes. Genomics 1997, 46, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Karnik, P.; Paris, M.; Williams, B.R.; Casey, G.; Crowe, J.; Chen, P. Two distinct tumor suppressor loci within chromosome 11p15 implicated in breast cancer progression and metastasis. Hum. Mol. Genet. 1998, 7, 895–903. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Reviews. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R.; Annese, T. Epithelial-Mesenchymal Transition in Cancer: A Historical Overview. Transl. Oncol. 2020, 13, 100773. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Yan, G.; Chen, Y.; Zhou, M.; Wu, Y.; Li, Y. Inflammation and tumor progression: Signaling pathways and targeted intervention. Signal Transduct. Target. Ther. 2021, 6, 263. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | Primer Seqeunce (5′ → 3′) | |
|---|---|---|---|
| CCND1 | NM_053056.3 | Forward: | ATCAAGTGTGACCCGGACTG |
| Reverse: | CTTGGGGTCCATGTTCTGCT | ||
| CD24 | NM_013230.3 | Forward: | GCTCCTACCCACGCAGATTT |
| Reverse: | GCCTTGGTGGTGGCATTAGT | ||
| CTNNB1 | NM_001098209.2 | Forward: | AATCAGCTGGCCTGGTTTGA |
| Reverse: | GCTTGGTTAGTGTGTCAGGC | ||
| EGFR | NM_005228.4 | Forward: | GAGCTCTTCGGGGAGCAG |
| Reverse: | TCGTGCCTTGGCAAACTTTC | ||
| FN1 | NM_212482.2 | Forward: | AACAAACACTAATGTTAATTGCCCA |
| Reverse: | TCTTGGCAGAGAGACATGCTT | ||
| FOS | NM_005252.4 | Forward: | GGGGCAAGGTGGAACAGTTA |
| Reverse: | AGTTGGTCTGTCTCCGCTTG | ||
| FOSL1 | NM_005438.5 | Forward: | GCCTTGTGAACAGATCAGCC |
| Reverse: | AGTTTGTCAGTCTCCGCCTG | ||
| ITGA4 | NM_000885.5 | Forward: | GCTGTGCCTGGGGGTC |
| Reverse: | CACTAGGAGCCATCGGTTCG | ||
| ITGB1 | NM_002211.4 | Forward: | GCCGCGCGGAAAAGATGAAT |
| Reverse: | CACAATTTGGCCCTGCTTGTA | ||
| ITGB8 | NM_002214.2 | Forward: | GGCAGCTGTCTGTGAAAGTC |
| Reverse: | CCGTCATTGGGCACCACTAT | ||
| JUN | NM_002228.4 | Forward: | CTTTTCAAAGCCGGGTAGCG |
| Reverse: | TTTCTCTAAGAGCGCACGCA | ||
| LAMA1 | NM_000546.5 | Forward: | CACTGTTCTGGAAAAGCCCG |
| Reverse: | TCAACAAGATGTTTTGCCAACTG | ||
| RPL4 | NM_000968.4 | Forward: | CACGCAAGAAGATCCATCGC |
| Reverse: | CCGGAGCTTGTGATTCCTGG | ||
| RPS18 | NM_022551.2 | Forward: | ATTAAGGGTGTGGGCCGAAG |
| Reverse: | GGTGATCACACGTTCCACCT | ||
| GO ID | Molecular Functions | DEGs | Total Genes | q-Value |
| GO:0005201 | extracellular matrix structural constituent | 11 | 89 | 0.00570 |
| GO:0005539 | glycosaminoglycan binding | 11 | 101 | 0.00975 |
| GO:0000981 | DNA-binding transcription-factor activity (RNA polymerase II) | 39 | 863 | 0.01125 |
| GO:0005178 | integrin binding | 10 | 95 | 0.01125 |
| GO:0030296 | protein tyrosine kinase activator activity | 4 | 11 | 0.01125 |
| GO:0005518 | collagen binding | 7 | 46 | 0.01125 |
| GO:0008009 | chemokine activity | 3 | 5 | 0.01125 |
| GO:0003700 | DNA-binding transcription-factor activity | 39 | 894 | 0.01125 |
| GO:0000978 | RNA polymerase II cis-regulatory DNA binding | 35 | 770 | 0.01125 |
| GO:0001664 | G protein-coupled receptor binding | 11 | 122 | 0.01125 |
| GO ID | Biological Processes | DEGs | Total Genes | q-value |
| GO:0051240 | positive regulation of multicellular organismal process | 45 | 789 | 0.00012 |
| GO:0051239 | regulation of multicellular organismal process | 68 | 1481 | 0.00012 |
| GO:0051094 | positive regulation of developmental process | 43 | 760 | 0.00016 |
| GO:0050793 | regulation of developmental process | 65 | 1466 | 0.00049 |
| GO:2000026 | regulation of multicellular organismal development | 42 | 783 | 0.00060 |
| GO:0048731 | system development | 103 | 2818 | 0.00069 |
| GO:0009605 | response to external stimulus | 62 | 1406 | 0.00069 |
| GO:0032501 | multicellular organismal process | 130 | 3920 | 0.00174 |
| GO:0050789 | regulation of biological process | 199 | 6873 | 0.00174 |
| GO:0042127 | regulation of cell population proliferation | 46 | 971 | 0.00257 |
| GO ID | Cellular Components | DEGs | Total Genes | q-value |
| GO:0030312 | external encapsulating structure | 26 | 267 | <0.00001 |
| GO:0031012 | extracellular matrix | 26 | 267 | <0.00001 |
| GO:0062023 | collagen-containing extracellular matrix | 19 | 210 | 0.00016 |
| GO:0071944 | cell periphery | 102 | 2819 | 0.00072 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-khayyat, W.; Pirkkanen, J.; Dougherty, J.; Laframboise, T.; Dickinson, N.; Khaper, N.; Lees, S.J.; Mendonca, M.S.; Boreham, D.R.; Tai, T.C.; et al. Overexpression of FRA1 (FOSL1) Leads to Global Transcriptional Perturbations, Reduced Cellular Adhesion and Altered Cell Cycle Progression. Cells 2023, 12, 2344. https://doi.org/10.3390/cells12192344
Al-khayyat W, Pirkkanen J, Dougherty J, Laframboise T, Dickinson N, Khaper N, Lees SJ, Mendonca MS, Boreham DR, Tai TC, et al. Overexpression of FRA1 (FOSL1) Leads to Global Transcriptional Perturbations, Reduced Cellular Adhesion and Altered Cell Cycle Progression. Cells. 2023; 12(19):2344. https://doi.org/10.3390/cells12192344
Chicago/Turabian StyleAl-khayyat, Wuroud, Jake Pirkkanen, Jessica Dougherty, Taylor Laframboise, Noah Dickinson, Neelam Khaper, Simon J. Lees, Marc S. Mendonca, Douglas R. Boreham, Tze Chun Tai, and et al. 2023. "Overexpression of FRA1 (FOSL1) Leads to Global Transcriptional Perturbations, Reduced Cellular Adhesion and Altered Cell Cycle Progression" Cells 12, no. 19: 2344. https://doi.org/10.3390/cells12192344





